In this review, Hermant and Torres-Padilla summarize and discuss the transcription factors known to be involved in the sequence-specific recognition and transcriptional activation of specific transposable element families or subfamilies.
Keywords: retrotransposons, transcriptional regulation, regulatory elements, co-option
Abstract
Transposable elements (TEs) are genetic elements capable of changing position within the genome. Although their mobilization can constitute a threat to genome integrity, nearly half of modern mammalian genomes are composed of remnants of TE insertions. The first critical step for a successful transposition cycle is the generation of a full-length transcript. TEs have evolved cis-regulatory elements enabling them to recruit host-encoded factors driving their own, selfish transcription. TEs are generally transcriptionally silenced in somatic cells, and the mechanisms underlying their repression have been extensively studied. However, during germline formation, preimplantation development, and tumorigenesis, specific TE families are highly expressed. Understanding the molecular players at stake in these contexts is of utmost importance to establish the mechanisms regulating TEs, as well as the importance of their transcription to the biology of the host. Here, we review the transcription factors known to be involved in the sequence-specific recognition and transcriptional activation of specific TE families or subfamilies. We discuss the diversity of TE regulatory elements within mammalian genomes and highlight the importance of TE mobilization in the dispersal of transcription factor-binding sites over the course of evolution.
Transposable elements (TEs) are DNA sequences that, in principle, have the ability to move from one location to another within the genome. While most TE sequences degenerate with evolutionary time, a substantial fraction of mammalian genomes is composed of remnants of TE insertions. Indeed, the initial sequencing of the human and mouse genomes, at the onset of the 21st century, identified ∼46% and 37% in humans and mice, respectively, as remnants of TE insertions (International Human Genome Sequencing Consortium 2001; Mouse Genome Sequencing Consortium 2002). Improvement in TE annotation in the following years have led to current estimates of TEs abundance of ∼48% in humans and 41% in mice (mm10). It has been suggested that the fraction of mammalian genomes derived from TEs is in fact underestimated, owing to significant sequence divergence occurring at the most ancient TE insertions, preventing their recognition in the modern genomes (de Koning et al. 2011; Hubley et al. 2016). Hence, TEs have been remarkably successful at colonizing mammalian genomes and they are increasingly recognized as significant players in the evolution of genomes and their regulatory networks (Feschotte 2008; Chuong et al. 2017).
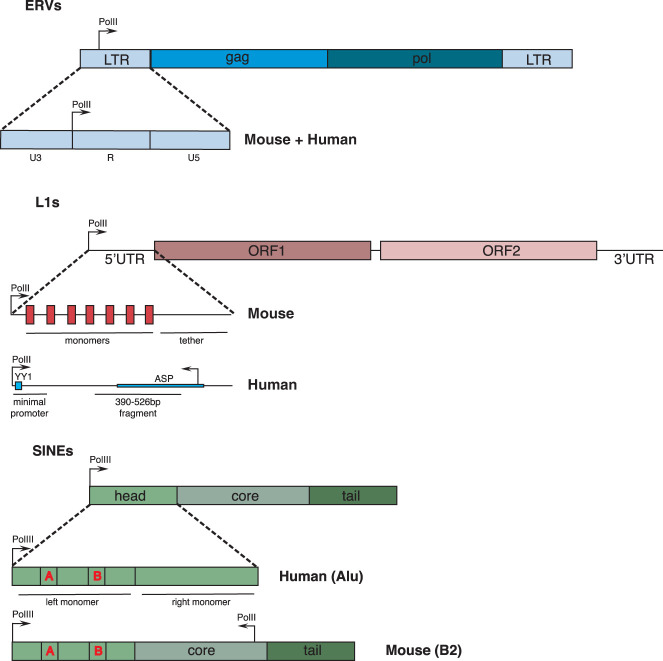
Depending on the mechanism used for transposition, TEs can be broadly divided into two main classes (Finnegan 1989; Wicker et al. 2007). Class I TEs, referred to as retrotransposons, mobilize via an RNA intermediate, which is reverse transcribed and subsequently reintegrated elsewhere in the genome. On the other hand, class II TEs (DNA transposons) mobilize and reintegrate directly as DNA molecules. Retrotransposons substantially dominate the mammalian TE repertoire (International Human Genome Sequencing Consortium 2001; Mouse Genome Sequencing Consortium 2002; Rodriguez-Terrones and Torres-Padilla 2018) likely as a result of their copy-and-paste mechanism of replication, possibly facilitating their expansion in number. Retrotransposons can be further divided into LTR and non-LTR elements. As the name indicates, LTR retrotransposons are characterized by the presence of two initially identical and equally oriented long terminal repeats (LTRs) at the 5′ and 3′ end of the element, which range from 100 bp to >5 kb in size (Mager and Stoye 2015). The non-LTR retrotransposons include two major orders displaying distinct structures: the long and short interspersed nuclear elements (LINEs and SINEs) (Goodier and Kazazian 2008).
The mobility of TEs depends on their ability to generate a full-length transcript. Autonomous TEs encode transcripts that promote their own replication within the host genome, typically independently from the host replication. However, they rely on the host machinery to orchestrate their transcription. In order to do so, TEs have evolved cis-regulatory sequences that function to recruit host-encoded factors, such as RNA polymerases or transcription factors (TFs), thereby ensuring their amplification within the host. Hence, all types of TEs encompass regulatory elements, which in the case of retrotransposons is either embedded within an LTR or contained within the 5′ region preceding the coding sequences in the case of non-LTR elements (Goodier and Kazazian 2008; Mager and Stoye 2015).
Given their mobile nature, TEs have long been conceived as threats to genomic integrity. Indeed, sustained TE activity is a hallmark of human diseases (Hancks and Kazazian 2016); hence, the mechanisms underlying their restriction have been extensively studied. It is believed that genomes have evolved several layers of “defense” mechanisms to suppress TE mobilization (for review, see Goodier 2016). At the transcriptional level, in most mammalian somatic cells, TE silencing is primarily dependent on DNA methylation and repressive histone modifications such as H3K9me3, two classical marks of constitutive heterochromatin. However, TE transcripts may constitute a fraction of the transcriptome of somatic tissues across vertebrates, out of which a high proportion derives from recent TE insertions (Pasquesi et al. 2020). While the work by Pasquesi et al. (2020) cannot discriminate read-through transcription from TE-driven transcription, it suggests that TE transcription is tightly regulated in somatic cells, and thus their impact on gene regulation might be more extensive than initially established, as previously suggested (Chuong et al. 2017). Notwithstanding, blastomeres during preimplantation development, germ cells as well as transformed cells are characterized by a robust expression of specific TEs. In these developmental and disease contexts, TE expression is not only restricted to the read-through transcription of TE-derived sequences within host genes, but it also includes transcription of truncated or full-length TEs driven by their own cis-regulatory elements (Evsikov et al. 2004; Peaston et al. 2004; Fadloun et al. 2013; Göke et al. 2015; Jang et al. 2019). Long thought to be a side effect of the extensive epigenetic reprogramming that is characteristic of these biological contexts, the transcriptional activation of TEs is emerging as a process that is tightly regulated and of key biological relevance to the host.
Therefore, identifying and characterizing molecular players that recognize, activate and regulate TE expression in a sequence-specific manner is of uttermost importance. While most of the research in the past decades has focused on how transcriptional repression of TEs is achieved (Imbeault and Trono 2014; Molaro and Malik 2016; Ecco et al. 2017; Yang et al. 2017), we review here the TFs that have been shown to directly activate specific TE subfamily expression, mainly the relatively young retrotransposons. In addition, we emphasize the diversity of TE repertoires within modern mammalian genomes and highlight the importance of motifs for pluripotency-related factors, which appear to have been embedded within TE regulatory elements throughout mammalian evolution. The review focuses primarily on mouse and human studies, which are the two species where most research has been conducted.
Overview of the regulatory regions of major human and mouse TEs
Endogenous retroviruses (ERVs)
As their name indicates, ERVs are genomic remnants of ancestral viral infections by retroviruses. Their presence in modern genomes stems from provirus integration in the germline, which was subsequently vertically transmitted and eventually endogenized within the host. ERVs constitute ∼8.5% and 11.5% of the human and mouse genomes, respectively (estimations from RepeatMasker and mm10 annotations), and exist in a range of different forms within contemporary mouse and human genomes, most of which are incomplete or truncated (Mager and Stoye 2015).
Complete ERVs are structurally closely related to proviruses: They display internal coding sequences for the viral proteins Gag, Pol, and in some instances, Env, which are flanked by two initially identical LTRs. The LTRs of these elements contain the cis-regulatory elements for their transcriptional regulation. LTRs can be subdivided into three parts: U3, R, and U5. The genesis of an LTR originates in the reverse transcription process of the viral RNA. This RNA contains the R region followed by U5 on the 5′ end, whereas on the 3′ end the R region is preceded by U3. Reverse transcription generates duplicates of both U5 and U3, giving rise to the two LTRs. Hence, U3 is the 5′-most fragment of the LTR, U5 is the 3′-most fragment of the LTR and the R region is included between U3 and U5 (Fig. 1; Vogt 1997; Mager and Stoye 2015). Based on these structural characteristics, transcription of a full-length RNA is expected to initiate at the U3/R boundary and to terminate at the R/U5 boundary (Boeke and Corces 1989; Vogt 1997). Accordingly, the U3 region contains the promoter and potential enhancer elements involved in the transcriptional control of ERVs.
Figure 1.
Schematic representation of the regulatory elements of major human and mouse TEs. Boxes represent the different parts of the elements, as indicated. For L1s, the light-blue box represents the YY1-binding site. For the SINE elements, A and B refer to the A and B boxes. (ERVs) Endogenous retroviruses, (LTR) long terminal repeat, (UTR) untranslated region, (ORF) open reading frame, (ASP) antisense promoter.
The ability of the LTR to drive transcription has been tested using several approaches. For example, early functional analyses using reporter assays concluded that the regulatory elements necessary for transcriptional activity are present within the U3 segment of the mouse-specific intracisternal A-type particle (IAP) LTR, including partial sequences homologous to the TATA-box motif (Christy and Huang 1988; Falzon and Kuff 1988). In contrast, 5′ rapid amplification of cDNA ends (RACE) analysis of the HERVK promoter in human cancer cell lines led to the identification of a major transcription start site (TSS) as well as minor variable TSSs ranging between 10 bp upstream of to 30 bp downstream from the major TSS (Fuchs et al. 2011). Accordingly, no functional TATA box was identified upstream of the major TSS. HERVK transcription was shown to be mediated by SP1 and SP3, knockdown of which significantly reduced the activity of a luciferase reporter. Specific recruitment of these two ubiquitous TFs to three GC boxes present in close proximity to the major TSS was demonstrated by chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA) (Fuchs et al. 2011). Hence, different ERV subfamilies harbor distinct core promoter elements involved in the control of their transcriptional regulation.
Long interspersed nuclear elements (LINEs)
In contrast to ERVs, LINEs do not harbor LTR sequences. LINEs account for about one fifth of the mouse and human genomes, with a consistent domination of L1 over L2 in both species. L1 accounts for 19.9% and 17.5% of the genomic content in mice and humans, respectively, according to mm10 annotation and RepeatMasker. However, L2 are more abundant in humans than in mice, with 3.7% versus only 0.4% of the genome, respectively. Hence, L1 is considered as the most successful and abundant retrotransposon in both species.
A full-length L1 accommodates two open reading frames, Orf1 and Orf2, themselves encoding proteins that are required for a full retrotransposition cycle, which takes place through target site-primed reverse transcription. The sequence composition of Orf1 and Orf2 is relatively conserved within the family and even to some extent between species (Fanning and Singer 1987a,b). Notwithstanding, while L1 regulatory regions share homologous features such as the presence of CpG islands and the lack of traditional binding motifs such as TATA boxes (Furano 2000), the composition and structure of the L1 regulatory elements differ considerably between mice and humans.
The regulatory element of the human L1 is composed of a single 900-bp-long GC-rich 5′UTR (Fig. 1), which was shown in early studies to be sufficient to drive L1 transcription in pluripotent teratocarcinoma cell lines (NTera2D1) (Skowronski and Singer 1985; Swergold 1990) as well as in HeLa cells (Moran et al. 1996). A typical RNA polymerase II promoter initiates transcription downstream from the preinitiation complex (PIC) site at the promoter. However, L1 requires a promoter that initiates transcription upstream of the promoter sequence elements, which would otherwise be lost as a result of transcription. To investigate this issue, the human L1 regulatory regions have been extensively characterized using heterologous systems and deletion assays, which have led to the identification of the first 100 bp within the 900-bp-long promoter segment as critical for transcription of a full-length L1 transcript (Swergold 1990; Minakami et al. 1992). The human L1 promoter has been shown to contain a binding site for Yin Yang 1 (YY1) (Minakami et al. 1992; Becker et al. 1993, 1994), a ubiquitous transcription factor, which interacts with components of the basal transcription machinery and can bind the initiator (INR) element in TATA-less promoters (Seto et al. 1991; Zawel and Reinberg 1995). Subsequently, YY1 was shown to be necessary for accurate transcriptional initiation of L1 full-length transcripts in HeLa cells and was therefore considered a core player in the generation of retrotransposition-competent L1s (Athanikar et al. 2004). While in the context of a full-length promoter mutating the YY1 binding site did not affect transcription nor retrotransposition in HeLa cells, mutations in the YY1 site affected L1 core promoter activity. Interestingly, in the context of a minimal promoter containing only the first 150 bp of the L1 5′UTR driving luciferase expression, mutation of the YY1 binding site induced a threefold to 10-fold decrease in luciferase activity, depending on the cell type used. This indicates that in the absence of the downstream regulatory elements of the 5′UTR, YY1 activity as L1 transcriptional activator is critical (Athanikar et al. 2004). A sustained variability in L1 TSSs, considerably more than previously thought, was subsequently documented and is reminiscent of TATA-less promoters (Lavie et al. 2004). Alexandrova et al. (2012) also significantly contributed to the understanding of L1 transcriptional initiation by providing explanations of TSS variability in L1s. They identified an additional regulatory region located between positions 390 bp and 526 bp, which contains the majority of the binding sites for TFs and can act both as an internal enhancer of full-length transcription from the conventional +1 TSS, as well as a promoter element driving transcription from alternative TSSs (Fig. 1; Alexandrova et al. 2012). Their work, based on a combination of 5′ RACE and heterologous reporter assays in HEK293 and neuronal NTera2 cells, suggested that this 390- to 526-bp fragment harbors essential elements for L1 promoter function.
The genomic landscape in which L1 insertions land in the host is also thought to play an important role in regulating L1 transcriptional activity, given that the 5′ flanking sequence can influence L1 transcription (Swergold 1990; Athanikar et al. 2004; Lavie et al. 2004; Philippe et al. 2016). In addition, pervasive transcription from a neighboring host gene can lead to the production of mature mRNAs containing parts of or full-length L1 sequences. Thus, transcriptional regulation of L1s must be regarded as a more complex process, which can potentially involve interaction with the host genome.
Last, L1 elements in humans also contain an antisense promoter (ASP) within the L1 regulatory region (Speek et al. 2001). The TSS of the ASP has been mapped to two regions between +400 bp and +600 bp (Fig. 1). The ASP activity is only between 1/10th and 1/20th that of the sense promoter activity (Yang et al. 2003) and was demonstrated to be able to drive the transcription of adjacent genes as well (Speek et al. 2001; Nigumann et al. 2002). Subsequently, the 5′UTR of human L1 was shown to contain a primate-specific ORF, referred to as ORF0, which influences L1 mobility (Denli et al. 2015). The promoter of ORF0 overlaps with the originally identified ASP, suggesting that antisense transcription in the primate L1 can initiate from several TSSs, similarly to the L1 sense transcription. Indeed, 5′ RACE analyses revealed that the initiation site of L1-ASP transcription is variable, and that antisense transcripts derived from both young and older L1 elements (Cruickshanks and Tufarelli 2009). Transcripts derived from the ASP promoter have been identified in both chimpanzee and human induced pluripotent stem (iPS) cells. These transcripts are capped and are predominantly cytoplasmic (Denli et al. 2015).
The mouse L1 regulatory region is bipartite and is composed of tandem repeats (the monomers), which are bound to Orf1 via a linker region termed the tether (Fig. 1; Padgett et al. 1988; Naas et al. 1998). While early studies tended to draw a consensus mouse L1 promoter with about seven monomers (Naas et al. 1998), young mouse L1, namely, L1MdA, L1MdGf and L1MdTf types (referred to here as A, G, and T types) were shown to have an average count of 2.7, 2.9, and 3.1 monomers per promoter, respectively (Zhou and Smith 2019). Some promoters were shown to be extremely long (with the longest constituted of 50 type A monomers), albeit 99% of the young L1 promoters had no more than 10 monomers. The regulatory role of the monomers has been established from transient expression assays, in which the monomers have been shown to be sufficient to drive the expression of reporter genes (Padgett et al. 1988; Naas et al. 1998; Furano 2000). Hence, the monomers provide mouse L1 with a recruitment platform for transcriptional regulators. Such a modular system of transcription in which each subunit contains the elements necessary for regulation enables L1s to preserve their promoter regions during transposition, even if some monomers are truncated as a result of inaccurate retrotransposition (Loeb et al. 1986). In fact, most L1s within the mouse genome are truncated at their 5′ end (Voliva et al. 1983).
A YY1 binding site is embedded within the T and G monomers (DeBerardinis and Kazazian 1999). YY1 binding to mouse L1 promoter was recently shown to occur in mouse ESCs devoid of DNA methylation and upon inhibition of histone deacetylase (HDAC) activity using trichostatin A (Cusack et al. 2020). In addition, cap analysis of gene expression (CAGE) sequencing data revealed a broad TSS in the mouse L1 promoters that contain a YY1-binding site (Zhou and Smith 2019). Finally, even though an antisense promoter has not been formerly established in mouse L1 promoters, G and T monomers were shown to display strong antisense TSS signals, hinting at a presence of a potential bidirectional promoter element in these monomers (Zhou and Smith 2019). Hence, despite human and mouse L1 major structural differences, it appears that young insertions share comparable promoter components.
Short interspersed nuclear elements (SINEs)
SINEs belong to the nonautonomous family of retrotransposons since they depend on the machinery of L1s in order to retrotranspose (Dewannieux et al. 2003; Dewannieux and Heidmann 2005). Nevertheless, the transcription of a full element is essential for SINE propagation. In contrast to most other TEs, SINEs are mainly transcribed by RNA polymerase III. SINEs are classified based on the origin of their 5′ sequence, which derive from different cellular RNA genes, such that SINE1 contains a head derived from 7SL RNAs while in SINE2, the 5′ fragment derives from tRNAs. SINE elements, similarly to their evolutionary predecessors, contain a “body” and a “tail.” The body of most SINEs varies between subfamilies of the same class while the tail is often composed of poly(A). The size of a full SINE varies between 100 and 600 bp (Kramerov and Vassetzky 2011). There exist three major types of RNA polymerase III promoters. SINEs are derived from type II RNA polymerase III promoters, which typically contain two internal motifs that are able to recruit RNA polymerase III: the conserved A and B boxes of ∼11 nt each (Schramm and Hernandez 2002). Essentially, as is the case for L1 propagation, an internal promoter is essential for SINE amplification, ensuring that the transcriptional regulatory regions are preserved throughout the full retrotransposition process. Biochemically, the transcriptional mechanism used by RNA polymerase III on type II promoters is characterized by its ability to start transcription upstream of its promoter, thereby in principle ensuring the integrity and the maintenance of the promoter within the element after SINE mobilization.
In humans, the predominant SINE family are Alu elements, comprising ∼11% of the genome, which derive from the 7SL RNA and hence belong to the SINE1 class of elements (Ullu and Tschudi 1984). Alu elements have a dimeric structure resulting from the fusion of two monomers, which themselves arose from the 7SL RNA gene (Deininger et al. 1981; Quentin 1992). The 7SL RNA gene only contains an A box; the 37 nt upstream of the 7SL gene are essential for its accurate and robust transcriptional initiation (Ullu and Weiner 1985). The evolution of Alu elements within the human genome, and of most primates analyzed, has not only involved the fusion of two monomers, but also the acquisition of a B box within the left monomer, and conversely, the loss of the A box in the right monomer (Fig. 1; Quentin 1992). Hence, the conversion of this initially cellular RNA gene into a SINE element has entailed important modifications in the promoter region, which enable Alu transcription irrespective of the 5′ flanking sequence (Kramerov and Vassetzky 2011). Nonetheless, the 5′ flanking sequence of a specific Alu element has also been demonstrated to stimulate its transcription (Chesnokov and Schmid 1996).
In rodents, SINE1 elements have also been identified and are commonly referred to as B1. SINE B1 elements contain the A and B boxes required for RNA polymerase III-dependent transcription, yet rodent SINE B1s display monomeric structures (Krayev et al. 1980; Quentin 1994). A rodent-specific, highly successfully propagated family of SINE elements is the B2 family, which belongs to the class II SINEs since their heads derive from tRNA genes and constitute ∼2.4% of the mouse genome. The SINE B2 promoter includes the two conserved regulatory elements present in a tRNA gene promoter: the A and B boxes (Fig. 1; Schramm and Hernandez 2002; Kramerov and Vassetzky 2011). Contrary to Alu elements, the conversion of a tRNA gene to a SINE B2 in rodents did not seem to require or undergo extensive modifications of their promoter (Kramerov and Vassetzky 2011). In addition, some SINE B2s contain an internal RNA polymerase II promoter located downstream within the body of the element, which induces transcription in the opposite direction (Fig. 1; Ferrigno et al. 2001; Allen et al. 2004).
In terms of their transcriptional initiation, the TSS is expected to be much like their cognate RNA genes for all SINEs: at the 5′ end of the element. However, pervasive transcription is a very common feature of SINEs. Many SINEs are located in the 3′UTR of host genes and are therefore transcribed together with the latter, most often by RNA polymerase II (Roy-Engel et al. 2005; Chen et al. 2009).
Despite the promoter changes that have led to the evolution of Alu elements described above, it is puzzling how such similar RNA polymerase III promoters can display such different expression patterns. For example, B2 promoters share regulatory elements with essential and ubiquitously expressed cellular RNAs, yet B2 themselves are only expressed in restricted developmental contexts. This may arise as a consequence of differential recruitment of RNA polymerase III subunits (Varshney et al. 2015), or alternatively, to differences in transcription factor-binding sites (TFBSs) involved in their transcriptional activation. In fact, Alu elements have been shown to harbor a number of TFBSs for TFs such as nuclear receptors and p53 (for review, see Deininger 2011), which could provide a basis for the specific expression patterns of Alus.
Transposable elements as ‘hubs’ for transcription regulatory signals
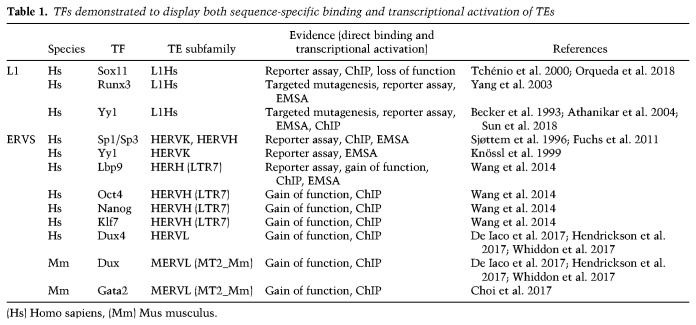
Upon her discovery of the transposable elements in the maize genome, McClintock (1950) conceptualized them as a source of regulatory sequences for host gene expression (McClintock 1956). Only a couple of years later, this hypothesis gained further support by Britten and Davidson (1969), who also proposed that TEs play a role in the evolution of genomes and regulatory mechanisms in many organisms. Both concepts are supported today by a large body of experimental evidence owing to the development of genomic studies (Rebollo et al. 2012; Chuong et al. 2017). Indeed, an enticing model was suggested, in which the expansion of a single TE and its associated cis-regulatory elements would spread TFBSs across the genome and result in evolutionary regulatory innovations (Jordan et al. 2003; van de Lagemaat et al. 2003; Wang et al. 2007; Bourque et al. 2008; Feschotte 2008; Cohen et al. 2009; Chuong et al. 2016). Hence, during the past 20 yr, perhaps initially prompted by the appreciation of the extent to which mammalian genomes are composed of TE-derived sequences, TEs have been increasingly shown to display hallmarks of active regulatory elements. For example, several studies have shown that a considerable proportion of host gene promoters coincide with TE-derived sequences (Jordan et al. 2003; Conley et al. 2008; Faulkner et al. 2009). In addition, DNaseI hypersensitivity analysis has revealed that in human embryonic stem cells (ESCs), fibroblasts and several cancer cell lines, 44% of open chromatin regions correspond to TE-related sequences (Jacques et al. 2013). Moreover, the analysis of several mouse and human tissues, and cell lines indicated that most of the species-specific DNaseI hypersensitive sites (DHS) are enriched in sequences deriving from all TE families (Vierstra et al. 2014), with up to 63% of the primate-specific hypersensitive regions occupied by TE remnants (Jacques et al. 2013). These observations suggest that TE-related regions are important constituents of regulatory regions across cell types. Furthermore, it raises interesting implications regarding genome evolution driven by TE co-option and species-specific diversification of TEs with respect to the regulation of transcriptional programs. To gain further insights into the impact of TEs and their remnants in shaping cell type specific gene regulation, several studies have focused on investigating TF occupancy across the genomes of a myriad of cell types. We discuss some of this work below. In addition, we have summarized the TFs, which have been shown both to bind to, as well as to activate transcription of TEs in vivo (Table 1).
Table 1.
TFs demonstrated to display both sequence-specific binding and transcriptional activation of TEs
In human colon carcinoma cells and other cancer cells, genomic p53 target sites are enriched in ERV1 sequences, more particularly from the LTR10 and MER61 families, which are two primate-specific ERV1 elements (Wang et al. 2007; Bourque et al. 2008). Other transcription factors associated with breast cancer such as C/EBPβ, E2F1 and MYC have also been shown to bind genomic TE sequences, and almost 55% of their genomic target sites in breast cancer cell lines overlap with TEs (Jiang and Upton 2019). Analysis of ChIP data for 26 pairs of homologous TFs in mouse and human leukemic lymphoblast cell lines, revealed that the ChIP-seq peaks for most of the TFs studied fall into a repeat region (e.g., a region annotated in repeat masker), with ∼20% of their genomic targets being composed of TEs in both species (Sundaram et al. 2014).
Beyond cancer or transformed cell lines, the binding profiles of pluripotency-associated transcription factors have been extensively studied in mouse and human ESCs. In an initial study using published ChIP-seq data, Bourque et al. (2008) showed that 23.8% of OCT4-SOX2 binding peaks fall into annotated ERVK repeats in mouse ESCs. In 2010, TEs were even suggested to have “rewired” the core pluripotency network, where 25% of the binding sites for OCT4 and NANOG was shown to fall into repeat-masker annotated regions in both humans and mice (Kunarso et al. 2010). More specifically, 20.9% and 14.6% of the binding regions for OCT4 and NANOG, respectively, were associated with repetitive elements in humans. In addition, ERV1 elements were the main contributors of these repeat-associated binding sites in humans. Indeed, alone they accounted for 7.2% and 8.3% of the NANOG and OCT4 binding regions, respectively. The proportion of OCT4 binding regions containing repeats was lower in mouse ESCs, representing ∼7% of the total binding sites, while TEs contributed to ∼17% of the NANOG binding regions. Even though ERVK (ERV2), not ERV1, dominated the percentage of repeat-associated binding sites of NANOG and OCT4 in mice, there was a clear overrepresentation of ERVs, and specifically their LTRs, in the contribution to NANOG- and OCT4-binding sites in both species. Specific TE subfamilies, such as the human LTR9B (ERV1) were found to be particularly frequently bound by one of the pluripotency-associated factors. For instance, 33.2% of the 767 LTR9B repeats were bound by OCT4. Hence, it appeared that there is a specific targeting of pluripotency-associated factors to TEs, especially to ERVs and their LTRs (Kunarso et al. 2010). These results suggest that TFs are able to target specific TE regions, potentially leading to cell-specific gene regulation. The degree to which this binding to the chromatin reflects actual functional transactivation activity remains to be worked out (de Souza et al. 2013). However, it is clear that the conservation of those binding sites as well as the abundance of such TFBS in TEs in the mouse and human genomes is indicative of strong evolutionary selection, reflecting their positive impact to the fitness of the TE but also, presumably, pointing toward functional advantages to the host.
CTCF, a conserved transcription factor involved in enhancer-promoter insulation and in maintaining topologically-associated domain (TAD) boundaries, also binds a significant proportion of TE sequences (Bourque et al. 2008; Kunarso et al. 2010). CTCF binding sites are primarily composed of B2 elements in mice, with B2 composing 33.8% of CTCF-binding regions in mouse ESCs (Bourque et al. 2008). The specific association of CTCF with mouse-specific amplified B2 repeats is appreciated when compared with the much lower representation of repeats among CTCF-binding peaks in human ESCs, which constitute only 11% of CTCF binding sites (Kunarso et al. 2010). Based on these findings, it was suggested that B2 elements may work together with CTCF to regulate 3D genome organization, particularly in mouse cells. This would imply that TEs and their derived sequences not only have the potential to regulate promoter and enhancer activity, but also the genome more globally, through regulating higher-order chromatin structure.
More recently, 519 ChIP-seq data sets for 97 sequence-specific TFs obtained using 94 human cell types were analyzed in a single study to comprehensively determine the contribution of ERVs to those TF binding sites (Ito et al. 2017). The TFs analyzed varied in terms of DNA binding motif and family, and included general TFs, ubiquitous TFs such as SP1 and YY1, but also more lineage-specific TFs such as GATA1, SPI1, and TAL1, specific to hematopoietic cells, or GATA4/6, SOX17, and FOXA1/A2 characteristic of endodermal cells. Several TFs were found to have their binding sites more frequently located within various types of HERV/LTRs than at “random,” considering the abundance of the corresponding LTR in the genome. Even though the proportion of TFBS overlapping with ERVs, LINEs and SINEs annotated regions was approximately equivalent (12%, 15%, and 16%, respectively), the number of TFs significantly binding to ERVs was substantially higher than in the two other TE classes. This is consistent with LTRs being more likely to retain their regulatory activity, as opposed to L1s for instance, which suffer 5′ truncations. By defining HERV/LTR-shared regulatory elements (HSRE), which consisted of binding motifs identified in a substantial fraction of HERVs at a consensus position, the authors aimed at identifying whether the binding motif was present in the genome prior to its insertion within the genome. Most HSRE were contained within LTR regions (87%) and the regulatory regions of ancient LTRs were more divergent than those of young LTRs. The “erosion” of HSRE on older elements resulting from sequence divergence suggests that the acquisition of TFBS within TE regulatory regions results from relics of selfish strategies adopted by the elements to achieve their own expansion within the host genome. These observations also go in hand with the hypothesis that TE genomic expansion results in the “spreading” of pre-established TFBS embedded within TE regulatory regions (Bourque et al. 2008; Feschotte 2008; Chuong et al. 2017). Regarding LINE elements, human L1 in particular has been extensively studied, and L1 transcriptional activity and regulators have also been a matter of intensive research. A recent study proposed a “molecular choreography” underlying TF binding to L1 across several human cell types, whereby the analysis of 512 TFs in 118 different cell types revealed a vast number of TFs binding specifically to L1, mostly to evolutionary young L1 elements (Sun et al. 2018). More than 80% of the TFs found to bind L1 were binding its 5′UTR, and the binding events were observed more frequently in cells expressing L1, such as cancer cells and ES cells than in other cell types. This suggests the existence of a combinatorial regulatory network involving several TFs regulating L1 expression in mammalian cells.
Furthermore, many of the regulatory networks involving TEs and/or the TFBS within them are regulated in a species-specific manner. In most cases, the TE-derived regulatory elements are in fact species-specific or have evolved within a given species after insertion in the host genome. Strikingly, the above mentioned studies performed across mouse and human cell types revealed a species-specific binding of TFs to TEs (Kunarso et al. 2010; Sundaram et al. 2014). Indeed, the binding sites for most TFs occur on genomic sites, which are not conserved between humans and mice: Only 2% and 1% of the TE-associated human and mouse-derived TF-bound peaks, respectively, were found syntenically in the other genome. In addition, even when the TE family in question is shared between the two species, the TF-binding site itself is not conserved. This suggests that a potential ancestral element present in both species would have been subject to species-specific retrotransposition. Notably though, a subset of TFs such as CTCF and the cohesin subunits Rad21 and SMC3 display an increased number of conserved binding events between mice and humans (Sundaram et al. 2014). These conserved binding events are more often found in internal TE sequences, rather than in their promoters or 5′ regions. Given the role of these 3 DNA binding proteins in genome organization, it is tempting to speculate that perhaps internal TE regions would have been co-opted for genome structure purposes, whereas the regulatory elements of TEs have been co-opted for gene regulatory purposes.
Thus, it appears that TEs, particularly LTR retrotransposons (Thompson et al. 2016; Ito et al. 2017), act as binding platforms for the recruitment of a multitude of TFs in a cell type-specific and species-specific manner. In fact, there is certain specificity for TF association to a given TE family, which once again supports the hypothesis that TEs originally containing TFBS functioned as spreaders of such TFBS throughout the whole genome (Bourque et al. 2008; Feschotte 2008; Chuong et al. 2017). Notwithstanding, in some cases TE expression does not necessarily correlate with the binding of the TF to the latter (Sun et al. 2018). In other cases, such as in mouse and human ESCs, the binding of TFs to their cognate TE does correlate with the expression pattern of the associated TE (Kunarso et al. 2010; Sun et al. 2018). Even though these correlative studies support a potential role of TE-derived TF binding sites in host regulatory networks, functional analysis whereby TEs at given locations are deleted will be required to substantiate these hypotheses. These experiments are now starting to emerge, facilitated by, e.g., Crispr/Cas9 approaches, even though they still pose a number of technical challenges related to the repetitive nature of TEs. Nevertheless, in the case of iPS cells, for example, a specific LTR5HS ERV was deleted, resulting in a significant reduction of expression of its coregulated gene, GDP1 (Fuentes et al. 2018). Likewise, deletion of an individual ERVK LTR, RLTR15 in mouse embryos, resulted in a partial loss of imprinting of the associated Gab1 gene in placenta and yolk sac (Hanna et al. 2019). While these experiments directly demonstrate a role for the LTRs themselves in gene regulation, investigating the role of the TFBS within them through motif mutagenesis, will provide additional information and mechanistic insights on the actual roles of TF-binding to TE-derived sequences.
TEs appear to extensively contribute to mammalian transcription regulation and provide TFBS for a number of tissue specific TFs, supporting their potential involvement in cell type-specific gene regulation. Nevertheless, it is worth mentioning that the repetitive nature of TEs makes the analysis and the mapping to the genome more complicated than unique gene analysis. Indeed, the precise origin of a transcript or a ChIP-seq peak may not be accurately definable, and could be mapped to several identical elements while it might be truly coming from an individual element. Hence, the extent of contribution of TE to the transcriptome or to the TFBS of a specific TF could be overestimated. While these considerations must be kept in mind, there is increasing amount of evidence that the immense sequence diversity of TEs, especially of their regulatory regions, across species, families, subfamilies and even as genetic variants within subfamilies, contributes to the propensity of TE as providers of TFBS.
Dynamic evolution of TE regulatory regions
Evolutionary dynamics of the L1 5′ UTR: the ‘5′ turnover’
The L1 element present in placental mammalian genomes derives from a common ancestor, which was present before the radiation of placental mammals, dating from ∼100 million years ago (Furano 2000; Khan et al. 2006; Richardson et al. 2015). The diversity of the structures observed at the 5′ end of the different mammalian L1s suggests that the acquisition of a new 5′ end has occurred repeatedly and independently in different species, and has been necessary to the successful propagation of the L1 family (Scott et al. 1987).
In humans, most L1 elements amplified after the divergence of the ancestral mouse and human lineages, ∼65 million to 75 million years ago (International Human Genome Sequencing Consortium 2001; Richardson et al. 2015). There are 16 identified human-specific L1 subfamilies (L1PA1 to L1PA16), which are believed to have emerged by subfamily succession as described above (Richardson et al. 2015). Among these, L1Hs is the most recent one, thought to have emerged ∼2 million years ago (Khan et al. 2006), and importantly is the only active subset of L1 in the human genome (with 80–100 active elements per individual) (Boissinot et al. 2000; Brouha et al. 2003). Hence, significant effort has been invested in understanding L1Hs transcriptional regulation. A series of functional assays of the impact of TFs on L1Hs expression have been performed.
L1 retrotransposition occurs during mouse and human neurogenesis, where it is thought to contribute to the genetic mosaicism of neuronal precursor cells (Muotri et al. 2005; Sanchez-Luque et al. 2019). Thus, additional efforts have been dedicated to identifying the factors involved in the specific transcription of L1 full-length transcripts during the process of neurogenesis. Using EMSA, an early study showed that SOX transcription factors bind to the L1 promoter. Two binding sites for SRY factors have been mapped in the human L1 promoter region, and both are required for the activation of an L1 reporter system in human rhabdomyosarcoma (RD) cells (Tchénio et al. 2000). SOX11 was specifically shown to transactivate a reporter gene in RD cells, which has been further supported in a recent study demonstrating the direct role of SOX11 in activating L1Hs during SH-SY5Y cells neuronal differentiation (Orqueda et al. 2018). SOX2 was shown to repress L1Hs transcription in neural stem cells. In agreement with this, decreasing SOX2 expression upon neuronal differentiation results in transient stimulation of L1Hs expression (Muotri et al. 2005). The family of Runx TFs was also added to the list of human L1 regulators (Yang et al. 2003). The 5′ UTR of 20 L1Hs elements known to be actively retrotransposing contain a conserved tripartite RUNX binding site. Mutating the first site leads to decreased L1 transcriptional and retrotransposition activity in 143B cells. More specifically, RUNX3 was shown to significantly increase L1Hs promoter activity and it is thought to do so by directly binding to L1Hs DNA, in a sequence-specific manner as shown by EMSA. In addition, several p53 responsive elements are present in L1Hs, which seem to have resulted from mutations occurring upon the emergence of the L1PA3 family ∼20 million years ago, and which have been then conserved until the emergence of L1Hs ∼2 million years ago (Harris et al. 2009). According to the analysis of L1 transcript levels in p53 mutant cells, which contain higher levels of L1 transcripts than the p53 wild-type counterparts, Wylie et al. (2016) established that p53 functions to restrain transposable element expression. Similarly, the oncoprotein MYC binds the L1 5′UTR in a cell-type specific manner and restricts its expression, as its knockdown in HEK293 cells results in sense as well as antisense promoter activity (Sun et al. 2018).
In human ESCs, the KRAB/Trim28(KAP1) pathway restricts L1 expression (Matsui et al. 2010; Rowe et al. 2010). The KRAB containing ZFP (KZFP) protein ZNF93 is recruited to the L1 5′UTR, leading to the subsequent recruitment of KAP1 and the repressive SETDB1 machinery. Remarkably, ZNF93 and its partner KAP1 binding is restricted to certain specific L1 subfamilies, excluding the older (L1PA7 and older) as well as the younger (L1Hs and L1PA2) insertions (Castro-Diaz et al. 2014; Jacobs et al. 2014). A 129-bp deletion within the 5′UTR of the two most recent L1 elements resulted in the abrogation of the ZNF93 binding site and could explain the lack of ZNF93 binding to L1Hs and L1PA2 families (Jacobs et al. 2014). These observations are in strong support of a model in which TEs and KRAB-ZFPs are participating in an arms race, whereby L1 evasions from host-mediated repression mechanisms through the acquisition of a different 5′UTR would in turn drive the expansion of the KZFP family of proteins (Thomas and Schneider 2011; Castro-Diaz et al. 2014; Jacobs et al. 2014; Ecco et al. 2017). The monophyletic subfamily origin, characteristic of the L1 family, provides further support to this model. Later on, additional KZFPs, including ZNF141, ZNF649 and ZNF765 were also found to bind specific L1PA subfamilies. The evolution of the most recent L1 insertions (L1Hs, L1PA2) involved mutations in the binding sites for these aforementioned TFs that can therefore no longer bind to the elements, providing an explanation to their “escape” from this repressive mechanism (Imbeault et al. 2017). As opposed to the KRAB/KAP1 pathway, the conserved YY1 binding site present in human L1 appears to be crucial to the specific repression of young L1 insertions (L1Hs and L1PA2) by mediating their DNA methylation in hESCs, NPCs and hippocampal neurons (Sanchez-Luque et al. 2019). The authors suggested that YY1 would have repressed new mobile L1 families repeatedly during evolution while the control was relocated to a KAP1-mediated repression as insertions age and become less likely to mobilize. One possible hypothesis is that the absence of KAP1-mediated heterochromatin formation on young L1 provides access to YY1 to the promoter of these elements specifically. A 5′ truncation leading to a deletion of the YY1 binding site in young elements enable these insertions to escape host-mediated repression and achieve retrotranposition. Hence, it appears that the extensive, linear 5′ turnover that characterizes human L1 resulted from a complex interplay between the action of host-encoded TFs and the evolutionary dynamics of the regulatory regions of TEs. Interestingly, both in mice and humans, the youngest L1 elements stand out as most active (DeBerardinis and Kazazian 1999; Boissinot et al. 2000; Goodier et al. 2001; Brouha et al. 2003, Beck et al. 2010). Furthermore, analysis of ChIP-seq data sets for 36 TFs in hESCs showed a high number of these TFs bound to young L1 promoters (L1Hs, L1PA2, and L1PA3) and this number significantly decreased with evolutionary age, down to only one TF for L1PA6 and L1PA7 (Sun et al. 2018). This raises the attractive possibility that, in addition to escaping the repressive host factors, new L1 subfamilies could be driven by the acquisition of activating transcription factors.
The mouse L1 promoter has also experienced extensive changes through evolution. Indeed, even though all mouse L1 promoters exhibit a tandem monomeric repeat form, there is an extensive heterogeneity among the members of the murine L1 family. Most of the TFs that are known to activate human L1 transcription have not been found in mouse L1, which is expected due to the lack of similarity between the murine and the human L1 sequence. Interestingly, the Wnt pathway, a key regulator of neurogenesis through the regulation of TCF/LEF transcription factors, has been involved in mouse L1 regulation (Kuwabara et al. 2009). Overlapping binding sites for TCF and SOX transcription factors (SOX/LEF) were identified in mouse, rat, and human L1. Using a combination of ChIP-seq, RT-qPCR and reporter assays, SOX2 was shown to associate with mouse L1 in undifferentiated neuronal stem cells and L1 was up-regulated upon stimulation by a Wnt ligand in mouse cells (Kuwabara et al. 2009). The involvement of the KRAB/Trim28(KAP1) pathway in restricting mouse L1 transcription has also been studied (Castro-Diaz et al. 2014). In particular, the KZFP Gm6871 was found to bind L1 in mouse ESCs. Gm6871 was specifically bound to older (L1MdF2 and L1MdF3) elements whereas it is nearly absent from younger elements such as the A and T subfamilies. This evolutionary dynamic pattern of KZFPs binding to mouse L1 suggests, just like human L1, a complex interplay between the evolution of the regulatory regions and the action of host-encoded sequence specific TFs.
LTR evolution reflects selfish colonization strategies
Tracing the evolutionary dynamics of TFBS embedded within the LTR region of ERVs is more challenging than it is for LINEs. Indeed, the pool of LTRs currently occupying mammalian genomes comes from a complex combination of genomic invasions from external sources, genomic expansions, and recombination events between different ERVs, as opposed to the “linear” evolution of L1 elements (Vargiu et al. 2016; Ecco et al. 2017). Notwithstanding, comparative evolutionary studies performed on ERVs have initiated the theory of the “arms race” in which ERV insertions and KZFPs appeared concomitantly and KZFPs would have evolved to control ERVs’ expression during development both in mice and humans (Emerson and Thomas 2009; Thomas and Schneider 2011; Ecco et al. 2017; Wolf et al. 2020). Later, 217 subfamilies of human ERVs were found to be bound by one or several KZFPs from 222 KZFPs ChIP-exo experiments in HEK293T cells (Imbeault et al. 2017). Corresponding KZFP-TE pairs could be established, most of which were highly conserved and suggestive of a pressure to keep the repressive activity of a KZFP and a given TE, even after the latter has lost retrotransposition capacities. Importantly, even closely related ERV subfamilies showed differential KZFPs recruitment. For example, the three subfamilies of MER11 (LTR of HERVK11) did not exhibit enrichment for the same KZFP: MER11A recruits ZFN433, ZNF808, ZNF440, and ZNF468, while MER11C is bound by ZNF808 and ZNF525.
While these observations concern repressive sequence-specific DNA binding proteins, which have been more extensively studied than activating proteins, many ERV subfamilies have been shown to be recognized by activating TFs and a distinct recruitment to evolutionary closely connected subfamilies is a recurrent conclusion (Kunarso et al. 2010; Sundaram et al. 2014; Wang et al. 2014; Grow et al. 2015; Chuong et al. 2016; Choi et al. 2017). Indeed, it appears that ERVs, particularly their associated LTR, have experienced waves of gains and losses of TF binding along their evolution and waves of genomic invasions within the host.
An influence of the ancestral retroviral tropism?
The tropism of a virus, or a retrovirus, resides in its ability to infect and replicate within specific cell lines or tissues. A well-known example of viral tropism of extant viruses is the human immunodeficiency virus (HIV), which is known to specifically target the cells of the immune system. Even among HIV isolates, different types can be distinguished according to the precise immune cell types the virus is able to infect (macrophages or T-cells) (Berger et al. 1998). The transcriptional regulation of HIV within the infected cells results from a complex interplay between host- and viral-encoded factors, which have been shown to involve host-encoded TFs such as IRF1 and NFKB. Indeed, HIV contains NFKB-binding sites embedded within its LTR (Roulston et al. 1995; Sgarbanti et al. 2008). Hence, the variety of TFBS embedded within ERV LTRs might derive from strategies of infection used by the ancestral retrovirus they descend from.
As a matter of fact, the HERVK LTR (ERV2) has been referred to as a “landing strip” for transcription factors involved in the immune response. It contains, for example two interferon-stimulated response elements (ISRE) that can be bound notably by IRF3, IRF5 and IRF9 (Manghera and Douville 2013; Csumita et al. 2020). The HERVK LTR consensus sequence contains a number of other binding sites for TFs involved in the immune response, which are reviewed elsewhere (Manghera and Douville 2013). However, none of these factors have been directly implicated in transcriptional activation of HERVK. Of all the binding sites present within the HERVK LTR, only the ubiquitous TFs, such as YY1 and SP1/SP3 have been experimentally demonstrated to regulate HERVK, mostly in teratocarcinoma cells such as GH, Tera2 or MelC9 cells, which are known to display high levels of expression of HERVs (Knössl et al. 1999; Fuchs et al. 2011). In an equivalent cellular context (NTera2D1 cells), SP1/SP3 were indeed shown to be able to drive the expression of HERVH (ERV3) as well (Sjøttem et al. 1996).
A subsequent study implicated MER41 (ERV1 LTR) as enhancers for the immunity-related transcriptional programme, by providing TFBS for the immunity-related TF, STAT1 (Chuong et al. 2016). Deletion of members of these elements in HeLa cells impaired the response to IFNγ treatment. Interestingly, the specific binding site for STAT1 within MER41 is present in the consensus sequence for MER41, as well as in the MER41B subfamily, but absent for the closely related MER41A. Indeed, MER41A underwent a 43-bp deletion resulting in the loss of the STAT1 binding site, and therefore to the inability of STAT1 to bind to it, as demonstrated by the lack of ChIP-seq enrichment. These results shed light into the level of specificity of the TFBS present within the MER41 family. In this specific case STAT1 binding to the LTR did not elicit transcriptional activation of the TE sequence itself, but was instead required for triggering the interferon response. However, the presence of the STAT1 binding site within many MER41 elements and notably, within the consensus (ancestral) sequence, hints at the presence of an ancestral binding site reflecting an ancestral strategy employed by this particular TE for self-replication and expansion. Of note, MER41 is primate specific; however, a mouse-specific ERV1 (RLTR30B) was found to be enriched in STAT1 signals in IFN-stimulated mouse macrophages (Chuong et al. 2016). RLTR30B displays enhancer activity, as shown using reporter assays in HeLa cells. This is particularly remarkable, since it illustrates how different TEs can end up performing a similar function in the host genome of two different species, in spite of a different sequence or evolutionary origin within either species.
When comparing the enrichment of TEs in active versus repressed regions across 24 tissues based on the analysis of the distribution of five histone modifications therein (H3K4me1, H3K4me3, H3K36me3, H3K9me3, and H3K27me3), Trizzino et al. (2018) showed that ancient TEs are mostly enriched in active regions across all 24 tissues. On the contrary, young TEs appeared mainly enriched in repressed regions across tissues, with the noteworthy exception of immune cells, which were the only cell types that displayed enrichment of young TEs in their active regulatory regions. This observation comes hand in hand with the results obtained by Chuong et al. (2016), explained above, and underlies once again the importance of the original selfish colonization strategies of TEs to their regulation within modern genomes.
A strong selection for vertical transmission
As mentioned above, the binding sites for many pluripotency-associated TFs in mice and humans are embedded within ERVs (Kunarso et al. 2010; Xie et al. 2010; Sundaram et al. 2014; Ito et al. 2017). Young, species-specific ERV subfamilies tend to possess most of these TFBS, and there is a strikingly poor conservation of these binding sites between mice and humans (Kunarso et al. 2010; Ito et al. 2017). In both species, TEs show a robust transcriptional activation during early development (Peaston et al. 2004; Svoboda et al. 2004; Macfarlan et al. 2012; Fadloun et al. 2013; Göke et al. 2015). Indeed, TE expression at this developmental stage is dynamic and abundant and appears to be tightly temporally regulated, with TE subfamilies showing distinct expression patterns (for review, see Rodriguez-Terrones and Torres-Padilla 2018). The transcriptional activation of TEs during preimplantation development coincides with the time in embryogenesis during which cells are highly plastic, and for the most, their fate is not fixed. In addition, a major epigenetic reprogramming wave occurs whereby DNA is largely demethylated (Smith et al. 2014), heterochromatic marks such as H3K9me3 remodeled, and the global chromatin structure is considered to be more “open,” as established by FRAP (Bošković et al. 2014), ATAC-seq (Wu et al. 2016) and DNaseI (Gao et al. 2018) analyses. Thus, this epigenetic and chromatin landscape could be considered as a “window of opportunity” for the transcriptional activation of TEs. However, because the patterns of expression of TEs display developmental stage and family specificity (Rodriguez-Terrones and Torres-Padilla 2018), it is likely that regulatory mechanisms specific to each subfamily exist, as opposed to just a spurious wave of transcription resulting as a side-effect of heterochromatin reprogramming. In fact, the dynamics of transcriptional activation and repression of TEs cannot merely be explained by the known patterns of DNA methylation and H3K9me3 (Smith et al. 2014; Wang et al. 2018). In addition, removal of heterochromatin modifiers during preimplantation development largely does not affect TE expression (Burton et al. 2020). The action of sequence specific TF-mediated transcriptional activation or repression could, indeed, clearly fulfil the requirement for family and temporal-specific transcriptional regulation and therefore govern this complete process.
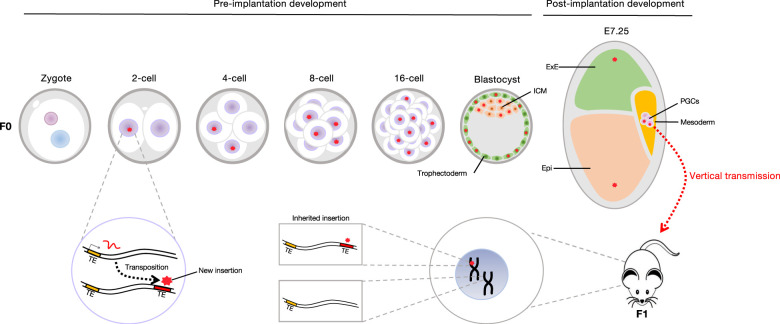
In addition, as genomic parasites, TEs have evolved under the selective pressure to efficiently amplify within the host genome. A critical moment for their expansion is during or prior to germ cell development, in order for the new insertions to be passed on to the next generations, by vertical transmission. The first cluster of primordial germ cells (PGCs) arises at E7.25 during mouse development. All cells during preimplantation development therefore contribute to the subsequent generation of germ cells (Fig. 2). Hence, it is possible that TEs experience a selective pressure to be expressed during this advantageous window of development. In fact, heritable endogenous L1 insertions arising in PGCs have recently been reported for the first time (Richardson et al. 2017). It is tempting to conceive that the acquisition of binding sites for TFs specifically expressed at these stages would have a positive impact on the fitness of the element.
Figure 2.
Preimplantation development is a window of opportunity for vertical transmission. All cells during preimplantation development will contribute to the germline; hence, a new insertion occurring during preimplantation development has increased chances to be transmitted to the next generation by vertical transmission. A new insertion will also be present in cells of the ExE and Epi, which is represented as a red star in the schematic representation of these tissues. (TE) Transposable element, (ICM) inner cell mass, (ExE) extraembryonic endoderm, (Epi) epiblast, (PGCs) primordial germ cells. Mouse drawing was adapted from https://www.clker.com/clipart-simple-black-and-whitemouse.html.
In humans, the expression of HERVK and its LTR, LTR5, is induced in 8-cell stage embryos and is maintained up to the blastocyst stage (Grow et al. 2015). LTR5Hs, as opposed to its evolutionary predecessors LTR5A and LTR5B, contain a binding site for OCT4, precisely at the position 692-699. This binding site is absent from LTR5A and LTR5B specifically, even though they share 88% sequence similarity with LTR5Hs. ChIP-seq in human embryonic carcinoma cells (hECCs) revealed an enrichment of OCT4 over LTR5Hs together with active histone marks (H3K27ac and H3K4me3) and p300. These enrichments were not observed in hESCs, which are not permissive to HERVK transcription. Knockdown of OCT4 resulted in a significant down-regulation of HERVK transcription, supporting a role for OCT4 in LTR5Hs/HERVK transcriptional activation. Knockdown of SOX2 using siRNA resulted in a comparable decrease of HERVK expression, involving SOX2 in the pathways regulating HERVK in hECCs, although whether this effect is direct or indirect was not established in this study (Grow et al. 2015). Subsequent clustering analysis of the human LTR5 elements, based on phylogeny and TFBS, has divided them into five classes, among which one of the clusters, the fourth one, was specifically associated with the pluripotency network of TFs (Ito et al. 2017). Importantly, this fourth cluster was also among the youngest elements, suggesting that the presence of pluripotency TFBS may be important for the genomic expansion of new TF families.
Another type of human-specific ERV, HERVH, is also expressed during preimplantation human development. LTR7 is the highest transcribed subfamily of HERVH in human pluripotent cells (ESCs and iPSCs) and its expression seems to be specific to pluripotent cells of the blastocyst, and naïve ESCs, as LTR7 transcripts have not been detected in other cell types (Ohnuki et al. 2014; Wang et al. 2014). LTR7 possesses TFBS, among which the direct binding of the TF LBP9 to its binding site was confirmed by EMSA and ChIP-qPCR (Wang et al. 2014). The LBP9 binding site is present exclusively on active LTR7 elements, which were deemed so based on the presence of activating histone marks. Ectopic expression of LBP9 in human primary fibroblasts resulted in the activation of an LTR7-GFP reporter system, as well as the endogenous HERVH expression. Analysis of ChIP-seq data from hESCs also identified binding sites for OCT4, NANOG and KLF4, overexpression of which induced HERVH expression in fibroblasts, albeit to a lesser extent than LBP9 overexpression. The role of these four TFs in HERVK regulation was confirmed by knockdown experiments in hESCs (Wang et al. 2014). In addition, HERVH and its associated LTR7 become hyperactivated during factor-induced reprogramming of somatic cells to iPSCs, which was shown to be mediated by OCT3/4, SOX2 and KLF4 (Ohnuki et al. 2014). Clustering analysis of LTR7 based on its phylogeny and TFBS identified three distinct groups (Ito et al. 2017). The youngest subgroup of LTR7 elements showed the highest enrichment of ChIP-seq reads for the pluripotency factors (NANOG, SOX2, and OCT4) in iPSCs or ESCs, suggesting once again that the acquisition of these TFBS might be important to the genomic expansion of new TE subfamilies. Interestingly, the LTR7B and LTR7, which are closely related HERVH LTR subfamilies, are characterized by substantially different temporal expression profiles during development, where LTR7B is specific to 8-cell and morula stage embryos, while LTR7 expression is only observed in the pluripotent inner cell mass (Rodriguez-Terrones and Torres-Padilla 2018). It is tempting to speculate that these significant differences in the temporal expression pattern, which these closely related subfamilies display, could be explained by the different TFBS that these LTRs contain.
In mice, retrotransposons and their remnants are also transcriptionally active in germ cells (Peaston et al. 2004) and in the early embryo (Peaston et al. 2004; Fadloun et al. 2013). The mouse-specific ERVL, MERVL, is expressed during a very short time window at the two-cell stage only (Svoboda et al. 2004; Macfarlan et al. 2012; Ishiuchi et al. 2015). The ERVL family was initially identified in humans, and subsequently observed in all placental mammals (Bénit et al. 1999). A common ERVL ancestor was present >70 million years ago and ERVLs seem to have been retained in both mice and humans, with independent expansions in both genomes. Because of their potential role in driving the transcriptional program of totipotent cells in the embryo (Evsikov et al. 2004; Peaston et al. 2004), several groups have sought to identify TFs that could function as ERVL regulators. For example, the microRNA mir34a was recently shown to regulate “two-cell stage-specific genes” in mouse ESCs (Choi et al. 2017). Specifically, down-regulation of mir34a results in the up-regulation of MERVL expression. Among the most strongly up-regulated MERVL elements following mir34a shRNA, 18 consistently contained three binding sites for the GATA2 TF within their LTR, MT2_mm. The expression pattern of GATA2 during preimplantation development correlates with that of MERVL. Mutation of the two most conserved GATA2 binding sites within MT2_mm reduced the effect of GATA2 on MERVL expression, as determined using a luciferase reporter assay in ESCs. In addition, specific binding of a Flag-tagged version of GATA2 to MT2_mm was documented by ChIP in mir34a-deficient mouse ESCs (Choi et al. 2017). MT2A, MT2B1, and MT2B2 are closely related to MT2_mm but do not contain binding sites for GATA2. In fact, the GATA2-binding sites as well as the robust expression at the 2-cell stage are in combination, unique features of MT2_mm.
In addition to GATA2, MT2 also contains a binding site for the TF DUX (Duxf3), a double homeodomain TF. The human ortholog, DUX4 was initially identified as aberrantly expressed in facioscapulohumeral muscular dystrophy (FSHD) cells, characterised by high transcriptional activity of ERVs, particularly of MaLR, a nonautonomous type of ERVL (Geng et al. 2012; Young et al. 2013). Through its C terminus, DUX4 can recruit the histone acetyltransferases p300/CBP enabling local chromatin remodeling at its target genes, primarily by p300/CBP-mediated deposition of H3K27ac (Choi et al. 2016). Therefore, DUX4 acts as a pioneer transcription factor, and its function is conserved by the mouse ortholog DUX, despite the relatively low homology in their homeodomains (Eidahl et al. 2016). The overexpression of either the mouse or the human DUX TF in the corresponding mouse ESCs or human iPSCs strongly induces the expression of their respective species-specific ERVL (De Iaco et al. 2017; Hendrickson et al. 2017; Whiddon et al. 2017). The first homeodomains of DUX and DUX4 share only 33% identity (Eidahl et al. 2016), yet they display similarly strong binding and consequent transcriptional activation of their respective species-specific ERVL. Notably, the sequence of MERVL and HERVL also substantially diverge. Hence, these two proteins have evolved independently in each species, but together with their species-specific target TEs, they orchestrate a similar function. This is an exceptional example of functional conservation without sequence conservation involving a TE and a host TF.
Transposable element co-option in mouse preimplantation development
The unleashing of TE expression, which characterizes mouse preimplantation development occurs concomitantly with developmental processes, all of which are essential to the formation of a multicellular organism. Indeed, the epigenome is extensively reprogrammed to establish totipotency (Burton and Torres-Padilla 2014), and the transcriptional machinery of the newly formed embryo must be activated for the first time. Studies during the past 20 yr have converged to establish that the transcriptional activation of TEs during preimplantation development is not solely a side effect of reprogramming, but plays a role in these crucial developmental processes.
For example, a substantial proportion of the two-cell transcriptome initiates transcription within the MERVL LTR (Peaston et al. 2004). The MERVL LTR appears to have been co-opted by the host genome to drive the expression of two-cell stage-related genes, acting as an alternative promoter of genes expressed during zygotic genome activation, and consequently for the establishment of totipotency (Evsikov et al. 2004; Peaston et al. 2004; Franke et al. 2017). Ectopic expression of DUX in mouse ESCs not only triggers the expression of MERVL, but also a significant portion of the two-cell-specific transcriptional program (De Iaco et al. 2017). Using sequence-specific targeting approaches in preimplantation mouse embryos, L1 transcription was also shown to be necessary for development to proceed, most likely by regulating global chromatin accessibility (Jachowicz et al. 2017). Even though the transcriptional activation of SINE B2 during development has not yet been found to play a specific biological function during this time window, they appear to play a role in chromatin organization in other developmental contexts, whereby their bi-directional transcription acts as a domain insulator element (Lunyak et al. 2007). Thus, future work on understanding the interplay with additional TE families as well as the regulatory machinery in the host will certainly provide exciting findings.
Concluding remarks: transposable elements as parasites, co-opts, or symbionts?
Above, we have reviewed the instances in which host TFs have been shown to bind retrotransposons or their remnants, in our genomes. A fraction of these TFs have been demonstrated to activate transcription of their cognate retrotransposon. While we focused primarily on relatively young elements, older retrotransposons such as CR1, L2, or MIR, which expanded earlier during mammalian evolution, might have laid the groundwork for mammalian-specific regulatory networks. While these older elements have not yet been extensively ascribed to gene regulatory functions, L2 has recently been found to be more frequently represented within enhancers than younger LINE elements (Zhou et al. 2020). Thus, assessing how much of the mammalian-specific regulatory networks arose from these ancient TEs will be extremely exciting.
We have also portrayed the biological and evolutionary context of these observations, which in several cases have been shown to impact key physiological processes of the host. Particularly important is the time window spanning preimplantation development, as it provides a golden opportunity for TE expansion (Fig. 2). The work discussed above documents the strong biological relevance for the transcription of TEs during early mammalian development. However, conceptually, a major question remains: Who actually exploited whom? One possibility is that the host itself evolves these TFBS, thereby using the multiple TE insertions to efficiently multiply a platform for coordinated TF activity across the genome. It could also be that the transcriptional activation in germ cells and perhaps preimplantation development represents a mechanism mediated by the host to sense TEs and consequently repress them. In fact, it has been suggested that the “loss” of silencing chromatin marks and the TE activation occurring in germ cells would constitute a way to sense and repress them, through the piRNA pathway (Zamudio and Bourc'his 2010). Finally, it could be that the expansion of TFBS within the TEs have resulted from TE selfish strategies to promote their own replication within the host and ensure their transmission to subsequent generations. Nevertheless, to date, there is no evidence of retrotransposition occurring prior to the blastocyst stage. Even if the answer to this question remains unresolved, it is reasonable to view TEs as symbionts, rather than co-opts or parasites, whereby a long-standing relationship involving a “give and take” between TEs and the host has shaped the modern genomes and the regulatory networks within them.
Acknowledgments
We thank Diego Rodriguez-Terrones and Adam Burton for critical reading of the manuscript. Work in the Torres-Padilla laboratory is funded by the Helmholtz-Gemeinschaft, the Deutsche Forschungsgemeinschaft (CRC 1064), and H2020 Marie Skłodowska-Curie Actions (ITN EpiSystem and ChromDesign).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.344473.120.
References
- Alexandrova EA, Olovnikov IA, Malakhova GV, Zabolotneva AA, Suntsova MV, Dmitriev SE, Buzdin AA. 2012. Sense transcripts originated from an internal part of the human retrotransposon LINE-1 5′ UTR. Gene 511: 46–53. 10.1016/j.gene.2012.09.026 [DOI] [PubMed] [Google Scholar]
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. 2004. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol 11: 816–821. 10.1038/nsmb813 [DOI] [PubMed] [Google Scholar]
- Athanikar JN, Badge RM, Moran JV. 2004. A YY1-binding site is required for accurate human LINE1 transcription initiation. Nucleic Acids Res 32: 3846–3855. 10.1093/nar/gkh698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker KG, Swergold G, Ozato K, Thayer RE. 1993. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet 2: 1697–1702. 10.1093/hmg/2.10.1697 [DOI] [PubMed] [Google Scholar]
- Becker KG, Jedlicka P, Templeton NS, Liotta L, Keiko O. 1994. Characterization of hUCRBP (YY1, NF-E1, δ): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene 150: 259–266. 10.1016/0378-1119(94)90435-9 [DOI] [PubMed] [Google Scholar]
- Bénit L, Lallemand J-B, Casella J-F, Philippe H, Heidmann T. 1999. ERV-L elements: a family of endogenous retrovirus-like elements active throughout the evolution of mammals. J Virol 73: 3301–3308. 10.1128/JVI.73.4.3301-3308.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EA, Doms RW, Fenyö E-M, Korber BTM, Littman DR, Moore JP, Sattentau QJ, Schuitemaker H, Sodroski J, Weiss RA. 1998. A new classification for HIV-1. Nature 391: 240–240. 10.1038/34571 [DOI] [PubMed] [Google Scholar]
- Boeke JD, Corces VG. 1989. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43: 403–434. 10.1146/annurev.mi.43.100189.002155 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol 17: 915–928. 10.1093/oxfordjournals.molbev.a026372 [DOI] [PubMed] [Google Scholar]
- Bošković A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EVS, Meshorer E, TorresPadilla M-E. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 28: 1042–1047. 10.1101/gad.238881.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew J-L, Ruan Y, Wei C-L, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762. 10.1101/gr.080663.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. 1969. Gene regulation for higher cells: a theory. Science 165: 349–357. 10.1126/science.165.3891.349 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla M-E. 2014. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 15: 723–735. 10.1038/nrm3885 [DOI] [PubMed] [Google Scholar]
- Burton A, Brochard V, Galan C, Ruiz-Morales ER, Rovira Q, Rodriguez-Terrones D, Kruse K, Le Gras S, Udayakumar VS, Chin HG, et al. 2020. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by nonrepressive H3K9me3. Nat Cell Biol 22: 767–778. 10.1038/s41556-020-0536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. 10.1101/gad.241661.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ara T, Gautheret D. 2009. Using Alu elements as polyadenylation sites: a case of retroposon exaptation. Mol Biol Evol 26: 327–334. 10.1093/molbev/msn249 [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Schmid CW. 1996. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol 42: 30–36. 10.1007/BF00163208 [DOI] [PubMed] [Google Scholar]
- Choi SH, Gearhart MD, Cui Z, Bosnakovski D, Kim M, Schennum N, Kyba M. 2016. DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res 44: 5161–5173. 10.1093/nar/gkw141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Lin C-P, Risso D, Chen S, Kim TA, Tan MH, Li JB, Wu Y, Chen C, Xuan Z, et al. 2017. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science 355: eaag1927 10.1126/science.aag1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy RJ, Huang RC. 1988. Functional analysis of the long terminal repeats of intracisternal aparticle genes: sequences within the U3 region determine both the efficiency and direction of promoter activity. Mol Cell Biol 8: 1093–1102. 10.1128/MCB.8.3.1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114. 10.1016/j.gene.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Conley AB, Miller WJ, Jordan IK. 2008. Human cis natural antisense transcripts initiated by transposable elements. Trends Genet 24: 53–56. 10.1016/j.tig.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Cruickshanks HA, Tufarelli C. 2009. Isolation of cancer-specific chimeric transcripts induced by hypomethylation of the LINE-1 antisense promoter. Genomics 94: 397–406. 10.1016/j.ygeno.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Csumita M, Csermely A, Horvath A, Nagy G, Monori F, Göczi L, Orbea H-A, Reith W, Széles L. 2020. Specific enhancer selection by IRF3, IRF5 and IRF9 is determined by ISRE half-sites, 5′ and 3′ flanking bases, collaborating transcription factors and the chromatin environment in a combinatorial fashion. Nucleic Acids Res 48: 589–604. 10.1093/nar/gkz1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack M, King HW, Spingardi P, Kessler BM, Klose RJ, Kriaucionis S. 2020. Distinct contributions of DNA methylation and histone acetylation to the genomic occupancy of transcription factors. Genome Res 30: 1393–1406. 10.1101/gr.257576.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Kazazian HH. 1999. Analysis of the promoter from an expanding mouse retrotransposon subfamily. Genomics 56: 317–323. 10.1006/geno.1998.5729 [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. 2017. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49: 941–945. 10.1038/ng.3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P. 2011. Alu elements: know the SINEs. Genome Biol 12: 236 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger PL, Jolly DJ, Rubin CM, Friedmann T, Schmid CW. 1981. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol 151: 17–33. 10.1016/0022-2836(81)90219-9 [DOI] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. 2011. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 7: e1002384 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MCN, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al. 2015. Primate-Specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163: 583–593. 10.1016/j.cell.2015.09.025 [DOI] [PubMed] [Google Scholar]
- de Souza FSJ, Franchini LF, Rubinstein M. 2013. Exaptation of transposable elements into novel cisregulatory elements: is the evidence always strong? Mol Biol Evol 30: 1239–1251. 10.1093/molbev/mst045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. 2005. L1-mediated retrotransposition of murine B1 and B2 SINEs recapitulated in cultured cells. J Mol Biol 349: 241–247. 10.1016/j.jmb.2005.03.068 [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48. 10.1038/ng1223 [DOI] [PubMed] [Google Scholar]
- Ecco G, Imbeault M, Trono D. 2017. KRAB zinc finger proteins. Development 144: 2719–2729. 10.1242/dev.132605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl JO, Giesige CR, Domire JS, Wallace LM, Fowler AM, Guckes SM, Garwick-Coppens SE, Labhart P, Harper SQ. 2016. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum Mol Genet 25: 4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325 10.1371/journal.pgen.1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, et al. 2004. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res 105: 240–250. 10.1159/000078195 [DOI] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. 2013. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol 20: 332–338. 10.1038/nsmb.2495 [DOI] [PubMed] [Google Scholar]
- Falzon M, Kuff EL. 1988. Multiple protein-binding sites in an intracisternal A particle long terminal repeat. J Virol 62: 4070–4077. 10.1128/JVI.62.11.4070-4077.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning T, Singer M. 1987a. The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res 15: 2251–2260. 10.1093/nar/15.5.2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning TG, Singer MF. 1987b. LINE-1: a mammalian transposable element. Biochim Biophys Acta 910: 203–212. 10.1016/0167-4781(87)90112-6 [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Virolle T, Djabari Z, Ortonne J-P, White RJ, Aberdam D. 2001. Transposable B2 SINE elements can provide mobile RNA polymerase II promoters. Nat Genet 28: 77–81. [DOI] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405. 10.1038/nrg2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan DJ. 1989. Eukaryotic transposable elements and genome evolution. Trends Genet 5: 103–107. 10.1016/0168-9525(89)90039-5 [DOI] [PubMed] [Google Scholar]
- Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, et al. 2017. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res 27: 1384–1394. 10.1101/gr.216150.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs NV, Kraft M, Tondera C, Hanschmann K-M, Löwer J, Löwer R. 2011. Expression of the human endogenous retrovirus (HERV) group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by transcription factors Sp1 and Sp3. J Virol 85: 3436–3448. 10.1128/JVI.02539-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes DR, Swigut T, Wysocka J. 2018. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. Elife 7: e35989 10.7554/eLife.35989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV. 2000. The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64: 255–294. 10.1016/S0079-6603(00)64007-2 [DOI] [PubMed] [Google Scholar]
- Gao L, Wu K, Liu Z, Yao X, Yuan S, Tao W, Yi L, Yu G, Hou Z, Fan D, et al. 2018. Chromatin accessibility landscape in human early embryos and Its association with evolution. Cell 173: 248–259.e15. 10.1016/j.cell.2018.02.028 [DOI] [PubMed] [Google Scholar]
- Geng LN, Yao Z, Snider L, Fong AP, Cech JN, Young JM, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al. 2012. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 22: 38–51. 10.1016/j.devcel.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göke J, Lu X, Chan Y-S, Ng H-H, Ly L-H, Sachs F, Szczerbinska I. 2015. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell 16: 135–141. 10.1016/j.stem.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Goodier JL. 2016. Restricting retrotransposons: a review. Mob DNA 7: 16 10.1186/s13100-016-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–35. 10.1016/j.cell.2008.09.022 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Du K, Kazazian HH. 2001. A novel active L1 retrotransposon subfamily in the mouse. Genome Res 11: 1677–1685. 10.1101/gr.198301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche D, Martin L, Ware C, Blish CA, Chang HY, et al. 2015. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature 522: 221–225. 10.1038/nature14308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Pérez-Palacios R, Gahurova L, Schubert M, Krueger F, Biggins L, Andrews S, Colomé Tatché M, Bourc'his D, Dean W, et al. 2019. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol 20: 225 10.1186/s13059-019-1833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CR, DeWan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. 2009. P53 responsive elements in human retrotransposons. Oncogene 28: 3857–3865. 10.1038/onc.2009.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim J-W, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. 2017. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 49: 925–934. 10.1038/ng.3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AFA, Wheeler TJ. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res 44: D81–D89. 10.1093/nar/gkv1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Trono D. 2014. As time goes by: KRABs evolve to KAP endogenous retroelements. Dev Cell 31: 257–258. 10.1016/j.devcel.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid P-Y, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543: 550–554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla M-E. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Ito J, Sugimoto R, Nakaoka H, Yamada S, Kimura T, Hayano T, Inoue I. 2017. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet 13: e1006883 10.1371/journal.pgen.1006883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Bošković A, Rando OJ, Torres-Padilla M-E. 2017. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet 49: 1502–1510. 10.1038/ng.3945 [DOI] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques P-É, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences Are derived from transposable elements. PLoS Genet 9: e1003504 10.1371/journal.pgen.1003504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 51: 611–617. 10.1038/s41588-019-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J-C, Upton KR. 2019. Human transposons are an abundant supply of transcription factor binding sites and promoter activities in breast cancer cell lines. Mob DNA 10: 16 10.1186/s13100-019-0158-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. 2003. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet 19: 68–72. 10.1016/S0168-9525(02)00006-9 [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res 16: 78–87. 10.1101/gr.4001406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knössl M, Löwer R, Löwer J. 1999. Expression of the human endogenous retrovirus HTDV/HERV-K is enhanced by cellular transcription factor YY1. J Virol 73: 1254–1261. 10.1128/JVI.73.2.1254-1261.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb) 107: 487–495. 10.1038/hdy.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev AS, Kramerov DA, Skryabin KG, Ryskov AP, Bayev AA, Georgiev GP. 1980. The nucleotide sequence of the ubiquitous repetitive DNA sequence B1 complementary to the most abundant class of mouse fold-back RNA. Nucleic Acids Res 8: 1201–1215. 10.1093/nar/8.6.1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia N-Y, Jeyakani J, Hwang C, Lu X, Chan Y-S, Ng H-H, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet 42: 631–634. 10.1038/ng.600 [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, Lie DC, Moore L, Nakashima K, Asashima M, Gage FH. 2009. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci 12: 1097–1105. 10.1038/nn.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. 2004. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res 14: 2253–2260. 10.1101/gr.2745804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison CA. 1986. The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol 6: 168–182. 10.1128/MCB.6.1.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Prefontaine GG, Núñez E, Cramer T, Ju B-G, Ohgi KA, Hutt K, Roy R, García-Díaz A, Zhu X, et al. 2007. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science 317: 248–251. 10.1126/science.1140871 [DOI] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, Stoye JP. 2015. Mammalian endogenous retroviruses. Microbiol Spectr 3: MDNA3-0009–2014 10.1128/microbiolspec.MDNA3-0009-2014 [DOI] [PubMed] [Google Scholar]
- Manghera M, Douville RN. 2013. Endogenous retrovirus-K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10: 16 10.1186/1742-4690-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- McClintock B. 1950. The origin and behavior of mutable loci in maize. PNAS 36: 344–355. 10.1073/pnas.36.6.344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. 1956. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol 21: 197–216. 10.1101/SQB.1956.021.01.017 [DOI] [PubMed] [Google Scholar]
- Minakami R, Kurose K, Etoh K, Furuhata Y, Hattori M, Sakaki Y. 1992. Identification of an internal cis-element essential for the human Li transcription and a nuclear factor(s) binding to the element. Nucleic Acids Res 20: 3139–3145. 10.1093/nar/20.12.3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Malik HS. 2016. Hide and seek: how chromatin-based pathways silence retroelements in the mammalian germline. Curr Opin Genet Dev 37: 51–58. 10.1016/j.gde.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. 1998. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17: 590–597. 10.1093/emboj/17.2.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Mätlik K, Speek M. 2002. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics 79: 628–634. 10.1006/geno.2002.6758 [DOI] [PubMed] [Google Scholar]
- Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, et al. 2014. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci 111: 12426–12431. 10.1073/pnas.1413299111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orqueda AJ, Gatti CR, Ogara MF, Falzone TL. 2018. SOX-11 regulates LINE-1 retrotransposon activity during neuronal differentiation. FEBS Lett 592: 3708–3719. 10.1002/1873-3468.13260 [DOI] [PubMed] [Google Scholar]
- Padgett RW, Hutchison CA, Edgell MH. 1988. The F-type 5′ motif of mouse L1 elements: a major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucleic Acids Res 16: 739–749. 10.1093/nar/16.2.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquesi GIM, Perry BW, Vandewege MW, Ruggiero RP, Schield DR, Castoe TA. 2020. Vertebrate lineages exhibit diverse patterns of transposable element regulation and expression across tissues. Genome Biol Evol 12: 506–521. 10.1093/gbe/evaa068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 7: 597–606. 10.1016/j.devcel.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, Corbin A, Nigumann P, Cristofari G. 2016. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 5: e13926 10.7554/eLife.13926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. 1992. Origin of the Alu family: a family of Alu-like monomers gave birth to the left and the right arms of the Alu elements. Nucleic Acids Res 20: 3397–3401. 10.1093/nar/20.13.3397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. 1994. A master sequence related to a free left Alu monomer (FLAM) at the origin of the B1 family in rodent genomes. Nucleic Acids Res 22: 2222–2227. 10.1093/nar/22.12.2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46: 21–42. 10.1146/annurev-genet-110711-155621 [DOI] [PubMed] [Google Scholar]
- Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr 3: MDNA3-0061–2014 10.1128/microbiolspec.MDNA3-0061-2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea G-O, Muñoz-Lopez M, Jesuadian JS, Kempen M-JHC, Carreira PE, Jeddeloh JA, et al. 2017. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res 27: 1395–1405. 10.1101/gr.219022.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Terrones D, Torres-Padilla M-E. 2018. Nimble and ready to mingle: transposon outbursts of early development. Trends Genet 34: 806–820. 10.1016/j.tig.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Roulston A, Lin R, Beauparlant P, Wainberg MA, Hiscott J. 1995. Regulation of human immunodeficiency virus type 1 and cytokine gene expression in myeloid cells by NF-κB/Rel transcription factors. Microbiol Mol Biol Rev 59: 481–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, LayardLiesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, El-Sawy M, Farooq L, Odom GL, Perepelitsa-Belancio V, Bruch H, Oyeniran OO, Deininger PL. 2005. Human retroelements may introduce intragenic polyadenylation signals. Cytogenet Genome Res 110: 365–371. 10.1159/000084968 [DOI] [PubMed] [Google Scholar]
- Sanchez-Luque FJ, Kempen M-JHC, Gerdes P, Vargas-Landin DB, Richardson SR, Troskie R-L, Jesuadian JS, Cheetham SW, Carreira PE, Salvador-Palomeque C, et al. 2019. LINE-1 evasion of epigenetic repression in humans. Mol Cell 75: 590–604.e12. 10.1016/j.molcel.2019.05.024 [DOI] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620. 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1: 113–125. 10.1016/0888-7543(87)90003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Shi Y, Shenk T. 1991. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature 354: 241–245. 10.1038/354241a0 [DOI] [PubMed] [Google Scholar]
- Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, et al. 2008. IRF-1 Is required for full NF-κB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J Virol 82: 3632–3641. 10.1128/JVI.00599-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjøttem E, Anderssen S, Johansen T. 1996. The promoter activity of long terminal repeats of the HERV-H family of human retrovirus-like elements is critically dependent on Sp1 family proteins interacting with a GC/GT box located immediately 3′ to the TATA box. J Virol 70: 188–198. 10.1128/JVI.70.1.188-198.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Singer MF. 1985. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc Natl Acad Sci 82: 6050–6054. 10.1073/pnas.82.18.6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. 2014. DNA methylation dynamics of the human preimplantation embryo. Nature 511: 611–615. 10.1038/nature13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol 21: 1973–1985. 10.1128/MCB.21.6.1973-1985.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang X, Tang Z, Grivainis M, Kahler D, Yun C, Mita P, Fenyö D, Boeke JD. 2018. Transcription factor profiling reveals molecular choreography and key regulators of human retrotransposon expression. Proc Natl Acad Sci 115: E5526–E5535. 10.1073/pnas.1722565115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. 2004. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol 269: 276–285. 10.1016/j.ydbio.2004.01.028 [DOI] [PubMed] [Google Scholar]
- Swergold GD. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10: 6718–6729. 10.1128/MCB.10.12.6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchénio T, Casella JF, Heidmann T. 2000. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res 28: 411–415. 10.1093/nar/28.2.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Schneider S. 2011. Coevolution of retroelements and tandem zinc finger genes. Genome Res 21: 1800–1812. 10.1101/gr.121749.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PJ, Macfarlan TS, Lorincz MC. 2016. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62: 766–776. 10.1016/j.molcel.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Kapusta A, Brown CD. 2018. Transposable elements generate regulatory novelty in a tissue-specific fashion. BMC Genomics 19: 468 10.1186/s12864-018-4850-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. 1984. Alu sequences are processed 7SL RNA genes. Nature 312: 171–172. 10.1038/312171a0 [DOI] [PubMed] [Google Scholar]
- Ullu E, Weiner AM. 1985. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. Nature 318: 371–374. 10.1038/318371a0 [DOI] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry J-R, Mager DL, Medstrand P. 2003. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19: 530–536. 10.1016/j.tig.2003.08.004 [DOI] [PubMed] [Google Scholar]
- Vargiu L, Rodriguez-Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J. 2016. Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13: 7 10.1186/s12977-015-0232-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney D, Vavrova-Anderson J, Oler AJ, Cairns BR, White RJ. 2015. Selective repression of SINE transcription by RNA polymerase III. Mob Genet Elements 5: 86–91. 10.1080/2159256X.2015.1096997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, Rynes E, Sandstrom R, Zhang M, Canfield T, Hansen RS, Stehling-Sun S, Sabo PJ, Byron R, Humbert R, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cisregulatory evolution. Science 346: 1007–1012. 10.1126/science.1246426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt VM. 1997. Retroviral virions and genomes. In: Retroviruses (ed. Coffin JM et al. ), p 27–69. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: https://www.ncbi.nlm.nih.gov/books/NBK19454 [PubMed] [Google Scholar]
- Voliva CF, Jahn CL, Comer MB, Hutchison CA, Edgell MH. 1983. The L1Md long interspersed repeat family in the mouse: almost all examples are truncated at one end. Nucleic Acids Res 11: 8847–8859. 10.1093/nar/11.24.8847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci 104: 18613–18618. 10.1073/pnas.0703637104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xie G, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. 2014. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 516: 405–409. 10.1038/nature13804 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu X, Gao Y, Yang L, Li C, Liu W, Chen C, Kou X, Zhao Y, Chen J, et al. 2018. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat Cell Biol 20: 620–631. 10.1038/s41556-018-0093-4 [DOI] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong C-J, Zhong JW, Tapscott SJ. 2017. Conservation and innovation in the DUX4-family gene network. Nat Genet 49: 935–940. 10.1038/ng.3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8: 973–982. 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Wolf G, de Iaco A, Sun M-A, Bruno M, Tinkham M, Hoang D, Mitra A, Ralls S, Trono D, Macfarlan TS. 2020. KRAB-zinc finger protein gene expansion in response to active retrotransposons in the murine lineage. Elife 9: e56337 10.7554/eLife.56337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. 2016. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534: 652–657. 10.1038/nature18606 [DOI] [PubMed] [Google Scholar]
- Wylie A, Jones AE, D'Brot A, Lu W-J, Kurtz P, Moran JV, Rakheja D, Chen KS, Hammer RE, Comerford SA, et al. 2016. p53 genes function to restrain mobile elements. Genes Dev 30: 64–77. 10.1101/gad.266098.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D, Chen C-C, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S. 2010. Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 20: 804–815. 10.1101/gr.100594.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zhang L, Zhang Y, Kazazian HH Jr. 2003. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res 31: 4929–4940. 10.1093/nar/gkg663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Y, Macfarlan TS. 2017. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet 33: 871–881. 10.1016/j.tig.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Whiddon JL, Yao Z, Kasinathan B, Snider L, Geng LN, Balog J, Tawil R, van der Maarel SM, Tapscott SJ. 2013. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet 9: e1003947 10.1371/journal.pgen.1003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamudio N, Bourc'his D. 2010. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity 105: 92–104. 10.1038/hdy.2010.53 [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. 1995. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem 64: 533–561. 10.1146/annurev.bi.64.070195.002533 [DOI] [PubMed] [Google Scholar]
- Zhou M, Smith AD. 2019. Subtype classification and functional annotation of L1Md retrotransposon promoters. Mob DNA 10: 14 10.1186/s13100-019-0156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Liang G, Molloy PL, Jones PA. 2020. DNA methylation enables transposable element-driven genome expansion. PNAS 117: 19359–19366. 10.1073/pnas.1921719117 [DOI] [PMC free article] [PubMed] [Google Scholar]