In this study, Muys et al. set out to elucidate new mechanisms responsible for p53-mediated tumor suppression. Using both in vivo and genomic analyses, the authors report splicing regulation as a direct effect of the p53 pathway via the activation of the splicing regulator ZMAT3.
Keywords: p53, RNA-binding protein, RBP, ZMAT3, splicing, CD44, colorectal carcinoma, PAR-CLIP
Abstract
p53 is an intensely studied tumor-suppressive transcription factor. Recent studies suggest that the RNA-binding protein (RBP) ZMAT3 is important in mediating the tumor-suppressive effects of p53. Here, we globally identify ZMAT3-regulated RNAs and their binding sites at nucleotide resolution in intact colorectal cancer (CRC) cells. ZMAT3 binds to thousands of mRNA precursors, mainly at intronic uridine-rich sequences and affects their splicing. The strongest alternatively spliced ZMAT3 target was CD44, a cell adhesion gene and stem cell marker that controls tumorigenesis. Silencing ZMAT3 increased inclusion of CD44 variant exons, resulting in significant up-regulation of oncogenic CD44 isoforms (CD44v) and increased CRC cell growth that was rescued by concurrent knockdown of CD44v. Silencing p53 phenocopied the loss of ZMAT3 with respect to CD44 alternative splicing, suggesting that ZMAT3-mediated regulation of CD44 splicing is vital for p53 function. Collectively, our findings uncover a p53–ZMAT3–CD44 axis in growth suppression in CRC cells.
The p53 protein is known as the “guardian of genome” and is the most frequently mutated gene in human cancers. Mechanistically, p53 directly activates the transcription of hundreds of genes by binding to p53 response elements at their promoter regions. When activated, its targets in turn mediate diverse functions, including but not limited to DNA repair, apoptosis, autophagy, cell cycle arrest, metabolism, and senescence (Vogelstein et al. 2000). Despite the critical role of p53 in tumor suppression and control of cell cycle arrest, apoptosis, and senescence, the mechanisms responsible for p53-mediated tumor suppression are not fully understood. To give an example, unlike p53-knockout mice that are highly prone to spontaneous tumor development, triple knockout mice for three major p53 target genes (p21, Noxa, and Puma) controlling cell cycle arrest and apoptosis, are not prone to spontaneous tumor development (Valente et al. 2013). This suggests that p53 targets regulating cellular processes other than cell cycle arrest and apoptosis could be important for p53-mediated tumor suppression. A focus on direct p53 targets could obscure its potential for broad rewiring of post-transcriptional gene regulatory pathways by controlling key RNA-binding proteins (RBPs), each of which can in turn regulate the expression of hundreds or thousands of mRNA targets (Gerstberger et al. 2014). While an unbiased survey of RBPs regulated by p53 is presently missing, there is evidence of p53 directly inducing specific RBPs, such as ZMAT3 (Israeli et al. 1997; Hellborg et al. 2001). A recent study uncovered a critical role of the ZMAT3 in p53-mediated tumor suppression (Janic et al. 2018). However, the molecular mechanism(s) by which ZMAT3 functions in an intact cell remains to be elucidated. Identifying the RNA targets of ZMAT3 is critical for better characterizing the genes and pathways it regulates downstream from p53 and for uncovering additional mechanisms contributing to p53-mediated tumor suppression.
ZMAT3 is a relatively small, 32-kDa zinc finger domain containing RBP highly conserved in vertebrates, from fish to humans (Israeli et al. 1997; Hellborg et al. 2001; Hellborg and Wiman 2004). ZMAT3 predominantly localizes to the nucleus and its three C2H2-type zinc finger domains prefer binding to RNA over DNA in vitro (Méndez-Vidal et al. 2002). Little is known about its target RNAs and regulatory impact on a transcriptome-wide scale. Nevertheless, based on RNA immunoprecipitation, biotin-pulldown, and reporter assays, ZMAT3 was previously suggested to interact with AU-rich elements in mRNA 3′ untranslated regions (UTR), either stabilizing its targets (e.g., p21 and p53) (Vilborg et al. 2009; Kim et al. 2012) or promoting their decay (FAS) (Bersani et al. 2014). Regulation of these key mRNAs has been linked to decreased clonogenicity and cell growth, suggesting a tumor suppressive role of ZMAT3 (Hellborg et al. 2001).
Here, we determined the set of RBPs up-regulated by p53 by integrating RNA-seq and GRO-seq after p53 induction with p53 ChIP-seq data. We found that ZMAT3 was the top of 11 p53-induced RBPs and we identified its RNA target sites in intact human colorectal cancer (CRC) cells on a transcriptome-wide scale and at nucleotide resolution by PAR-CLIP (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation) (Hafner et al. 2010). We focused on CD44, a potent oncogene and stem cell marker (Zöller 2011) because it was among the top PAR-CLIP targets and exhibited considerable changes in alternative splicing patterns upon ZMAT3 knockdown. Silencing ZMAT3 led to the inclusion of variant CD44 exons resulting in up-regulation of the oncogenic, longer CD44 variants (CD44v) and down-regulation of the standard CD44s isoforms. Importantly, silencing p53 phenocopied the loss of ZMAT3 with respect to alternative splicing of CD44, suggesting that ZMAT3-mediated regulation of CD44 splicing played a role in p53 function. Furthermore, the growth advantage after knockdown of ZMAT3 was rescued by concurrent knockdown of CD44v, suggesting that CD44v is a major downstream effector of ZMAT3. Collectively, our findings uncover a direct role of ZMAT3 in regulation of RNA splicing and demonstrate that expression of oncogenic CD44 splice variants is controlled by p53 via ZMAT3 to suppress growth and proliferation in human CRC cells.
Results
p53 directly induces the expression of specific RBPs, including ZMAT3
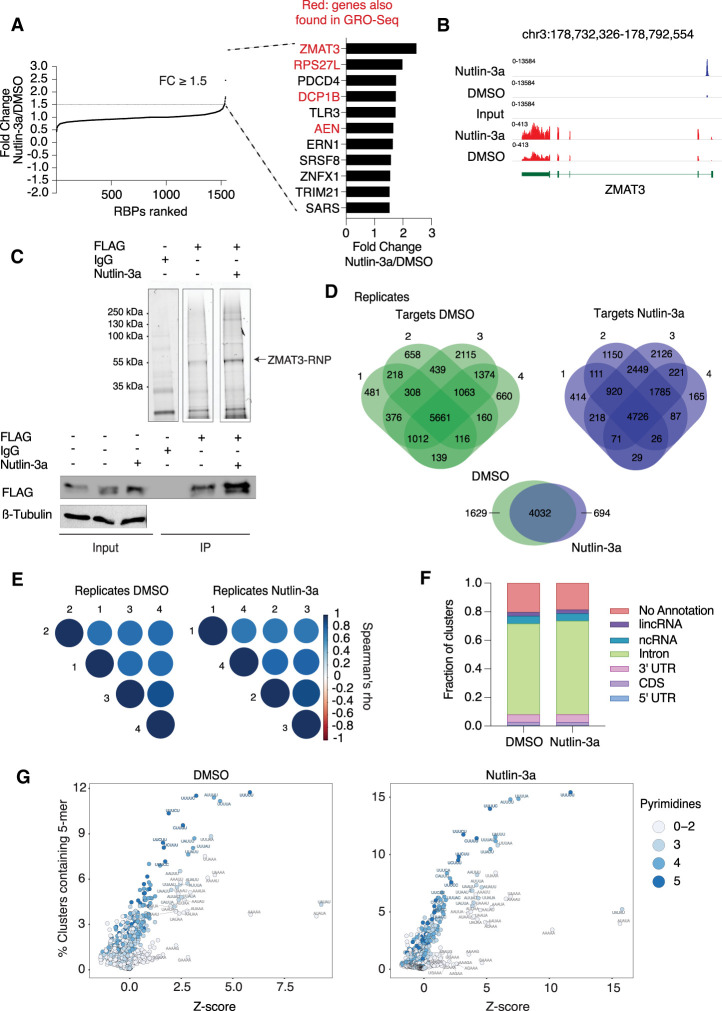
We aimed to determine the set of RBPs induced by p53 in an unbiased manner. To do this, we performed RNA-seq from HCT116 (CRC) cells treated with DMSO or Nutlin-3a, a molecule that induces p53 (Supplemental Table S1). Of the 1542 manually curated RBPs in the human genome (Gerstberger et al. 2014), 11 RBPs increased and 2 RBPs decreased in abundance more than 1.5-fold (P < 0.05) upon Nutlin-3a treatment (Supplemental Table S2). Among these, ZMAT3 was the most up-regulated and changed ∼2.5-fold (Fig. 1A), which is a modest increase but of similar order of magnitude as canonical p53 targets, such as CDKN1B, BTG2, GDF15, MDM2, and TP53INP1, that are induced ∼4.5-fold to sixfold upon p53 activation (Supplemental Table S1).
Figure 1.
ZMAT3 is the top RBP up-regulated by p53 and binds to thousands of pre-mRNAs at uridine rich sequences. (A) RBPs in HCT116 cells ranked according to their fold change upon activation of p53 by Nutlin-3a samples compared with DMSO. The 11 up-regulated RBPs sorted in decreasing order of up-regulation by p53 (fold change ≥ 1.5 and P < 0.05 as cutoff). Red font indicates whether their transcriptional activity increased upon p53 stimulation in GRO-seq experiments (Allen et al. 2014). (B) Genome browser snapshot from p53 ChIP-seq (blue tracks) and RNA-seq (red tracks) samples using HCT116 cells treated with Nutlin-3a or DMSO. ChIP-seq data were obtained from Andrysik et al. (2017). (C) Representative ZMAT3 PAR-CLIP images. (Top) Fluorescent image of SDS-PAGE separating cross-linked ZMAT3-FLAG RNPs ligated to a fluorescent 3′ adapter after immunoprecipitation (IP) using anti-FLAG or IgG antibodies from HCT116 lysates with or without Nutlin-3a treatment. The fluorescent band migrating at ∼55 kDa, corresponding to the expected size of the ZMAT3-FLAG-RNA-3′ adapter complex, is indicated. The bottom panel shows control immunoblots before (input) and after IP stained for FLAG. (D, top panel) Venn diagrams of intersection of genes bound by ZMAT3 recovered from four independent PAR-CLIP experiments each after DMSO or Nutlin-3a treatment. (Bottom panel) Venn diagram of intersection of genes reproducibly bound by ZMAT3 in all replicates of PAR-CLIP after DMSO and Nutlin-3a treatments. (E) Correlogram using Spearman's correlation coefficient of cross-linked reads among PAR-CLIP samples replicates upon DMSO (right panel) or Nutlin-3a treatment (left panel). (F) Average of distribution of cross-linked sequence reads from four biological replicates across different annotation categories. (G) The dot plots depict Z-scores (X-axis) and frequency (Y-axis) for the occurrence of all 1024 possible 5-mers in ZMAT3 PAR-CLIP binding sites from HCT116 cells treated with Nutlin-3a (left) or DMSO (right). Shade of blue indicates the number of pyrimidines in the respective 5-mer.
We argued that RBPs directly induced by p53 at the transcriptional level can be revealed by measuring transcriptional activity by GRO-seq, a method that captures nascent RNAs. We mined a previously published GRO-seq data set from HCT116 cells after 1 h of Nutlin-3a treatment (Allen et al. 2014). Considering that p53 is a transcriptional activator, we focused on the 198 genes that showed increased transcriptional activity after p53 activation. Among those, four of the 11 up-regulated RBPs showed increased transcriptional activity: ZMAT3, RPS27L, DCP1B, and AEN. Using publicly available p53 ChIP-seq data (Andrysik et al. 2017) we found strong enrichment of p53 at the promoter of ZMAT3 (Fig. 1B). Finally, we validated the activity of the previously reported p53 response element (RE) in the promoter of ZMAT3 (Wilhelm et al. 2002) with luciferase assays using the wild-type or mutant p53 RE (Supplemental Fig. S1A). Collectively, these lines of evidence indicated a potential role of p53 in indirectly reprograming post-transcriptional gene regulatory pathways by inducing a distinct set of RBPs.
ZMAT3 binds to thousands of precursor mRNA molecules at uridine-rich sequences
Considering that previous reports point to a role for ZMAT3 in tumor suppression and that ZMAT3 was the top RBP induced by p53, we decided to identify its RNA ligands, as well as its post-transcriptional gene regulatory impact in order to understand whether and how ZMAT3 could contribute to p53 function.
We used lentiviral infection to generate a stable HCT116 cell line expressing FLAG epitope-tagged ZMAT3 under the control of 13 p53 REs in its promoter (Supplemental Fig. S1B). Similar to the endogenous protein, transgenic ZMAT3-FLAG expression was indeed p53-dependent and increased by Nutlin-3a treatment approximately twofold and decreased upon p53 knockdown (kd) (Supplemental Fig. S1B,D).
In order to reliably capture ZMAT3 binding sites and characterize its RNA recognition elements (RREs) we mapped the RNA interactome of ZMAT3 in these cells on a transcriptome-wide scale at nucleotide resolution by 4-thiouridine (4SU) PAR-CLIP (Hafner et al. 2010; Benhalevy et al. 2017). For PAR-CLIP we used four biological replicates each from HCT116-ZMAT3-FLAG cells treated either with Nutlin-3a or DMSO. Fluorescence imaging of cross-linked, ribonuclease-treated, FLAG immunoprecipitate revealed a single ribonucleoprotein (RNP) complex at 55 kDa, corresponding to the ZMAT3-FLAG-RNP (37 kDa) ligated to a fluorescently labeled 3′ adapter (∼20 kDa) (Fig. 1C). After Nutlin-3a treatment the levels of the cross-linked and immunoprecipitated ZMAT3-FLAG-RNP increased in the PAR-CLIP experiments, consistent with the Nutlin-3a-dependent increase in ZMAT3-FLAG transgene expression (Supplemental Fig. S1D, left panel).
We recovered bound RNA fragments from ZMAT3-RNPs from four biological replicates each from the HCT116-ZMAT3-FLAG cells treated with either DMSO or Nutlin-3a and transformed them into small RNA cDNA libraries for next-generation sequencing. Next, we determined clusters of overlapping reads that harbor characteristic T-to-C conversions diagnostic of 4SU cross-linking events at higher frequencies than expected by chance (see Supplemental Table S3 for summary statistics; Corcoran et al. 2011). For all eight samples we found between 35,430 and 205,648 binding sites (data available at GEO: GSE150718) (Supplemental Table S3). These binding sites distributed on a set of 5561 and 4726 ZMAT3-bound target genes shared in all replicates from DMSO and Nutlin-3a treatment, respectively (Fig. 1D), suggesting that a large proportion of expressed transcripts in these cells are bound by ZMAT3. The biological replicates showed excellent correlation, with an R2 between 0.7 and 0.8 for both DMSO- and Nutlin-3a-treated samples (Fig. 1E). Furthermore, the correlation between DMSO and Nutlin-3a treated samples exceeded 0.7 indicating that the ZMAT3 binding profile was largely independent of its expression levels. Considering the high correlations between the biological replicates, we pooled the respective samples for DMSO- or Nutlin-3a-treated cells, which yielded altogether 66,671 and 44,589 binding sites (Supplemental Table S4). Approximately 80% of ZMAT3 binding sites were found on mRNA transcripts and of those, >60% were found in intronic sequences (Fig. 1F), indicating that ZMAT3 interacted with nascent transcripts, consistent with its nuclear localization (Fig. 2D; Hellborg et al. 2001).
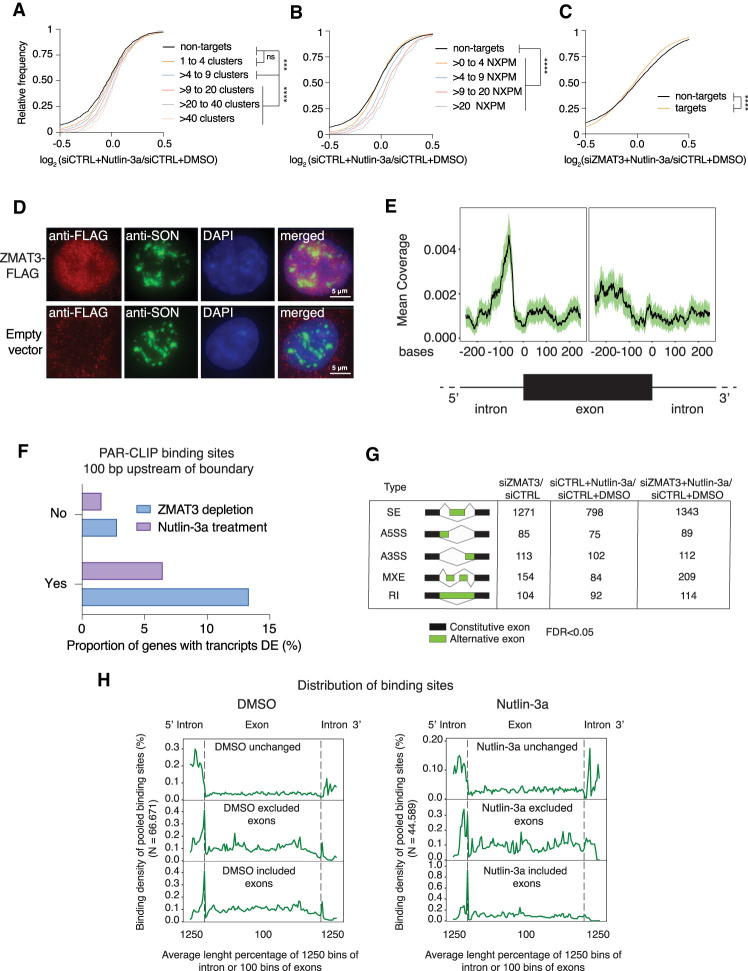
Figure 2.
ZMAT3 predominantly binds to regions close to 3′ splice sites and its knockdown induces widespread changes in splicing patterns. (A,B) mRNA expression changes upon ZMAT3 induction in HCT116 cells determined by RNA-seq in biological triplicates. The empirical cumulative distribution function of ZMAT3 PAR-CLIP targets (from HCT116 cells after Nutlin-3a treatment) binned by number of binding sites (A) or number of NXPM (cross-linked reads per million normalized to target gene expression levels) (B) compared with nontargets (FPKM > 5, black line). P-values were determined by Kolmogorov-Smirnov test (***) P < 0.001, (****) P < 0.0001, (ns) nonsignificant. (C) Same as A after knockdown of ZMAT3 and only separating ZMAT3 target genes (orange line) from nontargets (black line). (D) Fluorescent microscopy images of immunostaining experiments with HCT116-ZMAT3-FLAG (top images) and HCT116-FLAG cells stained with antibodies against FLAG (red) or the nuclear speckle marker protein SON (green). DNA is counterstained with DAPI (blue). (E) Plot of cross-linked sequence read coverage distributed along the intron-exon boundary from a representative sample of ZMAT3 PAR-CLIP from DMSO treated HCT116 cells. (F) Percentage of ZMAT3 target mRNAs with significantly changed abundance (P<0.05, Sleuth) in their transcripts that contained (Yes) or did not contain (No) ZMAT3-binding sites within 100 bp of the 3′ splice site. Note that the number of gene expression changes was significantly higher in HCT116 cells after ZMAT3 knockdown (kd) compared with ZMAT3 induction by Nutlin-3a. (DE) Differentially expressed. (G) Table summarizing alternative splicing events (FDR < 0.05, reads ≥ 5 and ΔPSI ≥ 10%) upon ZMAT3 kd, Nutlin-3a treatment, or ZMAT3 kd prior to Nutlin-3a treatment, compared with control HCT116 cells. SE, skipped exons; A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site; MXE, mutually exclusive exon; RI, retained introns. (H) Metagene plot of cross-linked sequence reads at the intron-exon boundary from pooled ZMAT3 PAR-CLIPs in DMSO- or Nutlin-3a-treated cells. The metagene plots were separated according to whether the exons were excluded, included, or unchanged upon ZMAT3 kd.
To determine the RNA recognition element of ZMAT3, we counted the occurrence of all possible 5-mer sequences in our binding sites and calculated their Z-scores over a background of shuffled sequences of the same nucleotide composition. Both PAR-CLIP data sets showed an enrichment of 5-mers that were containing uridines, with a statistically significant enrichment of those containing in addition As and to a lesser degree Cs (Fig. 1G; Supplemental Fig. S2A,B; Supplemental Table S5). In contrast, G-containing 5-mers were depleted (Supplemental Fig. S2A). Taken together our data indicate that ZMAT3 binds thousands of precursor mRNAs across the entire transcript body at U/A and pyrimidine-rich sequences.
ZMAT3 binding results in increased target mRNA abundance
In order to investigate the gene regulatory roles of ZMAT3, we next quantified mRNA abundance using total RNA sequencing (RNA-seq) from HCT116 cells upon knockdown of endogenous ZMAT3 using a pool of four specific siRNAs or negative control siRNA after induction of the p53 response with Nutlin-3a or DMSO vehicle.
Increasing levels of ZMAT3 by activating p53 led to an increase in target mRNA levels, dependent on the number of ZMAT3-binding sites (Fig. 2A) or the number of cross-linked reads per target mRNA normalized by overall mRNA abundance (normalized cross-linked reads per million, NXPM) (Fig. 2B). We previously found that both metrics correlated well with the occupancy of an RBP on its target (Hafner et al. 2010; Ascano et al. 2012; Yamaji et al. 2017). The positive effect of p53 activation on ZMAT3 target RNA levels was abolished by ZMAT3 knockdown (Fig. 2C). Finally, RNA levels of targets that contained the preferred sequence motifs in their ZMAT3-binding sites (Fig. 1G) were more likely to correlate with ZMAT3 levels compared with the rest of target genes (Supplemental Fig. S2C). Collectively, our data strongly suggest a direct effect of ZMAT3 binding on target RNA abundance.
ZMAT3 predominantly binds to introns near 3′ splice sites and affects splicing
ZMAT3 could affect target RNA levels via multiple mechanisms, either modulating their half-life, or interfering with their processing. Considering its preferential binding to pre-mRNAs, we asked whether ZMAT3 might be involved in splicing changes, which can then lead to mRNA abundance changes (Witten and Ule 2011). Immunostaining in HCT116-ZMAT3-FLAG cells revealed ZMAT3-FLAG localized predominantly in the nucleus where it was partially colocalized in nuclear speckles (Fig. 2D), structures also known as splicing factor-enriched compartments (Lamond and Spector 2003). Analysis of our binding sites suggested that the intronic region ∼100 nt upstream of mRNA 3′ splice sites was highly enriched for ZMAT3 binding (Fig. 2E). This region harbors the polypyrimidine tract that is crucial for recruitment of key splicing effector complexes to ensure the definition of the proper 3′ splice site. Furthermore, mRNAs with ZMAT3 binding sites in that region were more likely to be significantly differentially expressed upon changes in ZMAT3 levels (Fig. 2F). We used the rMATS software package (Shen et al. 2014) to predict alternative splicing (AS) events after ZMAT3 knockdown or Nutlin-3a treatment. Altogether, we identified 1727, 1151, and 1867 AS events (with at least five sequence reads covering the splice junction, FDR < 0.05 and ΔPSI (percent spliced in) ≥ 10%) upon ZMAT3 knockdown, Nutlin-3a, or combination of both treatments, respectively (Fig. 2G). The similarity of the AS event occurrence after ZMAT3 depletion with or without Nutlin-3a treatment suggests that changes in ZMAT3 may be the cause of the bulk of AS events mediated upon p53 activation, likely by direct regulation of its targets. Interestingly, we noticed an increase of ZMAT3 binding close to the 3′ splice site of exons that were differentially spliced following ZMAT3 knockdown, compared with unchanged exons (Fig. 2H). This is consistent with previous studies that found that splicing regulatory RBPs exhibit their strongest splicing promoting or suppressing effects when binding close to the splice sites (Dvinge et al. 2016).
Aberrant splicing events may cause the accumulation of damaged transcripts with premature termination codons and/or retained introns. Such transcripts are cleared by the nonsense-mediated decay (NMD) pathway (Ni et al. 2007) and we therefore tested whether mRNA abundance changes upon ZMAT3 depletion may be, at least partially, explained by NMD-dependent degradation of aberrantly spliced ZMAT3 target mRNAs. We performed RNA-seq after siRNA mediated knockdown of UPF1, a major factor for NMD (Supplemental Fig. S2D) and indeed found that alternatively spliced ZMAT3 target genes showed a modest increase in abundance compared with nontargets upon UPF1 depletion, suggesting that NMD could be at least one of the causes for their degradation.
Silencing ZMAT3 promotes expression of oncogenic CD44 isoforms
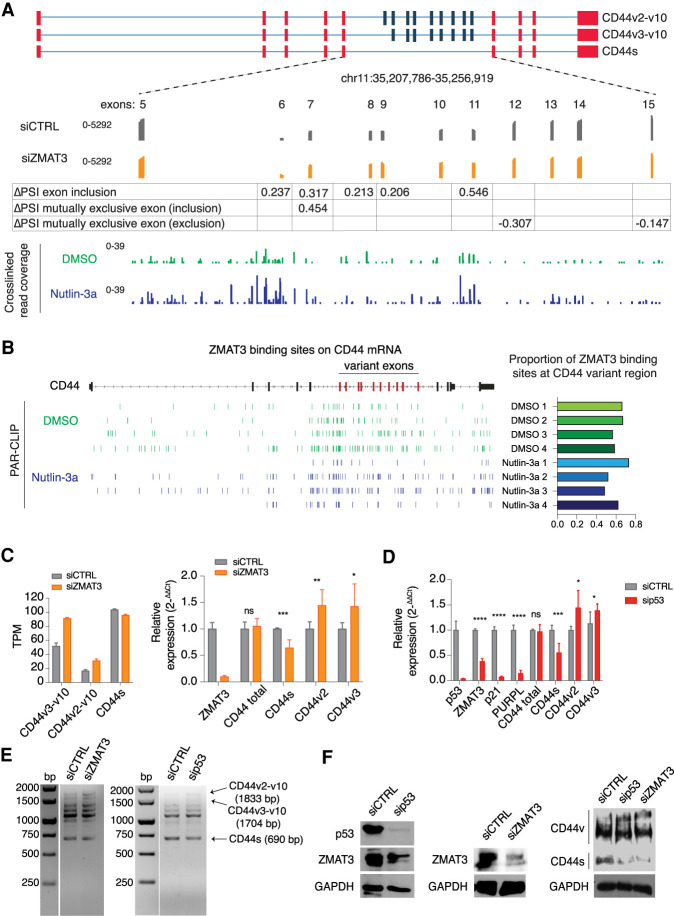
We noticed that the gene with the most significant splicing changes after ZMAT3 depletion was CD44, which was also among the top 200 PAR-CLIP targets (Supplemental Tables S4, S6). CD44 is a cell adhesion protein that is expressed in multiple isoforms from a pre-mRNA comprising 20 exons. Typically, the first and last five exons are retained in the mature CD44 mRNA to form the CD44s (standard) isoform. CD44 variant isoforms (CD44v) are generated by inclusion of some of the ten remaining exons (Zöller 2011). Importantly, increased CD44v expression is correlated with tumorigenesis in CRC (Wielenga et al. 1993; Bánky et al. 2012; Ozawa et al. 2014; Zeilstra et al. 2014).
The highest density of ZMAT3-binding sites on the CD44 pre-mRNA were found in the region containing the CD44 alternative exons (exons 6–14), both in cells treated with Nutlin-3a to induce ZMAT3 expression or cells treated with DMSO (Fig. 3A,B; Supplemental Table S7). Furthermore, depletion of ZMAT3 resulted in more frequent inclusion of CD44 alternative exons (Fig. 3A), resulting in increased expression of two CD44 variant transcripts (CD44v2-v10 and CD44v3-v10) and decreased levels of the standard transcript (CD44s) in RNA-seq and PCR experiments (Fig. 3C). We also tested whether silencing p53 would phenocopy the loss of ZMAT3. Indeed, knockdown of p53 resulted in increased expression of CD44v isoforms (Fig. 3D,E) and a reduction of known direct p53 targets, p21 and PURPL (Fig. 3E; Li et al. 2017). These changes in RNA expression translated to protein expression. Immunoblotting with a pan-CD44 antibody showed a marked decrease in CD44s levels but increase of the longer CD44v isoforms upon ZMAT3 or p53 depletion (Fig. 3F), likely including the longest ones corresponding to CD44v2-v10 and CD44v3-v10. Collectively, our data strongly suggest that ZMAT3 mediates the p53-dependent repression of oncogenic CD44 variants in CRC cells.
Figure 3.
ZMAT3 or p53 depletion promotes alternative splicing of the CD44 pre-mRNA, resulting in an increased expression of the oncogenic isoforms CD44v2-10 and CD44v3-v10, and decreased expression of the standard isoform CD44s. (A) Schematic representation of some CD44 isoforms, with invariant exons 1–5 and the last four exons colored in red and variant exons 6–10 colored in blue. The bottom track shows RNA-seq coverage from HCT116 cells treated with ZMAT3 or control siRNAs. The increase in PSI of each variant exon upon ZMAT3 silencing is indicated. The bottom track shows the distribution of cross-linked sequence reads from ZMAT3 PAR-CLIP experiments. (B) Genome browser track of ZMAT3 binding sites from all biological replicates across the CD44 gene showing the enrichment of ZMAT3 binding around the variant CD44 exons (left part) and proportion of binding sites at CD44 variant region/CD44 total RNA (right part). (C) Gene expression levels of CD44s, CD44v2-v10 or CD44v3-v10 upon ZMAT3 kd from RNA-seq in transcripts per million (TPM) (left), or RT-qPCR experiments (right). In the case of qPCR, we were only able to measure the expression of all CD44variants harboring v2 or v3 exons. (*) P < 0.05, (***) P < 0.001, (ns) nonsignificant. RT-qPCR values are the average of at least three biological replicates. (D) RT-qPCR quantification of CD44s, CD44v2, CD44v3, and known direct p53 targets (ZMAT3, p21, and the PURPL lincRNA) after p53 kd in HCT116 cells. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, (ns) nonsignificant. RT-qPCR values are the average of at least three biological replicates. (E) Representative images of semiquantitative PCR from HCT116 cells following ZMAT3 or p53 kd. The forward primer was designed on exon 2 and reverse on exon 18 of CD44 mRNA. Bands were Sanger-sequenced to confirm the isoforms. (F) Immunoblots from HCT116 lysates for p53, CD44, ZMAT3, and GAPDH following p53 or ZMAT3 kd.
ZMAT3 expression is anticorrelated with oncogenic CD44 isoforms in vivo
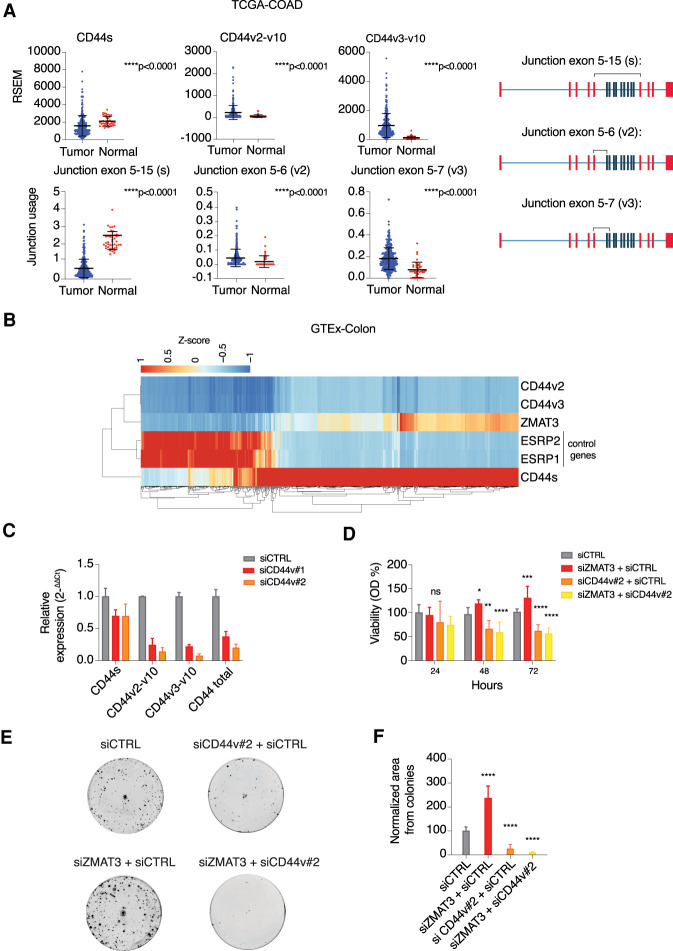
In order to extend the relevance of our findings to CRC tumors and normal human colon tissue, we turned to publicly available RNA-seq data collected from colon adenocarcinoma (COAD) and matched normal tissues deposited at the TSVdb database (Sun et al. 2018) and the GTEx portal (Lonsdale et al. 2013). We observed that both the expression of CD44s and exon 5–15 junction usage (its characteristic exon-junction) was decreased in COAD tumor samples compared with normal samples. In contrast, the expression of CD44v2-v10 and CD44v3-v10 was increased in the CRC (Fig. 4A).
Figure 4.
ZMAT3-levels anti-correlate with the expression of oncogenic CD44v isoforms in colon tissues and ZMAT3 and CD44v depletion lead to opposite phenotypes. (A) RSEM values of CD44s, CD44v3-v10, and CD44v2-10 derived from 285 primary TCGA-COAD solid tumor samples compared with 41 solid normal tissues (top panel), junction usage derived from exon junction quantification from the same samples (bottom panel), and schematic figures of CD44 mRNA showing each quantified junction (black lines above the CD44 mRNAs). Data were downloaded from the TSVdb database. (B) Heat map of gene expression of selected genes from 779 normal colon samples from the GTEx database. CD44s, CD44v3-v10, CD44v2-10, ZMAT3, ESRP1, and ESRP2 TPM values transformed into Z-scores. (C) RT-qPCR quantification of CD44v isoform depletion using specific siRNAs targeting exon 14. (D) Viability assay using Cell Counting Kit-8 upon kd of ZMAT3, CD44v, or both. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, (ns) nonsignificant. (E) Representative images derived from colony formation assays. (F) Cell colony formation assay after kd of ZMAT3, CD44v, or both using siRNAs. The values refer to the average of three biological replicates. Colony images were quantified using ImageJ software. (****) P < 0.0001.
Next, we wanted to investigate whether we could find correlations between CD44 splicing patterns and ZMAT3 expression levels in vivo. As a control, we first calculated the correlations of CD44s and CD44v isoforms and ESRP1 and ESRP2, two splicing factors that are known to regulate CD44 splicing by promoting CD44v formation at the expense of CD44s levels (Warzecha et al. 2009). ESRP1 or ESPR2 expression correlated negatively with CD44s (r = −0.47 and −0.48, respectively) and positively with CD44v3-v10 (r = 0.62 and r = 0.61, respectively) and CD44v2-v10 (r = 0.48 for both genes) in normal colon tissue (Fig. 4B; Supplemental Fig. S3A), supporting the validity of our approach. Next, we correlated the expression of ZMAT3 and CD44 isoforms in the same tissues. As predicted from our cell culture experiments, ZMAT3 levels correlated positively with CD44s (r = 0.69) and negatively with CD44v3-v10 (r = −0.52) and CD44v2-v10 (r = −0.40) (Fig. 4B; Supplemental Fig. S3A). Note the similarity of correlation coefficients for ZMAT3 or ESRP1/2 with CD44 isoforms, suggesting that the ZMAT3 effect on CD44 is similar to the well-characterized key splicing regulators ESRP1/2. These correlations hold true beyond colon tissue, as we found similar trends in normal small intestine and adipose tissue (Supplemental Fig. S3B,C), further supporting a role for ZMAT3 protein as a key splicing modulator of CD44 pre-mRNA.
Growth-suppressive effect of ZMAT3 is mediated via inhibition of oncogenic CD44 transcripts
Previous reports showed that ZMAT3 overexpression can inhibit tumor cell growth (Hellborg et al. 2001), and we first wanted to recapitulate this effect in our cell line. We carefully titrated transient overexpression of ZMAT3 in HCT116 cells to mirror ZMAT3 levels after p53 activation (Supplemental Fig. S1D) and found significant reduction of cell growth under these conditions (Supplemental Fig. S4A,B).
Next, we asked whether the regulation of CD44 splicing by ZMAT3 could be important in mediating the growth suppressive effects of ZMAT3. To test this, we decided to knockdown ZMAT3 or CD44v in HCT116 cells and compare the effects on cell viability and clonogenicity. First, we designed two different siRNAs against exon 14, present only in CD44 variant isoforms (siCD44v#1 and siCD44v#2) and quantified the expression of CD44s, CD44v2-v10, CD44v3-v10, and the sum of all CD44 isoforms using RT-qPCR. Both siRNAs knocked down CD44v, with siCD44v#2 showing greater knockdown efficiency (Fig. 4C) and thus, we used this siRNA for the functional assays.
Silencing ZMAT3 significantly increased cell viability whereas silencing CD44v alone or concurrent knockdown of ZMAT3 and CD44v significantly decreased cell viability after 48 or 72 h of transfection (Fig. 4D). Remarkably, the effect was more pronounced in colony formation assays, where we observed an ∼2.5-fold increase in clonogenicity upon ZMAT3 knockdown compared with the control. In contrast, silencing of CD44v alone or CD44v together with ZMAT3 dramatically reduced clonogenicity to approximately fourfold (Fig. 4E,F) revealing an epistatic interaction between the ZMAT3 and CD44. Together with our interactome and transcriptomic analyses, these data collectively uncover a previously unrecognized role of ZMAT3 in regulation of alternative splicing and reveal a growth suppressive function of the p53–ZMAT3–CD44 regulatory axis in CRC cells.
Discussion
Here, we showed that ZMAT3 was the most robustly induced RBP upon p53 activation in our CRC model. Besides ZMAT3, only a few other RBPs were suggested to be direct p53 targets, including PCBP4 (Zhu and Chen 2000), RNPC1 (Zhang et al. 2010), QKI (Chen et al. 2012), and RBM24 (Jiang et al. 2014). We were not able to find these RBPs up-regulated in our stringent analysis and a clear function of these RBPs in the p53 response has not been documented. Nevertheless, the integration of RNA-seq, GRO-seq, and ChIP-seq experiments from HCT116 cells after activation of the p53 pathway indicates the presence of a robust set of RBPs that are directly up-regulated by p53 and suggests that rewiring of post-transcriptional gene regulation is an underappreciated aspect of the otherwise well-characterized p53 response. In addition to RBPs, a number of other noncoding RNAs that could bind and affect target RNAs in a post-transcriptional manner, such as lncRNAs, miRNAs, and circRNAs were previously reported to be part of the p53 response (Goeman et al. 2017; Li et al. 2017; Chaudhary et al. 2020). We describe splicing regulation as a direct effect of the p53 pathway via the activation of the splicing regulator ZMAT3. That regulation of alternative splicing emerges as one aspect of p53-mediated tumor suppression may not be surprising considering that misregulation of alternative splicing is common in tumors (Zhang and Manley 2013).
ZMAT3 regulation of splicing may be related to its conspicuous binding at the polypyrimidine tract of pre-mRNA introns that is a crucial sequence element required for 3′ splice site definition. Generally, ZMAT3 binding sites most commonly consisted of Us, with additional significant contribution of A/U-rich sequences. A/U-rich elements (AREs) in mRNA 3′UTRs were previously predicted as ZMAT3-binding motif, based on sequence comparisons between mature mRNAs coimmunoprecipitated with ZMAT3 (Vilborg et al. 2009; Bersani et al. 2014, 2016). Binding to AREs in 3′UTRs—well-characterized sequence elements recruiting cytoplasmic trans-acting factors to promote rapid mRNA turnover (Shaw and Kamen 1986)—led to the interpretation that ZMAT3 modulated mRNA half-life (Vilborg et al. 2009; Bersani et al. 2014). However, these reports did not reveal the more widespread ZMAT3 binding across the entire pre-mRNA, which is consistent with its predominantly nuclear localization. In our analysis, the ZMAT3 binding pattern reminded less of the half-life changing ARE-binding proteins, such as the ZFP36 family (Carballo 1998), but more of other nuclear RBPs involved in splicing regulation, such as ELAVL1/HuR, which has a more pronounced affinity to A/U rich regions, but also to the 3′ splice site (Mukherjee et al. 2011).
On a mechanistic level, ZMAT3 might compete with the splicing machinery for binding to the polypyrimidine tracts and interfere with the proper recognition of 3′ splice sites. It thus might act as a splicing suppressor on some exons, but on the other hand also prevent binding of spliceosome components to weaker splice sites. ZMAT3 could thus prevent the formation of defective target mRNA isoforms containing, e.g., premature termination codons, which can make up to 40% of alternatively spliced transcripts (Tabrez et al. 2017).
Previous work demonstrated that p53 ablation resulted in increased expression of the adhesion protein and stem cell marker gene CD44 in mammary epithelial cell line/mouse mammary fat pad and mouse liver, respectively (Godar et al. 2008; Dhar et al. 2018), possibly due to direct binding of p53 to the CD44 promoter (Dhar et al. 2018). Our analysis revealed that rather than direct transcriptional regulation, in HCT116 and in human colon tissue, control of alternative splicing of the CD44 pre-mRNA is one of the major post-transcriptional regulatory events controlled by p53 or ZMAT3. Loss of either of these tumor-suppressive proteins resulted in higher abundance of the CD44v variants associated with many kinds of tumors (Zöller 2011) and a concomitant reduction of the shorter standard CD44s, the predominant isoform in adult normal tissues. Nevertheless, CD44s itself is also associated with the promotion of metastasis in breast (Brown et al. 2011), liver (Mima et al. 2012), and pancreatic (Li et al. 2014) cancers. Interestingly, CD44 seems to be a common target for a number of splicing regulatory RBPs, like ESPR1 and ESPR2 (Warzecha et al. 2009), HNRNPM (Xu et al. 2014), and HNRNPA1 (Loh et al. 2015), which reinforces the importance of balanced expression among its isoforms.
We found that CD44v silencing caused a decreased clonogenicity, further corroborating a body of literature describing the importance of CD44v to colorectal cancer and its association with increased tumorigenesis and worse prognosis (Wielenga et al. 1993; Bánky et al. 2012; Ozawa et al. 2014; Zeilstra et al. 2014). These effects are thought to be caused in part by expansions in the extracellular domain of CD44v introduced in the variant exons, which allow the protein to bind the growth factors HGF and FGF2 in addition to hyaluronan, the major CD44 ligand (Zöller 2011). This highlights the observation that different isoforms of the same gene can have different or opposing functions (Zhang and Manley 2013).
In summary, we present mechanistic evidence that the tumor-suppressive role of ZMAT3 (Hellborg et al. 2001) may be due to regulation of alternative splicing, and we furthermore suggest that at least a part of the p53 tumor suppressor pathway depends on post-transcriptional gene regulation via a p53–ZMAT3–CD44 axis.
Materials and methods
High-throughput data
The PAR-CLIP and RNA-seq data generated here are available at GEO under accession code GSE150718 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150718).
The custom K-mer/motif analysis is available on request. The in-house algorithm used to create pooled PAR-CLIP samples is available on request.
Cell lines
HCT116 colorectal carcinoma and 293T cell lines were purchased at American Type Culture Collection (ATCC) and were derived from adult male and fetus, respectively. They were cultivated in DMEM (Gibco 11965118) containing 10% fetal bovine serum (FBS) (Gibco 10082147) and 100 units/mL penicillin and 0.1 mg/mL streptomycin (Gibco 15140122). Cells were grown at 37°C and 5% CO2. Cell lines were routinely checked for mycoplasma contamination using Venor GeM mycoplasma detection kit (Sigma-Aldrich-Aldrich Co. MP0025-1KT). The cell lines have not been authenticated.
Plasmids construction and transduction
The expression vector containing ZMAT3-FLAG under control of p53 promoter was constructed using pLVX-Puro (Clontech 632164) as the backbone. The pCMV IE promoter was removed and 13 p53RE-Prom (p53-responsive promoter) sequences (5′-CCAGGCAAGTCCAGGCAGG-3′) were inserted upstream of the multiple cloning site (MCS). The ZMAT3 ORF containing three sequences of FLAG at its downstream portion was cloned at MCS using EcoRI and BamH1 restriction enzymes. The empty vector only contained the p53RE-Prom. Both vectors were generated by GenScript.
The vectors were expanded using DH5α cells (Invitrogen 18265017) and the plasmids were purified with Monarch plasmid miniprep kit (NEB T1010L). The lentiviruses were produced in 293T cells after cotransfection with a third generation of lentivirus packaging system using Lipofectamine 2000 (Thermo Fisher Scientific 11668027). HCT116 cells were transduced at MOI close to 1 and after 2 d transformants were selected with 2 μg/mL puromycin (Gibco A1113803) for 1 wk.
Wild-type (pGL3-Basic-ZMAT3-WT-promoter) and mutant (pGL3-Basic-ZMAT3-MUT-promoter) constructs containing the wild-type (5′-AGGCATGTCTGGACTTGTT-3′) and mutant (5′-CGTAAGGTCATGAGTTCTT-3′) p53 RE element, respectively, found in intron 1 from ZMAT3, were cloned into pGL3-Basic vector (Promega E2231) for luciferase assays. We used gBlocks gene fragments (IDT) at region chr3:179,069,907–179,070,507 (hg38) containing restriction sites from XhoI (NEB R0146S) and HindIII (NEB R0104S) at 5′ and 3′, respectively. We digested pGL3-Basic vector and DNA fragments with those enzymes, purified the products using Monarch DNA gel extraction kit (NEB T1020S) or QIAquick PCR purification kit (Qiagen. 28106) and ligated the inserts with the vector using T4 DNA ligase (NEB M0202S). The vectors were expanded and purified as aforementioned.
pcDNA3.1+/C-(K)DYK vector containing ZMAT3 ORF (OE ZMAT3; Genscript OHu10406/NM_022470.4) or empty vector was used for cell count assays.
Luciferase assay
The day before the transfection, 105 cells from 293T cell line were seeded in a 24-well plate. The next day, the cells were cotransfected using 250 ng of pGL3-Basic-ZMAT3-WT-promoter or pGL3-Basic-ZMAT3-MUT-promoter and 25 ng of pRL-TK (Promega E2231) vectors with 20 nM siCTRL (Allstars negative control siRNA; Qiagen 1027281) or sip53 (Horizon Discovery L-003329-00-0005) siRNAs. The cotransfection was made using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific 11668027) according to the manufacturer's protocol. Briefly, a mixture that contained 1 μL of lipofectamine, DNA, and siRNA in 100 μL of Opti-MEM (Gibco 31985062) was preincubated for 20 min at room temperature and added to the seeded cells. After 2 d, firefly and Renilla luminescences from pGL3 and pRL-TK vectors, respectively, were measured using dual-luciferase reporter assay system (Promega E1910) according to the manufacturer's protocol on EnSight Multimode plate reader (PerkinElmer). Firefly luminescence was normalized with Renilla luminescence for transfection efficiency.
PAR-CLIP
PAR-CLIP method was performed in four biological replicates as previously described in Hafner et al. (2010) and Benhalevy et al. (2017) but using a few modifications to track the ZMAT3 ribonucleoprotein complex.
Cell culture and UV cross-linking
HCT116 ZMAT3-FLAG cells were seeded in 10 15-mm cell culture dishes at 6 × 106 cells per plate. Forty-eight hours later, 10 μM final concentration Nutlin-3a (Sigma-Aldrich SML0580) dissolved in DMSO or equal volume of DMSO alone was added to the medium. After 2 h, 100 μM final concentration 4-thiouridine (Sigma-Aldrich T4509) was added to all cells. Cells were incubated for another 16 h followed by 5 min of cross-linking with 312-nm UV light using a Spectrolinker XL-1500 (Spectronics Corporation). Cells were scraped off on ice using a rubber policeman, collected by centrifugation at 500g, washed with 5 mL of 1× DPBS, and centrifuged again at 500g. Cell pellets were kept frozen at −80°C until further use. Pellets were thawed on ice and cells lysed in 1.5 vol of RIPA buffer (150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris at pH 8.0) made with DEPC water. Next, the cells were sonicated three times for 30 sec (power set to 60%; VirTis VIRSONIC 100) to shear chromatin followed by addition of 1.5 vol of 20 mM Tris-HCl buffer (pH 7.5). Lysates were cleared by centrifugation in a tabletop centrifuge at maximum speed for 15 min and the supernatant was kept for the next steps.
RNA digestion and dephosphorylation
RNase T1 (Thermo Fisher Scientific EN0541) was added to the lysates at a final concentration of 1 U/μL and the reaction mix was incubated at room temperature for 15 min. Reactions were cooled by incubating the lysates for 5 min in ice. RNPs were immunoprecipitated from the predigested lysates for 4 h using 80 μL of Pierce Protein A/G magnetic beads (Thermo Fisher Scientific 88803) coupled with 20 μg of anti-FLAG (Sigma-Aldrich. F1804) or anti-IgG (Santa Cruz Biotechnology sc-2025) antibodies for control samples. Beads were collected on a magnetic rack and washed three times with 1 mL of IP buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% [v/v] NP40, 0.5 mM DTT). After the final wash, beads were resuspended in 80 μL of IP buffer and RNase T1 was added to a final concentration of 10 U/μL. The reaction mix was incubated for 15 min at room temperature with mild agitation and washed three times with 1 mL of IP buffer.
Next, the beads were washed in 1 mL of dephosphorylation buffer (50 mM Tris-HCl at pH 7.9, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) without DTT. Washed beads were taken up in 80 μL dephosphorylation buffer and treated with 0.5 units/μL alkaline phosphatase, calf intestinal (CIP; NEB M0290S) for 10 min at 37°C with shaking. Subsequently, the beads were washed twice with 1 mL of dephosphorylation buffer and twice with 1 mL of ligation buffer (50 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 1 mM DTT) without DTT.
3′ adapter ligation and phosphorylation
After the final wash beads were taken up in 80 μL of ligation buffer once, followed by addition of 0.5 μL of 0.5 mM fluorescent 3′ adapter and 10 U/μL Rnl2(1-249)K227Q ligase (NEB M0351S) (adapters and primers sequences used for PAR-CLIP are listed in Supplemental Table S8). The reaction mix was incubated overnight at 4°C with gentle agitation. Beads were washed twice with 1 mL of ligation buffer without DTT and then twice with 1 mL of PNK buffer without DTT (50 mM Tris-HCl at pH 7.5, 50 mM NaCl, 10 mM MgCl2). After the final wash, beads were taken up in 80 μL of phosphorylation buffer and RNA 5′ ends were phosphorylated with 1 mM final concentration of ATP using 10 U/μL T4 PNK kinase (NEB M0201S) for 30 min at 37°C with shaking. Following this reaction, beads were washed twice with 1 mL of PNK buffer without DTT.
SDS-PAGE purification and proteinase K digestion
The cross-linked ribonucleoproteins were dissociated from beads by resuspending and incubating them in 65 μL of SDS loading buffer for 5 min at 95°C. The samples were loaded onto 4%–12% SDS–polyacrylamide gel NuPAGE, 1.0-mm Bis-Tris, and 12 Midi protein gel two-wells (Thermo Fisher Scientific WG1401BOX) and the band corresponded to the size of 55 kDa: The ZMAT3-FLAG-RNP (37 kDa) ligated to a fluorescently labeled 3′ adapter (∼20 kDa) was visualized on a fluorescent imager and cut from the gel. The recovered ribonucleoproteins were digested for 30 min initially in 200 μL of Proteinase K buffer (50 mM Tris-HCl at pH 7.5, 75 mM NaCl, 6.25 mM EDTA, 1% [w/v] SDS) containing 1.2 mg/mL Proteinase K (Roche 03450376103) and then 150 μL of the same buffer including 0.75 mg/mL Proteinase K was added twice at each 30 min, completing 1.5 h of reaction. The recovered RNA was purified with phenol-chloroform extraction and ethanol precipitation.
5′ primer adapter ligation and cDNA synthesis
Next, the recovered RNA was ligated to the 5′ chimeric DNA-RNA adapter 1.25 μM (final concentration) using 10 units/μL Rnl1 ligase (NEB M0204S) in a reaction volume of 20 μL for 1 h at 37°C. The ligated RNA was purified using Oligo Clean & Concentrator (Zymo Research D4061) and reverse transcribed with SuperScript IV reverse transcriptase (Invitrogen 18090010) using the 3′ RT primer according to the manufacturer's protocol. Adapters and primers sequences used for PAR-CLIP are listed in Supplemental Table S8.
PCR, purification, size selection, and sequencing
PCR amplification was done with Platinum Taq DNA polymerase (Thermo Fisher Scientific 10966018) using 0.5 μM 3′ RT primer and 5′ SHORT PCR primer in a 100-μL reaction volume. After eight cycles from PCR (10 sec at 94°C, 30 sec at 60°C, and 15 sec at 72°C), the samples were concentrated and purified using DNA Clean & Concentrator-5 (Zymo Research, Catalog no. D4013). Products of a size between 75 and 100 bp were isolated using a 3% agarose PippinPrep cassette (Sage Science CSD3010) on a BluePippin device.
The second PCR was done with the purified and size-selected samples using Platinum Taq DNA polymerase together with 0.5 μM 5′ long PCR primer and 3′ RNA index primer (barcoded primer) in 100-μL reaction volume for 15 cycles. The PCRs were again purified, and size-selected for a range between 147 and 173 bp. The resulting library was sequenced on a HiSeq 3000 system using a single-end 50 cycle protocol. Analysis was performed as described previously using PARalyzer (version 1.5; Corcoran et al. 2011) built into the PARpipe (Corcoran et al. 2011) pipeline mapping the reads to human genome hg19. Pooled version of reads was mapped on human genome hg38.
siRNAs transfection
HCT116 cells were reverse transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen 13778075) and siRNAs at 20 nM according to the manufacturer's protocol. Briefly, 3 × 105 cells were seeded in six-well plates, adding a mixture that was preincubated for 20 min at room temperature and contained 5 μL of lipofectamine and an amount sufficient for 20 nM siRNA in 500 μL of Opti-MEM (Gibco 31985062). Cells used for expression analysis were washed using 1× DPBS (Gibco 14190250) and lysed using 500 μL of TRIzol reagent (Invitrogen 15596018) 48 h after transfection. For p53 knockdown the cells were transfected one more time after 48 h and then, 2 d later, the RNA was extracted. Allstars negative control siRNAs (Qiagen 1027281) were used as control siRNAs. We used SMARTPool siRNAs (Horizon Discovery L-017558-00-0005, L-003329-00-0005, and L-011763-00-0005) against ZMAT3, p53, and UPF1, respectively. With the intention of targeting most of CD44v, we designed siRNAs against exon 14, which was included in most of the CD44 variants from Ensemble (https://useast.ensembl.org/index.html). The aforementioned siRNAs, siCD44v#1 (5′-rArArArUrCrArUrUrCrUrGrArArGrGrCrUrCrArArCrUrACT-3′ and 5′-rArGrUrArGrUrUrGrArGrCrCrUrUrCrArGrArArUrGrArUrUrUrGrG-3′) and siCD44v#2 (5′-rGrGrArArGrArArGrArGrArCrCrCrArArArUrCrArUrUrCTG-3′ and 5′-rCrArGrArArUrGrArUrUrUrGrGrGrUrCrUrCrUrUrCrUrUrCrCrArC-3′), were purchased from Integrated DNA Technologies (IDT). For colony formation and viability assays, cells were transfected using siRNAs against more than one target (e.g., siCTRL and siZMAT3). Consequently, the final concentration of siRNAs was 40 nM.
RNA extraction, RT-qPCRs, and RT-PCR
Total RNA for samples used for RT-qPCR had their total RNA extracted using TRIzol reagent (Invitrogen 15596018) according to the manufacturer's protocol. RNA (500 or 1000 ng) was used for cDNA synthesis using iScript reverse transcription supermix, (Bio-Rad 1708841) following the manufacturer's instructions. For RT-qPCR, 45 or 90 μL of H20 (for 500 ng or 1000 ng of initial RNA, respectively) was added to 10 μL of cDNA. Diluted cDNA (2.5 μL) was used in a reaction mixture containing 0.2 μM (final concentration) each primer, 5 μL of FastStart Universal SYBR Green Master (Rox; Millipore Sigma 4913914001), and H2O enough for 10 μL. All reactions were done on StepOnePlus real-time PCR (Applied Biosystems). GAPDH was used to normalize the expression and the relative expression was calculated using the 2−ΔΔCt method.
RT-PCRs for CD44 were made using 1 μL of nondiluted cDNA, 1 μL of each primer (CD44 total PCR, 10 μM stock), 12.5 μL of Phusion High-Fidelity PCR Master Mix with HF buffer (NEB M0531S), and H2O enough for 25 μL of reaction. The PCR cycle used was the following: once for 5 min at 94°C and 28 times for 10 sec at 98°C, 30 sec at 57°C, and 2 min at 72°C,. PCR products were resolved using 1.2% agarose gels.
The primers for RT-qPCRs and RT-PCRs are listed in Supplemental Table S8.
RNA-seq
RNA-seq was performed in biological triplicates from HCT116 cells treated for 6 h with Nutlin-3a (10 μM final concentration) or DMSO, 48 h after transfection with siRNAs against ZMAT3 or negative control as described above. Total RNA was isolated using the RNeasy Plus mini kit (Qiagen 74134) following the manufacturer's instructions. Samples were sequenced on HiSeq4000 using Illumina TruSeq stranded mRNA library preparation (Illumina 20020594) and paired-end sequencing.
RNA-seq following 48 h after transfection of UPF1 or control siRNAs in HCT116 cells was also performed in biological triplicates, as above. Total RNA was extracted using TRIzol Reagent (Invitrogen 15596026) according to manufacturer's instructions. We used the NEBNext Ultra Directional RNA library preparation kit for Illumina (NEB E7760) with NEBNext rRNA depletion kit (NEB E6318). The samples were sequenced on an Illumina HiSeq 3000 machine using the 50 cycles single end sequencing protocol.
Sequence reads were aligned to the human genome hg19 build using TopHat (version 2.1.1; https://ccb.jhu.edu/software/tophat/index.shtml) or STAR (version 2.5.4a; https://github.com/alexdobin/STAR). Differential gene expression was quantified using RSEM (version 1.2.31; https://deweylab.github.io/RSEM) and DESeq2 (version 1.26.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html).
For analysis at transcript level, raw fastq files were trimmed using Trimmomatic (version 0.36) (http://www.usadellab.org/cms/?page=trimmomatic) and Trim Galore (version 0.4.5) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Expression levels for gencode v19 version transcripts were quantified using Salmon (version 0.14.1; Patro et al. 2017). The output of Salmon was converted to Sleuth (version 0.30.0; Pimentel et al. 2017) compatible format using the prepare_fish_for_sleuth() function from the wasabi package (version 1.0.1; https://github.com/COMBINE-lab/wasabi). Normalized TPM expression values were obtained from the Sleuth package.
Western blots
Cells were lysed in 200–500 μL of RIPA buffer (Thermo Fisher Scientific 89901) containing protein inhibitors cOmplete, Mini, EDTA-free protease inhibitor cocktail (Millipore Sigma 11836170001). Subsequently, samples were sonicated twice for 5 sec (power set to 50%) while samples were on ice. Cleared supernatant from lysates were recovered by centrifugation and the protein was quantified using Pierce BCA protein assay kit (Thermo Scientific 23225) according to the manufacture's protocol. Protein (15 or 20 μg) was loaded into 6%, 10%, or 12% SDS-polyacrylamide gels and, subsequently, transferred onto a PVDF membrane in a semi-dry apparatus according to standard Western Blot protocols. Membranes were blocked and later they were incubated with primary antibodies overnight. We used GAPDH antibody (1:6000 dilution; Cell Signaling 5174S) or β-tubulin (1:1000 dilution; Cell Signaling 2128S) as a normalizer for protein expression. The other primary antibodies used are the following: FLAG (1:1000 dilution; Sigma F1804), p53 (DO-I) (1:1000 dilution; Santa Cruz Biotechnology sc-126), CD44 (1:1000; dilution Cell Signaling, Catalog no. 3570S), mouse IgG (Santa Cruz Biotechnology sc-2025) and ZMAT3 (1:250 dilution), that was generated from Abgent. Membranes were developed using ECL Prime Western blotting detection reagent (Fisher Scientific RPN2232).
Immunostaining
Immunostaining analysis was done in HCT116 ZMAT3-FLAG or empty vector control cells as described earlier (Tripathi et al. 2012). Briefly, cells were washed with 1× DPBS, fixed with 4% of freshly prepared formaldehyde solution for 15 min at room temperature and washed twice with 1× DPBS. Next, cells were permeabilized with 0.5% Triton X-100 in 1× PBS solution for 5 min at room temperature and washed twice with 1× PBS. The blocking was done using 2% bovine serum albumin for 30 min at room temperature and cells were washed twice with 1× PBS. Cells were incubated with anti-FLAG (1:200 dilution) or anti-SON (1:1000 dilution) (Sigma-Aldrich HPA023535) antibodies in 2% bovine serum albumin for 2 h at room temperature in a wet chamber, and then washed three times with 1× PBS. The cells were stained with DAPI for 3 min and the coverslip was mounted onto the slides with mounting medium. The Z-stack images were taken using Axioimager Z1 microscope (Zeiss) equipped with 63×/1.4 NA oil immersion objective and Zeiss Axiocam 506 monocamera. Images were processed through deconvolution and maximum intensity projections using ZEN software (version 3.1).
Cell count assays
For this experiment, 2 × 105 HCT116 cells were seed at 12-well plates. On the next day, cells were transfected with 100, 500, or 1000 ng of pcDNA3.1+/C-(K)DYK vector containing ZMAT3 ORF (OE ZMAT3) (Genscript OHu10406/NM_022470.4) or empty vector using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, 11668027), according to the manufacturer's protocol. After 48 h, the cells were counted using Trypan Blue staining.
Colony formation assay
In this experiment, 103 HCT116 cells were seeded in 6 well plates 24 h after siRNA transfections (see above). Cells were kept growing for 8 d and then were fixed using 100% ice-cold methanol for 15 min and stained with crystal violet 0.5% in methanol (10%) for 15 min. The ImageJ software package (version 2.0.0-rc-43/1.52n) was used to analyze images of the resulting colonies pictures.
Cell viability assays
We seeded 1.5 × 103 HCT116 cells in 96 well plates 24 h after siRNA transfections (see above). Cell counting kit 8 (Dojindo Molecular Technologies, Inc. CK04-13) was used for this assay according to the manufacturer's protocol at time points 24, 48, and 72 h after cells were seeded. Absorbance at measured at 450 nm on EnSight Multimode plate reader (PerkinElmer).
Alternative splicing analysis
For splice pattern analysis, raw FASTQ files were trimmed using trimGalore (version 0.4.5, https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) and Trimmomatic (version 0.36). Surviving reads were aligned to the hg19 genome with the STAR aligner (version 2.5.4a) using the GENCODE v19 gene annotation using the two-pass mode and “best recall at base and read level” settings as shown in Supplemental Table S37 of Baruzzo et al. (2017). Alternative splicing events were identified using the rMATS (version 4.0.2; Shen et al. 2014) package. Data from rMATS were filtered for events supported by at least five sequence reads, with ΔPSI (percent spliced in) ≥ 0.1 and FDR (false discovery rate) < 0.05 using the maser package (version 1.4.0; https://rdrr.io/bioc/maser/) for R (version 3.6.2; R Core Team 2019).
Venn diagrams
The venn diagram from Figure 1B was generated using VennDiagram package (Chen and Boutros 2011; https://CRAN.R-project.org/package=VennDiagram). The graph at Figure 1C graph was made with “Venn Diagrams” web site (http://bioinformatics.psb.ugent.be/webtools/Venn).
Average coverage of binding sites
Visualization of binding site coverage (Fig. 2A) was done using the RCAS tool (version 1.12.0; Uyar et al. 2017) with R software (version 3.6.2).
Correlograms
The correlograms from Supplemental Fig. S3 was generated using the corrplot (version 0.84) package (https://github.com/taiyun/corrplot) with R software (version 3.6.2).
Heat maps
The heat maps from Supplemental Figs. S3 and s4 were made using pheatmap package (version 1.0.12; https://cran.r-project.org/web/packages/pheatmap/index.html) with R software (version 3.6.2).
K-mer/motif analysis
The K-mer or motif analysis was done using an in-house script. Basically, it was performed by counting all 5-mer occurrences within the binding site sequences and comparing these values with 5-mer abundances within a background file. This background file was built by matching each binding site with random continuous sequences of equal length taken from the region of the gene where that binding site was found, like intron, 3′UTR, CDS, etc.
Quantification and statistical analysis
All the statistical analysis was performed using at least three replicates and bar or box plots from Figures 3 and 4 and Supplemental Figure 2 indicate mean ± SD. Statistical significance of differences were performed using the Student's t-test when comparing two groups or one-way ANOVA when comparing more than two groups. Spearman's coefficient correlation was used to analyze the expression and junction usage from data derived from TSVdb or GTEx. Kolmogorov-Smirnov test was used for comparisons of cumulative distributions. The data were considered significant when P < 0.05. Prism software (version 8) was used to make the analysis.
Acknowledgments
This work was supported by the Intramural Research Programs of the National Cancer Institute (NCI), the Center for Cancer Research (CCR), and the National Institute for Arthritis and Musculoskeletal and Skin Disease (NIAMS). We thank the NIAMS Genomics Core Facility as well as Gustavo Gutierrez-Cruz and Dr. Stefania Dell'Orso (NIAMS/National Institutes of Health [NIH]) for sequencing support. We also thank the NCI core sequencing facility for performing part of the RNA-seq. Work in K.V.P.'s laboratory was supported by grants from NIH (R21AG065748 and R01GM132458), a Cancer Center at Illinois seed grant, the Prairie Dragon Paddlers, and the National Science Foundation (EAGER; MCB1723008). We thank Dr. Glenn Merlino (NCI) for his insightful comments on our manuscript.
Author contributions: A.L., M.H., and B.R.M. conceived the study. B.R.M., D.G.A., X.L.L., I.G., X.W., and M.L. developed the methodology. D.G.A., D.C., L.P., and B.R.M. analyzed the data. D.G.A., X.L.L., I.G., X.W., K.V.P., and M.L. assisted with the experiments. B.R.M, M.I.A., K.V.P., A.L., and M.H. wrote the manuscript. A.L. and M.H. acquired the funding. A.L. and M.H. supervised the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.342634.120.
References
- Allen MA, Andrysik Z, Dengler VL, Mellert HS, Guarnieri A, Freeman JA, Sullivan KD, Galbraith MD, Luo X, Lee Kraus W, et al. 2014. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 3: e02200 10.7554/eLife.02200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrysik Z, Galbraith MD, Guarnieri AL, Zaccara S, Sullivan KD, Pandey A, MacBeth M, Inga A, Espinosa JM. 2017. Identification of a core TP53 transcriptional program with highly distributed tumor suppressive activity. Genome Res 27: 1645–1657. 10.1101/gr.220533.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánky B, Rásó-Barnett L, Barbai T, Tímár J, Becságh P, Rásó E. 2012. Characteristics of CD44 alternative splice pattern in the course of human colorectal adenocarcinoma progression. Mol Cancer 11: 83 10.1186/1476-4598-11-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruzzo G, Hayer KE, Kim EJ, DI Camillo B, Fitzgerald GA, Grant GR. 2017. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods 14: 135–139. 10.1038/nmeth.4106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhalevy D, McFarland HL, Sarshad AA, Hafner M. 2017. PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites. Methods 118–119: 41–49. 10.1016/j.ymeth.2016.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersani C, Xu LD, Vilborg A, Lui WO, Wiman KG. 2014. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene 33: 4407–4417. 10.1038/onc.2013.594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersani C, Huss M, Giacomello S, Xu LD, Bianchi J, Eriksson S, Jerhammar F, Alexeyenko A, Vilborg A, Lundeberg J, et al. 2016. Genome-wide identification of Wig-1 mRNA targets by RIP-seq analysis. Oncotarget 7: 1895–1911. 10.18632/oncotarget.6557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial–mesenchymal transition and breast cancer progression. J Clin Invest 121: 1064–1074. 10.1172/JCI44540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E. 1998. Feedback inhibition of macrophage tumor necrosis factor- production by tristetraprolin. Science 281: 1001–1005. 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- Chaudhary R, Muys BR, Grammatikakis I, De S, Abdelmohsen K, Li XL, Zhu Y, Daulatabad SV, Tsitsipatis D, Meltzer PS, et al. 2020. A circular RNA from the MDM2 locus controls cell cycle progression by suppressing p53 levels. Mol Cell Biol 40: e00473-19 10.1128/MCB.00473-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Boutros PC. 2011. Venndiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 12: 35 10.1186/1471-2105-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AJ, Paik JH, Zhang H, Shukla SA, Mortensen R, Hu J, Ying H, Hu B, Hurt J, Farny N, et al. 2012. STAR RNA-binding protein quaking suppresses cancer via stabilization of specific miRNA. Genes Dev 26: 1459–1472. 10.1101/gad.189001.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Georgiev S, Mukherjee N, Gottwein E, Skalsky RL, Keene JD, Ohler U. 2011. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol 12: R79 10.1186/gb-2011-12-8-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar D, Antonucci L, Nakagawa H, Kim JY, Glitzner E, Caruso S, Shalapour S, Yang L, Valasek MA, Lee S, et al. 2018. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell 33: 1061–1077.e6. 10.1016/j.ccell.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. 2016. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 16: 413–430. 10.1038/nrc.2016.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, Liu A, Miu K, Watnick RS, Reinhardt F, et al. 2008. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell 134: 62–73. 10.1016/j.cell.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman F, Strano S, Blandino G. 2017. MicroRNAs as key effectors in the p53 network. Int Rev Cell Mol Biol 333: 51–90. 10.1016/bs.ircmb.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp A-C, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellborg F, Wiman KG. 2004. The p53-induced Wig-1 zinc finger protein is highly conserved from fish to man. Int J Oncol 24: 1559–1564. [PubMed] [Google Scholar]
- Hellborg F, Qian W, Mendez-Vidal C, Asker C, Kost-Alimova M, Wilhelm M, Imreh S, Wiman KG. 2001. Human wig-1, a p53 target gene that encodes a growth inhibitory zinc finger protein. Oncogene 20: 5466–5474. 10.1038/sj.onc.1204722 [DOI] [PubMed] [Google Scholar]
- Israeli D, Tessler E, Haupt Y, Elkeles A, Wilder S, Amson R, Telerman A, Oren M. 1997. A novel p53-inducible gene, PAG608, encodes a nuclear zinc finger protein whose overexpression promotes apoptosis. EMBO J 16: 4384–4392. 10.1093/emboj/16.14.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, Yang H, Tai L, Vandenberg CJ, Kueh AJ, et al. 2018. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med 24: 947–953. 10.1038/s41591-018-0043-5 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Zhang M, Qian Y, Xu E, Zhang J, Chen X. 2014. Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. J Biol Chem 289: 3164–3175. 10.1074/jbc.M113.524413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Lee HC, Lee JJ, Choi CM, Kim DK, Lee JC, Ko YG, Lee JS. 2012. Wig1 prevents cellular senescence by regulating p21 mRNA decay through control of RISC recruitment. EMBO J 31: 4289–4303. 10.1038/emboj.2012.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. 2003. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612. 10.1038/nrm1172 [DOI] [PubMed] [Google Scholar]
- Li L, Hao X, Qin J, Tang W, He F, Smith A, Zhang M, Simeone DM, Qiao XT, Chen ZN, et al. 2014. Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology 146: 1108–1118.e12. 10.1053/j.gastro.2013.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Subramanian M, Jones MF, Chaudhary R, Singh DK, Zong X, Gryder B, Sindri S, Mo M, Schetter A, et al. 2017. Long noncoding RNA PURPL suppresses basal p53 levels and promotes tumorigenicity in colorectal cancer. Cell Rep 20: 2408–2423. 10.1016/j.celrep.2017.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh TJ, Moon H, Cho S, Jang H, Liu YC, Tai H, Jung DW, Williams DR, Kim HR, Shin MG, et al. 2015. CD44 alternative splicing and hnRNP A1 expression are associated with the metastasis of breast cancer. Oncol Rep 34: 1231–1238. 10.3892/or.2015.4110 [DOI] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, et al. 2013. The genotype-tissue expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG. 2002. The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res 30: 1991–1996. 10.1093/nar/30.9.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima K, Okabe H, Ishimoto T, Hayashi H, Nakagawa S, Kuroki H, Watanabe M, Beppu T, Tamada M, Nagano O, et al. 2012. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res 72: 3414–3423. 10.1158/0008-5472.CAN-12-0299 [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Tuschl T, Ohler U, Keene JD. 2011. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 43: 327–339. 10.1016/j.molcel.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M. 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718. 10.1101/gad.1525507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M, Ichikawa Y, Zheng YW, Oshima T, Miyata H, Nakazawa K, Guan HB, Shiozawa M, Akaike M, Watanabe K, et al. 2014. Prognostic significance of CD44 variant 2 upregulation in colorectal cancer. Br J Cancer 111: 365–374. 10.1038/bjc.2014.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Shaw G, Kamen R. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46: 659–667. 10.1016/0092-8674(86)90341-7 [DOI] [PubMed] [Google Scholar]
- Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y. 2014. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci 111: E5593–E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Duan T, Ye P, Chen K, Zhang G, Lai M, Zhang H. 2018. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics 19: 405 10.1186/s12864-018-4775-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabrez SS, Sharma RD, Jain V, Siddiqui AA, Mukhopadhyay A. 2017. Differential alternative splicing coupled to nonsense-mediated decay of mRNA ensures dietary restriction-induced longevity. Nat Commun 8: 1–13. 10.1038/s41467-017-00370-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, Dundr M, Prasanth KV. 2012. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell 23: 3694–3706. 10.1091/mbc.e12-03-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyar B, Yusuf D, Wurmus R, Rajewsky N, Ohler U, Akalin A. 2017. RCAS: an RNA centric annotation system for transcriptome-wide regions of interest. Nucleic Acids Res 45: e91 10.1093/nar/gkx120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LJ, Gray DHD, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A. 2013. P53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep 3: 1339–1345. 10.1016/j.celrep.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Vilborg A, Glahder JA, Wilhelm MT, Bersani C, Corcoran M, Mahmoudi S, Rosenstierne M, Grandér D, Farnebo M, Norrild B, et al. 2009. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc Natl Acad Sci 106: 15756–15761. 10.1073/pnas.0900862106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. 2000. Surfing the p53 network. Nature 408: 307–310. 10.1038/35042675 [DOI] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 33: 591–601. 10.1016/j.molcel.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielenga VJM, Heider KH, Offerhaus GJA, Adolf GR, van den Berg FM, Ponta H, Herrlich P, Pals ST. 1993. Expression of CD44 variant proteins in human colorectal cancer is related to tumor progression. Cancer Res 53: 4754–4756. [PubMed] [Google Scholar]
- Wilhelm MT, Méndez-Vidal C, Wiman KG. 2002. Identification of functional p53-binding motifs in the mouse wig-1 promoter. FEBS Lett 524: 69–72. 10.1016/S0014-5793(02)03004-1 [DOI] [PubMed] [Google Scholar]
- Witten JT, Ule J. 2011. Understanding splicing regulation through RNA splicing maps. Trends Genet 27: 89–97. 10.1016/j.tig.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Gao XD, Lee JH, Huang H, Tan H, Ahn J, Reinke LM, Peter ME, Feng Y, Gius D, et al. 2014. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev 28: 1191–1203. 10.1101/gad.241968.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Jishage M, Meyer C, Suryawanshi H, Der E, Yamaji M, Garzia A, Morozov P, Manickavel S, McFarland HL, et al. 2017. DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs. Nature 543: 568–572. 10.1038/nature21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilstra J, Joosten SPJ, Van Andel H, Tolg C, Berns A, Snoek M, Van De Wetering M, Spaargaren M, Clevers H, Pals ST. 2014. Stem cell CD44v isoforms promote intestinal cancer formation in Apc(min) mice downstream of Wnt signaling. Oncogene 33: 665–670. 10.1038/onc.2012.611 [DOI] [PubMed] [Google Scholar]
- Zhang J, Manley JL. 2013. Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 3: 1228–1237. 10.1158/2159-8290.CD-13-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cho SJ, Chen X. 2010. RNPC1, an RNA-binding protein and a target of the p53 family, regulates p63 expression through mRNA stability. Proc Natl Acad Sci 107: 9614–9619. 10.1073/pnas.0912594107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen X. 2000. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol Cell Biol 20: 5602–5618. 10.1128/MCB.20.15.5602-5618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöller M. 2011. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer 11: 254–267. 10.1038/nrc3023 [DOI] [PubMed] [Google Scholar]