Abstract
Inositol phospholipids are low-abundance regulatory lipids that orchestrate diverse cellular functions in eukaryotic organisms. Recent studies have uncovered involvement of the lipids in multiple steps in autophagy. The late endosome–lysosome compartment plays critical roles in cellular nutrient sensing and in the control of both the initiation of autophagy and the late stage of eventual degradation of cytosolic materials destined for elimination. It is particularly notable that inositol lipids are involved in almost all steps of the autophagic process. In this review, we summarize how inositol lipids regulate and contribute to autophagy through the endomembrane compartments, primarily focusing on PI4P and PI(4,5)P2.
Keywords: autophagy, late endosome/lysosome, phosphoinositide
Inositol lipids are relatively minor phospholipid components of eukaryotic membranes but they are essential for controlling the assembly and functions of many integral or peripheral membrane proteins to maintain cellular homeostasis. Inositol lipids are formed by phosphorylation of D3, D4 and D5 position on the inositol ring in phosphatidylinositol (PI) in some combination (1). In addition to the kinases and phosphatases that control the levels of these lipids, lipid transfer proteins also play critical roles in shaping membrane lipid composition and dynamics (1, 2). Distribution of individual inositol lipids is not uniform, each membrane compartment having a unique inositol lipid signature that ensures that their effector proteins properly work at those specific intracellular locations. The many well-documented roles of inositol lipids include regulation of actin cytoskeleton and vesicular transport, contribution to the proper function of ion channels and control of signal transduction in the plasma membrane (1). In addition to these important roles, increasing evidence suggest that inositol lipids are also involved in the control of autophagy (3). Autophagy serves two kind of purposes: (i) it helps recycle important cellular material from dispensable various cellular components and (ii) it clears defective macromolecules and organelles (4). Although autophagy has been thoroughly characterized in the last 10 years, there are still many unanswered questions that remain to be answered. When autophagy is triggered by specific stimuli, such as amino acids starvation, autophagosome precursors, termed phagophores sequester cytoplasmic materials and form spherical double-membranous structures, called omegasomes (4, 5). Once these structures are closed and form vesicles, they fuse with lysosomes creating autolysosomes for degradation of their content. The process of recovering lysosomes from autolysosomes is also important and is called lysosome reformation (5). A subpopulation of autophagosomes undergoes fusion with late endosomes before fusion with lysosomes (6).
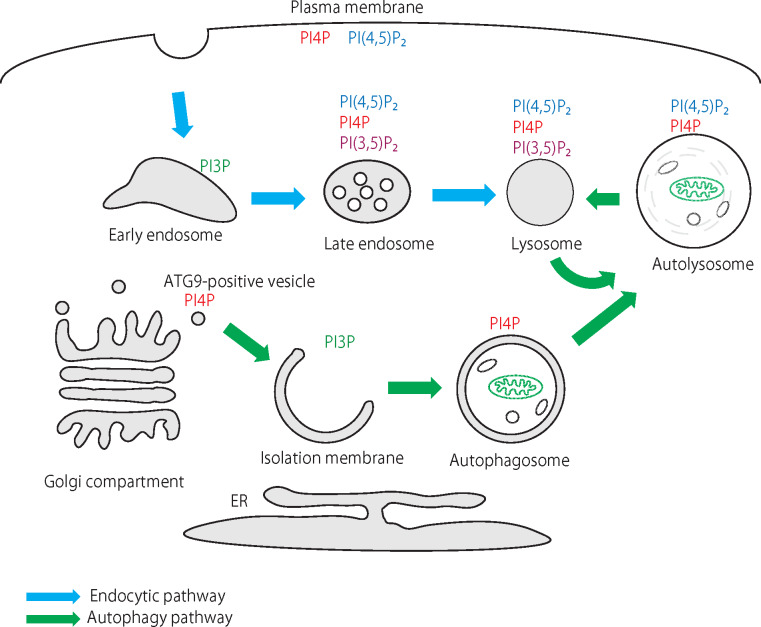
The origin of membranes during the early phase of autophagosome formation is highly debated and there are many questions about the precise sequence and the entities of the molecular events that guide each phase of the process. Inositol lipids have emerged as critical membrane lipid components that orchestrate many if not all of the steps in the autophagic maturation process. It has been now well established that PI3P plays a pivotal role in the initiation of autophagy (7) and that PI(3,5)P2 is important in the autophagosome–lysosome fusion step and subsequent acidification of this organelle (8). Recent studies, however, began to highlight the roles of PI4P and PI(4,5)P2 at various stages of autophagy (Fig. 1). PI4P and PI(4,5)P2 have been known as important precursors of second messengers in the plasma membrane and also to control endosome formation and maturation as well as Golgi function, but their involvement in autophagy has not been suspected until recently.
Fig. 1.
Distribution of various phosphoinositides in organelles related to endocytic trafficking and autophagy pathways. While the bulk of PI4P and PI(4,5)P2 are mainly found in the plasma membrane, small but functionally important pools are also localized to late endosomes and lysosomes as well as autophagosomes. In addition to the well-established roles of PI3P and PI(3,5)P2 in both endocytic trafficking and autophagy, PI4P and PI(4,5)P2 are emerging as critical regulators at several steps along the autophagy pathway.
Therefore, in this review, we will mainly focus on recent developments on endosomal PI4-kinases and PIP-kinases and their lipid products in the control of the autophagy process. Excellent reviews have been written about the roles of PI3P and PI(3,5)P2 in autophagy (3, 7, 9), so they will not be covered in this review.
PI4P is an important regulator of endomembrane trafficking
PI4P and PI(4,5)P2 are the two most abundant phosphorylated phosphoinositides in mammalian cells, mostly known to localize to the plasma membrane where they play an important role in signal transduction events initiated from cell surface receptors. The role of PI4P as a lipid with additional endomembrane functions was revealed by important studies carried out in yeast (10, 11) and also in mammalian systems. These studies identified the PI 4-kinase enzymes (PI4Ks) that produce PI4P by phosphorylating the D4 position on PI. PI4Ks are classified as type II PI4Ks (PI4K2A and PI4K2B) and type III PI4Ks (PI4KA and PI4KB) (1). The localization of the various PI4Ks is not identical. PI4KA is localized to the plasma membrane with the help of the adaptor proteins, EFR3, TTC7 and Fam126A (12) although its presence in the ER has also been reported by early studies (13). Undoubtedly, however, the plasma membrane pool of PI4P is primarily generated by PI4KA (12). PI4KB, on the other hand, is localized to the Golgi compartment, whereas type II PI4Ks are found on both the TGN and endosomes and some of the PI4K2B in the plasma membrane (1). Therefore, endosomal PI4P is mostly formed by the type II PI4Ks. These enzymes are peripheral membrane proteins and their strong binding to the cytoplasmic leaflet of membranes is provided by palmitoylation of cysteine residues within their catalytic region (14, 15). The type II PI4Ks have been shown to localize both in early (Rab4- or Rab5-positive) and late (Rab7-positive) endosomes, but their presence was also reported in Rab11-positive recycling compartments (16–18). Although the enzymes are located throughout the endolysosomal membrane system, the relative levels of PI4P have not been determined in those endosomal compartments. Our recent studies, applying the bioluminescence resonance energy transfer method for assessment of the relative levels of endosomal PI4P showed that PI4P is more abundant in the late endosomal, Rab7-positive compartment than in early endosomes marked by the presence of Rab5 (19). Moreover, this analysis also showed that PI4K2A is the major source of the late endosomal PI4P with only minor contribution from PI4K2B (19). This apparent discrepancy between the prominent localization of the PI4Ks in compartments where PI4P is less abundant could reflect the different activity of the enzymes themselves or of the metabolic processes that eliminate PI4P. For example, dephosphorylation of PI4P by the 4-phosphatase Sac2 enzyme is more important in Rab5 and Rab11 endosomes and less so in the Rab7-positive compartment (20, 21). Moreover, PI4P has been shown to be transferred from endosomes to the ER by the oxysterol binding protein, OSBP and degraded in the ER by the Sac1 phosphatase enzyme (22).
PI4P recruits several proteins to specific endomembrane compartments, including clathrin adaptors, AP1, AP3 and the GGAs (1), Arf regulators, such as ARFAPTINs (23) and some forms of spectrins (24). It also regulates the assembly of the Wiskott Aldrich syndrome protein and scar homologue complex to control actin polymerization and determine the targeting of the R-SNARE, VAMP3 (22, 25). PI4P also controls the dissociation of SNX4-associated vesicles from dynein–dynactin complex (26) and the recruitment of the exocyst tethering complex to endosome membranes (27). Detailed discussion of these PI4P effectors and their roles in trafficking is beyond the scope of this review and can be found elsewhere (28).
PI4Ks and PI4P are emerging as regulators of several steps in autophagy
PI4KB
PI4KB has been known as a main regulator of post-Golgi secretion but it has also been found in the nucleus (1, 29). Recent studies have identified PI4KB as an important component and regulator of the initiation phase of the early autophagosome membrane. These studies found that upon amino acid starvation, the autophagy protein, ATG9 marks a membrane compartment that is originated from the Golgi and is enriched in PI4KB and PI4K2A, to produce PI4P. This early autophagy compartment also contains Arf1, Arl1 and ARFIP2 as well as PKD2, known regulators of the activity of PI4KB in the Golgi. Importantly, these early steps involving ATG9 appear to precede and play a role in the recruitment of the ULK1 complex, WIPI2B, LC3B proteins adding a new element to the very early phases of autophagosome formation complementing the already well-documented roles of PI3P in the process (30, 31). These new studies also highlighted the Golgi as a major contributor to that autophagy maturation process that has traditionally been linked to ER (32).
PI4KB-generated PI4P has also been implicated at later stages of autophagosome maturation, namely in the process of lysosome reformation (33). Sidhar et al. (33) have found that PI4KB-depleted cells form tubulation of their LAMP1-positive lysosomes that corresponded to late-stage autolysosomes rather than autophagosomes. They showed that an enhanced efflux of lysosomal components in PI4KB-depleted cells led to the depletion of lysosomes in important regulatory proteins such as the V-ATPase. These studies postulated that PI4KB is critical for the proper control of lysosome reformation from autolysosomes.
PI4K2A
The first indication that PI4K2A is linked to lysosomal sorting and degradation was the finding that PI4K2A promoted EGF-receptor degradation (17). More recently, the Albanesi and Yin groups showed that PI4K2A is localized to autophagosomes under conditions of amino acid starvation by association and recruitment by the γ-aminobutyric acid receptor-associated protein (GABARAP), a member of the small ubiquitin-like autophagy modulatory mammalian ATG8 proteins. They showed that siRNA-mediated knockdown of PI4K2A or overexpression of a dominant negative, kinase-dead version of PI4K2A caused accumulation of abnormally large autophagosomes, suggesting a defect in autophagosome–lysosome fusion (34). Our recent studies using CRISPR/Cas9-mediated inactivation of the PI4K2A gene in HEK293 cells also confirmed that such cells showed major impairment in autophagosome–lysosome fusion (19). Our studies also showed the PI4K2A deletion caused massive tubulation of Rab7-positive late endosomes, but surprisingly did not reproduce the EGFR degradation defect reported by PI4K2A knock-down studies (17).
Is it PI4P or its phosphorylated products, PI(4,5)P2 and PI(3,4)P2 that control autophagy flux?
Although the role of PI4Ks is now well documented, the question arises whether PI4P is acting on its own right or it is converted to another phosphoinositide, such as PI(4,5)P2 or PI(3,4)P2. For example, PI4P produced by PI4K2A was found to act on its own right and not through PI(4,5)P2 when associating with the GABARAP protein and promote phagosome-lysosome fusion (34). However, several studies have shown a role of PI(4,5)P2 and PIP kinases in autophagy. First, it has been shown that PI4P 5-kinase γ isoform 5 (PIP5K1Ci5) regulates the lysosomal degradation of E-cadherin (35) and the EGF receptor (36). Recently, we found that PI4P generated by PI4K2A is converted to PI(4,5)P2 by PIP5K1C in Rab7-positive endosomes. PI(4,5)P2 then drives the hydrolysis of GTP within Rab7 turning it from its GTP- to a GDP-bound form and releasing the Rab7 effector PLEKHM1 from the late endosomal membrane (19). PLEKHM1 has been reported to work as a tether for stabilizing autophagosome-lysosome contact with the homotypic fusion and vacuole protein sorting (HOPS) complex (37). Inactivation of PI4K2A or silencing of PIP5K1C lead to accumulation of non-acidifying autophagosomes (19). Our studies suggested that PI(4,5)P2 activates a Rab7-specific GTPase activating GAP protein. Among the many members of the TBC domain family of Rab7-GAPs several has been shown to play important roles in autophagy including Armus/TBC1D2A (38, 39) and TBC1D15 (40). In fact, a recent study showed that out of the five splice variants of PIP5K1C, specifically, PIP5K1Ci5, directly interacts with Rab7 and recruits the Rab7 GTPase-activating protein TBC1D5 to the late endosome and lysosome (41). This finding is consistent with our hypothesis that increased formation of PI(4,5)P2 causes Rab7 inactivation through stimulation of a Rab7 GAP protein.
Other studies also suggested the role of PI(4,5)P2 in both early and late stages of autophagosome formation and maturation. PIP5K1Ci5 was shown to be responsible for an early step of autophagy. Tan et al. (42) showed that PIP5K1Ci5 and its product PI(4,5)P2 exist at ER-late endosome contact sites and controls autophagy initiation through PI(4,5)P2 interaction with the C-terminal BATS domain of ATG14L. This interaction promotes assembly of the Beclin1–ATG14L–Vps34 complex. The complexity of this process and its regulation is highlighted by another set of studies, which showed that another group of lipid kinases, the type-II PIP kinases or PI5P 4-kinases also play a role in the control of autophagy (43). PI5P 4-kinases (α and β forms, the γ form showing very low activity) convert the minor inositol lipid PI5P to PI(4,5)P2 representing a minor but functionally important route to PI(4,5)P2 synthesis tightly linked to regulation of cancer and metabolism (44). Lundquist et al. (43) have shown that double loss of PI5P4Kα and β causes abnormal accumulation of lipid droplets and autophagosomes in liver or in mouse embryonic fibroblasts derived from PI5P4Kα- and β-deficient mice and that loss of these kinases is especially toxic in p53 deficient conditions. Since type II PIP kinases use PI5P as a substrate, the question also arises how PI5P is formed and whether its production is also necessary for autophagy. The source of PI5P is somewhat controversial, as it is mostly produced indirectly through dephosphorylation of PI(3,5)P2 but also can be directly generated from PI by the lipid kinase PIKfyve. In either case, the PIKfyve enzyme is necessary for PI5P generation. In one study, it was shown that PI5P can initiate a non-canonical autophagosome pathway that bypasses the classical, VPS34-PI3P mediated mechanism (45).
As pointed out earlier, after autophagosome–lysosome fusion, autolysosomes elongate their membranes and form tubules for recovery of nascent lysosomes, termed autophagic lysosome reformation (ALR) (46). Rong et al. has shown that two PIP5Ks (PIP5K1A and PIP5K1B) are involved in this process. Using a combination of screening using large-scale siRNA knockdown and proteomic analysis of purified ALR, they found that PIP5K1B acts on ALR initiation while PIP5K1A acts to pinch off the proto-lysosome membranes by recruitment of a vesicle coat protein (47). These two PIP5Ks probably do not localize to lysosomes under nutrient-rich condition, but autophagy induction was found to stimulate their recruitment to lysosome membranes. The substrate of the PIP5Ks, namely PI4P in lysosomes could be provided by PI4KB, a type III PI4K. PI4KB primarily localizes to the Golgi, but was also found in lysosomes to control the lysosomal vesiculation or efficient excision as well as the sorting of lysosomal constituents (33).
In other studies, PI(4,5)P2 elimination was found critical for normal autophagosome lysosome fusion. OCRL, one of the inositol lipid 5-phosphatases, the mutation of which causes the human disease, oculo-cerebro-renal syndrome of Lowe, converts PI(4,5)P2 to PI4P. OCRL is localized to various endosomal compartments including the Golgi, but its loss also leads to PI(4,5)P2 accumulation in lysosomes. In OCRL-deficient cells, the increased PI(4,5)P2 in lysosomes causes inhibition of the calcium channel Mucolipin1 and hence interferes with autophagosome-lysosome fusion (48). Another study showed that loss of another inositol lipid 5-phosphatase, synaptojanin-1 in zebrafish photoreceptor cells leads to accumulation of autophagosomes (49).
In addition to PI(4,5)P2, PI4P can also be converted to PI(3,4)P2 by Class II PI 3-kinases (50). However, Class II PI3Ks can also generate PI3P, a lipid that has been found critical for autophagy initiation and progression (reviewed in detail elsewhere, 3, 7, 9). Indeed, it has been shown that PI3KC2A and B can substantially contribute to initiation of autophagy through PI3P formation, especially when the Class III PI3K, Vps34 is depleted (51, 52). However, Marat et al. (53) has shown that nutrient deprivation induces recruitment of the class II PI3KC2B to lysosomes and prompts PI(3,4)P2 synthesis, leading to suppression of mTOR complex 1 (mTORC1) and here the lipid effector is PI(3,4)P2 rather than PI3P.
Contribution of lipid transfer proteins to autophagy control orchestrated by inositol lipids
It has been recently recognized that PI4P and PI(4,5)P2 are important regulators of non-vesicular lipid transfer between membranes of various organelles (2). OSBP) and several members of its related proteins, the ORPs have been found to use PI4P gradients to fuel the transport of newly synthesized lipids from the ER to their cellular destinations or to distribute lipids taken up by the cells through endocytosis (2). Indeed, abolition of one of the VAMP-associated proteins, VAPB, which anchors OSBP and many of the ORPs to the ER, causes accumulation of autophagic vesicles, attributed to the aberrant accumulation of PI4P in the Golgi and endosomes and impeding lysosome acidification (54). PI4P accumulation may not be the sole problem when OSBP and ORP members do not function properly. Given the known connection between cholesterol content of late endosomes/lysosomes and autophagosome biology (55, 56), PI4P could play an important role through the regulation of the cholesterol transfer processes. For example, it has been reported that lysosomal cholesterol facilitates recruitment and activation of mTORC1 through the lysosomal arginine sensor SLC38A9 and that the lysosomal cholesterol is provided by OSBP with cholesterol transfer activity from ER to the limiting membrane of lysosomes (57, 58). Since OSBP is regulated by PI4P through its N-terminal PH domain that binds PI4P and active Arf1 and also transports PI4P as a cargo (59), depletion of PI4P can cause a defect in autophagy flux following loss of the cholesterol transfer to late endosomes and lysosomes.
Cholesterol transport from the PM can be equally important for autophagy regulation. Cholesterol is not only synthesized in the ER but is also provided by endocytosis of low-density lipoproteins (LDL). After its endocytosis, LDL is transported into the lysosome where cholesterol ester is hydrolysed to free cholesterol. Free cholesterol exits from the limiting membrane of lysosome with the help of the NPC1 and NPC2 proteins (60). ORP1L, a ORP family protein, mediates transfer of free cholesterol from the limiting membrane of lysosomes to ER (61, 62). ORP1L binds to PI4P via its PH domain for localization on late endosomes and lysosomes and other inositol lipids, namely PI(4,5)P2 and PI(3,4)P2 allosterically enhance ORP1L transport activity (61, 62). ORP1L has been found to regulate autophagosome lysosome fusion by controlling the membrane attachment of PLEKHM1 and RILP and contributing to the recruitment of the HOPS complex to autophagic vacuoles (63). Another ORP protein, ORP5 has been reported to mediate cholesterol exit from the late endosomes and lysosomes together with NPC1 and ORP5 knock-down causes cholesterol accumulation in the lysosome limiting membrane (64). However, recent studies have implicated ORP5 in a variety of membrane contacts, including ER-mitochondria (65), ER-lipid droplets (66) and ER-PM, and it has been shown to transport phosphatidylserine in exchange for PI4P across these membrane contacts (67, 68). Another cholesterol transport protein, StAR-related lipid transfer domain-3 is known as a sterol binding protein transferring cholesterol from ER to endosomes and its overexpression results in cholesterol accumulation in endosomes (69).
Concluding remarks
In this review, we summarize how two of the inositol lipids, PI4P and PI(4,5)P2 that have been classically associated with receptor triggered plasma membrane signalling, regulate autophagy from autophagosome initiation through late endosome and lysosomes fusion. Almost all of the seven phosphoinositide species have been known to control several steps in the endomembrane dynamics of eukaryotic cells. These include the trafficking steps originating from the plasma membrane as well as those that are initiated in the ER through the Golgi and are destined for the plasma membrane. An important destination of many of these trafficking steps is the lysosome that processes both the endocytosed nutrients and membrane components and the misfolded proteins originated from the ER. Under starvation, these processes are also utilized to recycle damaged cellular organelles or cytoplasmic material and the mTOR complex is the main orchestrator of these pathways. Since PI3P and PI(3,5)P2 have been known as key phosphoinositides that control endocytic flux and its direction toward the lysosomes, it is not surprising that they have been found to be also key players in autophagy. Similarly, the Rab proteins, especially Rab7 that functions in the late endosome–lysosome interface has been important components of the late autophagy events, such as the autophagosome-lysosome fusion and lysosome reformation. It appears that PI4P and PI(4,5)P2 are most closely associated with these processes determining of how Rab7 interacts with its numerous effectors. Not surprisingly, Rab7 has a large number of GAP and still a few GEF regulators and many of them have been linked to mTOR regulation and autophagy control. Deciphering the delicate steps that each one of these Rab7 regulators and effectors control in the autophagy machinery and their local control by PI4P and PI(4,5)P2 will be an exciting direction for the near future.
Funding
The research of Dr Balla has been supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health of the USA. The work of Dr Baba was supported by Grant-in-Aid for Research Activity Start-up (19K23) and the Takeda Science Foundation.
Conflict of Interest
None declared.
References
- 1. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balla T., Kim Y.J., Alvarez-Prats A., Pemberton J. (2019) Lipid dynamics at contact sites between the endplasmic reticulum and other organelles. Annu. Rev. Cell Dev. Biol. 35, 85–109 [DOI] [PubMed] [Google Scholar]
- 3. Palamiuc L., Ravi A., Emerling B.M. (2019) Phosphoinositides in autophagy: current roles and future insights. FEBS J. 287, 222–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mizushima N., Yoshimori T., Ohsumi Y. (2011) The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 5. Yu L., Chen Y., Tooze S.A. (2018) Autophagy pathway: cellular and molecular mechanisms. Autophagy 14, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klionsky D.J., Eskelinen E.-L., Deretic V. (2014) Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I’m confused. Autophagy 10, 549–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Noda T., Matsunaga K., Taguchi-Atarashi N., Yoshimori T. (2010) Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin. Cell Dev. Biol. 21, 671–676 [DOI] [PubMed] [Google Scholar]
- 8. Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M.W., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.-P., Brech A., Bilder D., Stenmark H. (2007) ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 17, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 9. Wirth M., Joachim J., Tooze S.A. (2013) Autophagosome formation—the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin. Cancer Biol. 23, 301–309 [DOI] [PubMed] [Google Scholar]
- 10. Flanagan C.A., Schnieders E.A., Emerick A.W., Kunisawa R., Admon A., Thorner J. (1993) Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262, 1444–1448 [DOI] [PubMed] [Google Scholar]
- 11. Audhya A., Foti M., Emr S.D. (2000) Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell 11, 2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakatsu F., Baskin J.M., Chung J., Tanner L.B., Shui G., Lee S.Y., Pirruccello M., Hao M., Ingolia N.T., Wenk M.R., De Camilli P. (2012) PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 199, 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wong K., Meyers R., Cantley L.C. (1997) Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 272, 13236–13241 [DOI] [PubMed] [Google Scholar]
- 14. Barylko B., Gerber S.H., Binns D.D., Grichine N., Khvotchev M., Südhof T.C., Albanesi J.P. (2001) A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J. Biol. Chem. 276, 7705–7708 [DOI] [PubMed] [Google Scholar]
- 15. Barylko B., Mao Y.S., Wlodarski P., Jung G., Binns D.D., Sun H.Q., Yin H.L., Albanesi J.P. (2009) Palmitoylation controls the catalytic activity and subcellular distribution of phosphatidylinositol 4-kinase IIα. J. Biol. Chem. 284, 9994–10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balla A., Tuymetova G., Barshishat M., Geiszt M., Balla T. (2002) Characterization of type II phosphatidylinositol 4-kinase isoforms reveals association of the enzymes with endosomal vesicular compartments. J. Biol. Chem. 277, 20041–20050 [DOI] [PubMed] [Google Scholar]
- 17. Minogue S., Waugh M.G., De Matteis M.A., Stephens D.J., Berditchevski F., Hsuan J.J. (2006) Phosphatidylinositol 4-kinase is required for endosomal trafficking and degradation of the EGF receptor. J. Cell Sci. 119, 571–580 [DOI] [PubMed] [Google Scholar]
- 18. Henmi Y., Morikawa Y., Oe N., Ikeda N., Fujita A., Takei K., Minogue S., Tanabe K. (2016) PtdIns4 KIIα generates endosomal PtdIns(4)P and is required for receptor sorting at early endosomes. Mol. Biol. Cell 27, 990–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baba T., Toth D.J., Sengupta N., Kim Y.J., Balla T. (2019) Phosphatidylinositol 4,5-bisphosphate controls Rab7 and PLEKMH1 membrane cycling during autophagosome–lysosome fusion. EMBO J. . 38, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakatsu F., Messa M., Nández R., Czapla H., Zou Y., Strittmatter S.M., de Camilli P. (2015) Sac2/INPP5F is an inositol 4-phosphatase that functions in the endocytic pathway. J. Cell Biol. 209, 85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu F.S., Hu F., Mao Y. (2015) Spatiotemporal control of phosphatidylinositol 4-phosphate by Sac2 regulates endocytic recycling. J. Cell Biol. 209, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong R., Saheki Y., Swarup S., Lucast L., Harper J.W., De Camilli P. (2016) Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cruz-Garcia D., Ortega-Bellido M., Scarpa M., Villeneuve J., Jovic M., Porzner M., Balla T., Seufferlein T., Malhotra V. (2013) Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. . 32, 1717–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salcedo-Sicilia L., Granell S., Jovic M., Sicart A., Mato E., Johannes L., Balla T., Egea G. (2013) βIII spectrin regulates the structural integrity and the secretory protein transport of the Golgi complex. J. Biol. Chem. 288, 2157–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jovi M., Kean M.J., Dubankova A., Boura E., Gingras A.-C., Brill J.A., Balla T. (2014) Endosomal sorting of VAMP3 is regulated by PI4K2A. J. Cell Sci. 127, 3745–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Niu Y., Zhang C., Sun Z., Hong Z., Li K., Sun D., Yang Y., Tian C., Gong W., Liu J.J. (2013) PtdIns(4)P regulates retromer-motor interaction to facilitate dynein-cargo dissociation at the trans-Golgi network. Nat. Cell Biol. . 15, 417–429 [DOI] [PubMed] [Google Scholar]
- 27. Ketel K., Krauss M., Nicot A.S., Puchkov D., Wieffer M., Müller R., Subramanian D., Schultz C., Laporte J., Haucke V. (2016) A phosphoinositide conversion mechanism for exit from endosomes. Nature 529, 408–412 [DOI] [PubMed] [Google Scholar]
- 28. Clayton E.L., Minogue S., Waugh M.G. (2013) Mammalian phosphatidylinositol 4-kinases as modulators of membrane trafficking and lipid signaling networks. Prog. Lipid Res. 52, 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson C., Venditti R., Rega L.R., Colanzi A., D'Angelo G., De Matteis M.A. (2011) The Golgi apparatus: an organelle with multiple complex functions. Biochem. J. 433, 1–9 [DOI] [PubMed] [Google Scholar]
- 30. Judith D., Tooze S.A. (2019) ATG9A supplies PtdIns4P to the autophagosome initiation site. Autophagy 15, 1660–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Judith D., Jefferies H.B.J., Boeing S., Frith D., Snijders A.P., Tooze S.A. (2019) ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIβ. J. Cell Biol. 218, 1634–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De Tito S., Hervás J.H., van Vliet A.R., Tooze S.A. (2020) The Golgi as an assembly line to the autophagosome. Trends Biochem. Sci. 45, 484–496 [DOI] [PubMed] [Google Scholar]
- 33. Sridhar S., Patel B., Aphkhazava D., Macian F., Santambrogio L., Shields D., Cuervo A.M. (2012) The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. . 32, 324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang H., Sun H.Q., Zhu X., Zhang L., Albanesi J., Levine B., Yin H. (2015) GABARAPs regulate PI4P-dependent autophagosome: lysosome fusion. Proc. Natl. Acad. Sci. USA . 112, 7015–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schill N.J., Hedman A.C., Choi S., Anderson R.A. (2014) Isoform 5 of PIPKIγ regulates the endosomal trafficking and degradation of E-cadherin. J. Cell Sci. . 127, 2189–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Y., Hedman A.C., Tan X., Schill N.J., Anderson R.A. (2013) Endosomal type Iγ PIP 5-kinase controls EGF receptor lysosomal sorting. Dev. Cell. 25, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., Kirchof A., Gatti E., Helfrich M.H., Wakatsuki S., Behrends C., Pierre P., Dikic I. (2015) PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57, 39–54 [DOI] [PubMed] [Google Scholar]
- 38. Frasa M.A.M., Koessmeier K.T., Ahmadian M.R., Braga V.M.M. (2012) Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat. Rev. Mol. Cell Biol. . 13, 67–73 [DOI] [PubMed] [Google Scholar]
- 39. Jaber N., Mohd-Naim N., Wang Z., DeLeon J.L., Kim S., Zhong H., Sheshadri N., Dou Z., Edinger A.L., Du G., Braga V.M.M., Zong W.-X. (2016) Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J. Cell Sci. . 129, 4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamano K., Fogel A.I., Wang C., van der Bliek A.M., Youle R.J. (2014) Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 3, e01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun M., Luong G., Plastikwala F., Sun Y. (2020) Control of Rab7a activity and localization through endosomal type Igamma PIP 5-kinase is required for endosome maturation and lysosome function. FASEB J. 34, 2730–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tan X., Thapa N., Liao Y., Choi S., Anderson R.A. (2016) PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc. Natl. Acad. Sci. USA . 113, 10896–10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lundquist M.R., Goncalves M.D., Loughran R.M., Possik E., Vijayaraghavan T., Yang A., Pauli C., Ravi A., Verma A., Yang Z., Johnson J.L., Wong J.C.Y., Ma Y., Hwang K.S.K., Weinkove D., Divecha N., Asara J.M., Elemento O., Rubin M.A., Kimmelman A.C., Pause A., Cantley L.C., Emerling B.M. (2018) Phosphatidylinositol-5-phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol. Cell. 70, 531–544.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mathre S., Reddy K.B., Ramya V., Krishnan H., Ghosh A., Raghu P. (2019) Functional analysis of the biochemical activity of mammalian phosphatidylinositol 5 phosphate 4-kinase enzymes. Biosci. Rep. 39, BSR20182210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vicinanza M., Korolchuk V.I., Ashkenazi A., Puri C., Menzies F.M., Clarke J.H., Rubinsztein D.C. (2015) PI(5)P regulates autophagosome biogenesis. Mol. Cell. 57, 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., Hailey D.W., Oorschot V., Klumperman J., Baehrecke E.H., Lenardo M.J. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rong Y., Liu M., Ma L., Du W., Zhang H., Tian Y., Cao Z., Li Y., Ren H., Zhang C., Li L., Chen S., Xi J., Yu L. (2012) Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. . 14, 924–934 [DOI] [PubMed] [Google Scholar]
- 48. De Leo M.G., Staiano L., Vicinanza M., Luciani A., Carissimo A., Mutarelli M., Di Campli A., Polishchuk E., Di Tullio G., Morra V., Levtchenko E., Oltrabella F., Starborg T., Santoro M., Di Bernardo D., Devuyst O., Lowe M., Medina D.L., Ballabio A., De Matteis M.A. (2016) Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat. Cell Biol. . 18, 839–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. George A.A., Hayden S., Stanton G.R., Brockerhoff S.E. (2016) Arf6 and the 5’phosphatase of synaptojanin 1 regulate autophagy in cone photoreceptors. Bioessays 38, S119–35 [DOI] [PubMed] [Google Scholar]
- 50. Gulluni F., De Santis M.C., Margaria J.P., Martini M., Hirsch E. (2019) Class II PI3K functions in cell biology and disease. Trends Cell Biol. 29, 339–359 [DOI] [PubMed] [Google Scholar]
- 51. Devereaux K., Dall’Armi C., Alcazar-Roman A., Ogasawara Y., Zhou X., Wang F., Yamamoto A., De Camilli P., Di Paolo G. (2013) Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS One 8, e76405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Merrill N.M., Schipper J.L., Karnes J.B., Kauffman A.L., Martin K.R., MacKeigan J.P. (2017) PI3K-C2α knockdown decreases autophagy and maturation of endocytic vesicles. PLoS One 12, e0184909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marat A.L., Wallroth A., Lo W.T., Müller R., Norata G.D., Falasca M., Schultz C., Haucke V. (2017) mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356, 968–972 [DOI] [PubMed] [Google Scholar]
- 54. Mao D., Lin G., Tepe B., Zuo Z., Tan K.L., Senturk M., Zhang S., Arenkiel B.R., Sardiello M., Bellen H.J. (2019) VAMP associated proteins are required for autophagic and lysosomal degradation by promoting a PtdIns4P-mediated endosomal pathway. Autophagy 15, 1214–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheng J., Ohsaki Y., Tauchi-Sato K., Fujita A., Fujimoto T. (2006) Cholesterol depletion induces autophagy. Biochem. Biophys. Res. Commun. 351, 246–252 [DOI] [PubMed] [Google Scholar]
- 56. Jaishy B., Abel E.D. (2016) Lipids, lysosomes, and autophagy. J. Lipid Res. 57, 1619–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Castellano B.M., Thelen A.M., Moldavski O., Feltes M., Van Der Welle R.E.N., Mydock-McGrane L., Jiang X., Van Eijkeren R.J., Davis O.B., Louie S.M., Perera R.M., Covey D.F., Nomura D.K., Ory D.S., Zoncu R. (2017) Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lim C.Y., Davis O.B., Shin H.R., Zhang J., Berdan C.A., Jiang X., Counihan J.L., Ory D.S., Nomura D.K., Zoncu R. (2019) ER–lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann–Pick type C. Nat. Cell Biol. . 21, 1206–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Antonny B., Bigay J., Mesmin B. (2018) The oxysterol-binding protein cycle: burning off PI(4)P to transport cholesterol. Annu. Rev. Biochem. 87, 809–837 [DOI] [PubMed] [Google Scholar]
- 60. Meng Y., Heybrock S., Neculai D., Saftig P. (2020) Cholesterol handling in lysosomes and beyond. Trends Cell Biol. 30, 452–466 [DOI] [PubMed] [Google Scholar]
- 61. Zhao K., Ridgway N.D. (2017) Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep. 19, 1807–1818 [DOI] [PubMed] [Google Scholar]
- 62. Dong J., Du X., Wang H., Wang J., Lu C., Chen X., Zhu Z., Luo Z., Yu L., Brown A.J., Yang H., Wu J.W. (2019) Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P 2/PI(3,4)P 2. Nat. Commun. 10, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wijdeven R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., Neefjes J. (2016) Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7, 11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Du X., Kumar J., Ferguson C., Schulz T.A., Ong Y.S., Hong W., Prinz W.A., Parton R.G., Brown A.J., Yang H. (2011) A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Galmes R., Houcine A., Vliet A.R., Agostinis P., Jackson C.L., Giordano F. (2016) ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function. EMBO Rep. . 17, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Du X., Zhou L., Aw Y.C., Mak H.Y., Xu Y., Rae J., Wang W., Zadoorian A., Hancock S.E., Osborne B., Chen X., Wu J.-W., Turner N., Parton R.G., Li P., Yang H. (2020) ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J. Cell Biol. 219, e201905162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L.B., Narayanaswamy P., Wenk M.R., Nakatsu F., De Camilli P. (2015) PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sohn M., Ivanova P., Brown H.A., Toth D.J., Varnai P., Kim Y.J., Balla T. (2016) Lenz-Majewski mutations in PTDSS1 affect phosphatidylinositol 4-phosphate metabolism at ER-PM and ER-Golgi junctions. Proc. Natl. Acad. Sci. USA . 113, 4314–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilhelm L.P., Wendling C., Védie B., Kobayashi T., Chenard M., Tomasetto C., Drin G., Alpy F. (2017) STARD 3 mediates endoplasmic reticulum‐to‐endosome cholesterol transport at membrane contact sites. EMBO J. . 36, 1412–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]