Abstract
Background
Prophylactic cotrimoxazole treatment is recommended in human immunodeficiency virus (HIV)–exposed, uninfected (HEU) infants, but the effects of this treatment on developing HEU infant gut microbiotas and resistomes are largely undefined.
Methods
We analyzed whole-metagenome sequencing data from 163 longitudinally collected stool samples from 63 HEU infants randomized to receive (n = 34; CTX-T) or to not receive (n = 29; CTX-N) prophylactic cotrimoxazole treatment. We generated taxonomic, functional pathway, and resistance gene profiles for each sample and compared microbiome signatures between the CTX-T and CTX-N infants.
Results
Metagenomic analysis did not reveal significant differences in taxonomic or functional pathway α-diversity between CTX-T and CTX-N infants. In contrast, resistance gene prevalence (P = .00719) and α-diversity (P = .0045) increased in CTX-T infants. These differences increased over time for both resistance gene prevalence measured by log-normalized abundance (4-month mean, 0.71 [95% confidence interval {CI}, .2–1.2] and 6-month mean, 0.85 [95% CI, .1–1.7]) and α-diversity (P = .0045). Unlike α-diversity, interindividual gut microbiome taxonomic (mean, −0.11 [95% CI, −.15 to −.077]), functional taxonomic (mean, −0.050 [95% CI, −.084 to −.017]), and resistance gene (mean, −0.13 [95% CI, −.17 to −.099]) β-diversity decreased in CTX-T infants compared with CTX-N infants. These results are consistent with persistent antibiotic selection pressure.
Conclusions
Cotrimoxazole prophylaxis in HEU infants decreased gut microbiome β-diversity and increased antibiotic resistance gene α-diversity and prevalence. Antibiotic resistance is a growing threat, especially in low- and middle-income countries where the higher perinatal HIV exposure rates result in cotrimoxazole prophylaxis. Understanding effects from current HEU infant antibiotic prophylaxis guidelines will inform guideline revisions and efforts to reduce increasing antibiotic resistance.
Keywords: cotrimoxazole prophylaxis, antibiotic resistance, HIV-exposed, uninfected infant, microbiome
Cotrimoxazole treatment in HIV-exposed, uninfected (HEU) infants increases resistance gene prevalence and α-diversity, and decreases microbial taxonomic, functional pathway, and resistance gene β-diversity. Future HEU infant prophylaxis recommendations should consider these findings given rising global antibiotic resistance.
(See the Editorial Commentary by Bourke and Evans on pages 2869–71.)
Infants perinatally exposed to human immunodeficiency virus (HIV) have higher infection risk after birth than HIV-unexposed infants [1–8]. This higher infection risk exists regardless of the infant’s HIV status [4, 8]. The World Health Organization (WHO) recommends prophylactic cotrimoxazole therapy for all HIV-positive adults and infants in areas with high prevalence of severe bacterial infections and malaria, and for HIV-exposed, uninfected (HEU) infants to prevent Pneumocystis jirovecii infection [9]. Cotrimoxazole is a broad-spectrum antibiotic coformulation of trimethoprim and sulfamethoxazole that inhibits bacterial tetrahydrofolic acid synthesis [10]. In HIV-infected infants, cotrimoxazole therapy reduces early-life infection due to pneumonia, diarrhea, and malaria [11] and may reduce systemic inflammation [12]. In malaria-endemic areas, cotrimoxazole therapy improves HEU infant survival, but in nonmalaria areas with routine HIV testing, cotrimoxazole therapy’s benefits may be outweighed by risk of gut dysbiosis and increased antimicrobial resistance [13–15].
WHO guidelines recommend that HEU infants receive prophylactic cotrimoxazole from 4–6 weeks after birth until breastfeeding ceases [9]. The guidelines say this is a “strong recommendation” with “very-low-quality evidence.” Since breast feeding is recommended until children are at least 2 years old [16], HEU infants receive cotrimoxazole prophylaxis through the critical period when the gut microbiota transitions from its nascent state to its mature composition [17–22]. Gut microbiota perturbations during infancy are linked to negative health consequences [23–25]. The WHO guidelines [9] note that cotrimoxazole prophylaxis in HEU infants “might cause gut perturbations and affect the gut microbiome,” but the details of these gut microbiome changes remain undefined.
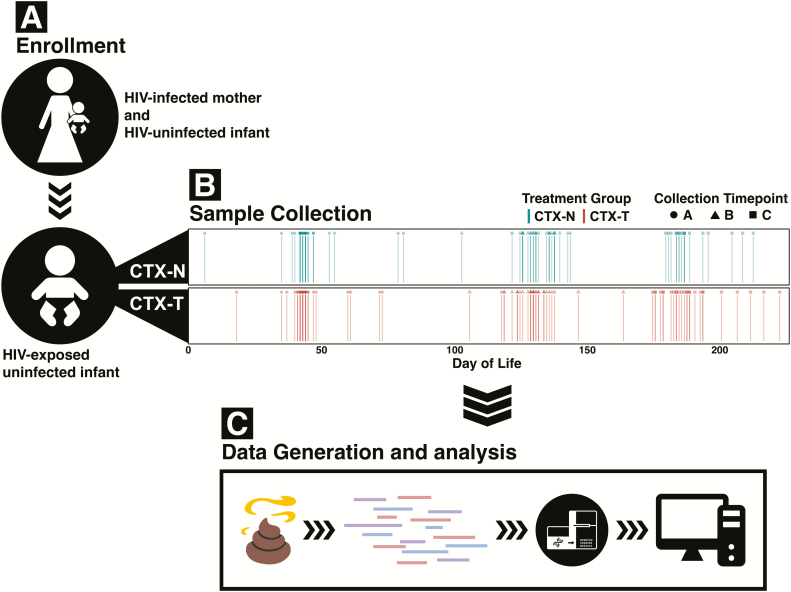
To address concerns about effects of cotrimoxazole prophylaxis on developing HEU infant gut microbiota, we sequenced 163 stool samples from a randomized controlled trial of cotrimoxazole treatment in 63 HEU infants [26]. Stool collections were concentrated during the first 6 months of life, referred to as the first stage of microbiome development [18]. Several studies suggest that infants are most vulnerable to gut microbiome perturbations during this period [18, 23]. We analyzed HEU infant stool metagenomes to determine their gut microbial taxonomic composition, microbiome functional genes, and antibiotic resistomes (Figure 1). We then compared these microbiome characteristics between cotrimoxazole-treated HEU infants (CTX-T infants) and HEU infants not treated with cotrimoxazole (CTX-N infants).
Figure 1.
Stool from human immunodeficiency virus (HIV)–exposed, uninfected infants randomized to receive or not receive cotrimoxazole prophylaxis was collected, sequenced, and analyzed. A, HIV-positive mothers with HIV-uninfected infants were enrolled in the study. Their HIV-negative infants were randomized to either receive cotrimoxazole prophylaxis according to World Health Organization guidelines (red, CTX-T) or to not receive cotrimoxazole prophylaxis (blue, CTX-N). B, Three stool samples were collected per infant at 6 weeks (timepoint A), 4 months (timepoint B), and 6 months (timepoint C). Spread in collection time is due to variability in patient visit time. C, Infant stool samples were then shotgun whole-metagenome sequenced.
MATERIALS AND METHODS
Sample and Metadata Collection
Ethical approval was obtained from the Biomedical Research Ethics Committee, University of KwaZulu-Natal, South Africa (BFC212/13). Before enrollment, written informed consent was obtained from mothers visiting the Cato Manor and Lancers Road Clinics in Durban, South Africa, for routine clinic appointments. Infants with birthweights >2000 g born to HIV-infected mothers were included and followed for 1 year. Infants and their mothers had to be receiving antiretroviral prophylaxis prior to enrollment. Infants had to have a negative HIV DNA polymerase chain reaction result by COBAS AmpliPrep/COBAS TaqMan HIV-1 qualitative test prior to enrollment and their first sample collection timepoint at approximately 6 weeks of age. Infants were excluded if they had any illnesses, antibiotics (including cotrimoxazole), or traditional medicines, and all infants were exclusively breastfed. HEUs were randomized at their first timepoint (6-week) clinic visit to receive or not receive cotrimoxazole therapy. Cotrimoxazole was administered once daily during the study period; infants <5 kg received 20 mg trimethoprim/100 mg sulfamethoxazole orally and infants 5–15 kg received 40 mg trimethoprim/200 mg sulfamethoxazole orally. Stool samples were collected from infants at 3 timepoints (6 weeks, 4 months, and 6 months). Fresh stool from plastic-lined diapers was transferred to 2 mL BD cryovials using disposable wooden sticks. Samples were frozen within 6–8 hours of collection and stored at −80°C until analysis as described previously by Dan et al and Agapova et al [27, 28]. Two hundred thirteen stool specimens were collected from 70 infants during the study. Infants with an initial 6-week stool and at least 1 subsequent sample (63 infants) were shotgun-sequenced and included.
Metagenomic DNA Extraction
PowerSoil DNA Isolation Kit (MoBio Laboratories) was used to extract metagenomic DNA from approximately 150 mg of each stool sample. Manufacturer protocol was used with minor modifications for sample lysis: 2 rounds of 2 minutes of bead beating at 2.5 k oscillations per minute for 2 minutes followed by 1 minute on ice and 2 additional minutes of beadbeating using a Mini-Beadbeater 24 (Biospec Products). DNA was quantified using a Qubit fluorometer dsDNA HS Assay (Invitrogen) and stored at −20°C.
Metagenomic Sequencing Library Preparation
Metagenomic DNA was diluted to 0.5 ng/µL before sequencing library preparation. Sequencing libraries were prepared with a Nextera DNA Library Prep Kit (Illumina) as described in Baym et al [29]. Libraries were purified using the Agencourt AMPure XP system (Beckman Coulter) and quantified using the Quant-iT PicoGreen dsDNA assay (Invitrogen). For each sequencing lane, 10 nM of approximately 96 samples was pooled 3 independent times. These pools were quantified using the Qubit dsDNA BR Assay and combined in an equimolar fashion. Samples were submitted for 2 × 150-bp paired-end sequencing on an Illumina NextSeq High-Output platform with a target sequencing depth of 3 million reads per sample.
Metagenomic Profiling
Illumina paired-end reads were binned by index sequence. Adapter and index sequences were trimmed and sequences were quality-filtered using Trimmomatic version 0.38 [30] with these parameters: java -Xms2048m -Xmx2048m -jar trimmomatic-0.38.jar PE -phred33 ILLUMINACLIP: NexteraPE-PE.fa:2:30:10:1:TRUE SLIDINGWINDOW:4:15 LEADING:10 TRAILING:10 HEADCROP:15 MINLEN:60. Microbial taxa relative abundance was calculated using MetaPhlAn2 [31] (repository tag 2.6.0). Metabolic pathway abundance was determined using HUMAnN2 [32] (repository tag 0.11.2). Raw count values were normalized for sequencing depth and collapsed by ontology, and tables were merged using HUMAnN2 utility scripts [32].
Resistance gene abundance including for dfr/sul genes was calculated using ShortBRED [33]. Marker sequences were built using the Comprehensive Antibiotic Resistance Database [34] with shortedbread_identify.py. Default parameters were used with exception for -clustid 0.95. Uniref90 [35] was the reference masking protein database. Efflux pumps and regulatory subunits were removed due to their lower specificity to antimicrobial resistance compared to other genes in the database.
Statistical Analysis
Where possible, day of life rather than timepoint was used to account for variation in patient visit dates. Statistical analysis was conducted in R software version 3.5.3 [36]. Visualizations were made using ggplot2 version 3.1.0 [37], ggpubr version 0.2 [38], and cowplot version 0.9.4 [39]. The vegan: Community Ecology Package version 2.5–4 [40] was used for canonical analysis of principal coordinates [41] (capscale function), α- and β-diversity calculations, and permutational multivariate analysis of variance tests (adonis2 function). Linear mixed-effects models were implemented with lme4 version 1.1–21 (lmer function) [42]. Dabestr version 0.2.0 [43] was used for bootstrapping samples and for calculating confidence intervals (CIs). Stats (base R) version 3.5.3 was used for statistical calculations. The wilcox.test function was applied with paired = T/F depending on if comparisons were inside or outside of the treatment group. The fisher.test function was for clinical metadata comparisons. The p.adjust function was applied where appropriate to correct for multiple hypothesis testing with method=“fdr” (Benjamini-Hochberg [44]). Log-transformation was implemented using the log function with default parameters.
Data Availability
Shotgun metagenomic reads (quality-checked and filtered for human reads) generated for this study were uploaded to the National Center for Biotechnology Information (NCBI) under the project code PRJNA549787 (see BioSample accession metadata file for individual sample accession codes). Metadata files for these shotgun metagenomic reads are also included in this submission: (1) SACTX_HEU_Infant_metadata.txt; (2) SACTX_DOL_metadata_mod.txt; (3) SACTX_HEU_Infant_bodyMeasurements.txt; (4) SACTX_HEU_Infant_Bloodwork.txt; (5) NCBI_BioSample_AccessionList.txt.
RESULTS
Maternal CD4 Count, Infant Physical Traits, Biological Markers, and Reported Illnesses Were Not Different Between CTX-T and CTX-N Infants
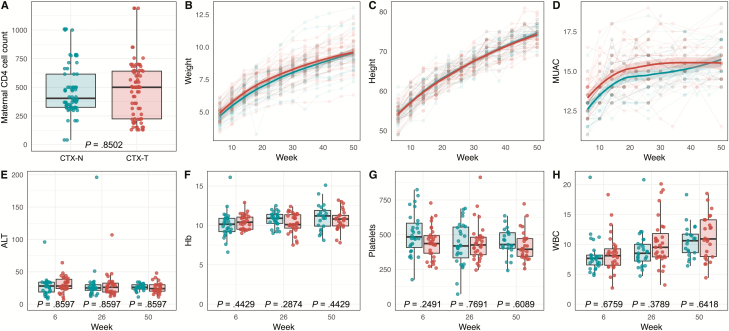
Maternal CD4 T-cell counts, anthropometric measurements (height, weight, and mid-upper arm circumference), biological markers (alanine aminotransferase, hemoglobin, platelets, and white blood cell count), and reported illnesses did not differ by cotrimoxazole treatment (Figure 2, Supplementary Notes E, and Supplementary Figure 1). As these metrics were indistinguishable between CTX-T and CTX-N infants, we investigated microbiome characteristics for intergroup differences.
Figure 2.
Maternal CD4 cell count, physical traits, and blood testing results are not different between cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T; red) and HEU infants not treated with cotrimoxazole (CTX-N; blue). Points represent individual measurements for infants. A, Comparison (unpaired Wilcoxon rank-sum test) of maternal CD4 count between CTX-T infants and CTX-N infants. Loess regression (bold line) with 95% confidence interval (shaded area) for CTX-T infants and CTX-N infants for weight (B), height (C), and mid-upper arm circumference (MUAC) (D). Boxplots (E–H) show median values (dark middle line) and first and third quartiles (lower and upper lines). Between-group comparisons for E–H are made using unpaired Wilcoxon tests (rank-sum) with Benjamini-Hochberg correction. Comparison between CTX-T infants and CTX-N infants at 3 separate collection times for alanine aminotransferase (ALT) (E), hemoglobin (Hb) (F), platelets (G), and white blood cell (WBC) count (H).
CTX-T Infants and CTX-N Infants Separated by Resistance Gene Content Following Cotrimoxazole Treatment
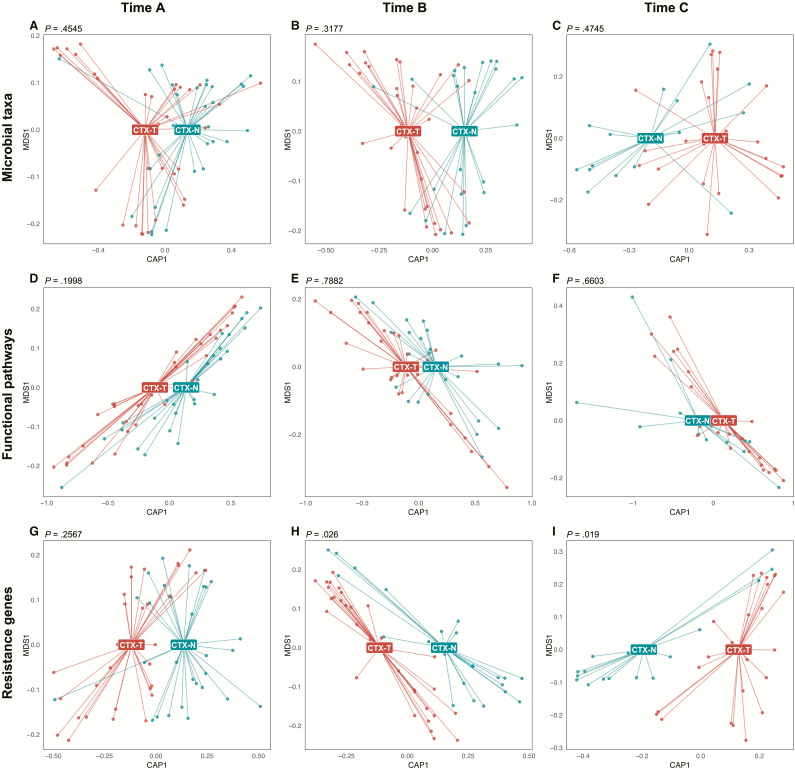
While microbial taxa and functional pathways did not have significantly detectable differences (Figure 3A–F), resistance gene differences between CTX-T and CTX-N infants manifested in timepoints B and C (Figure 3H and 3I; P = .026 and P = .019). This suggests that cotrimoxazole treatment in HEU infants affects resistance genes more than microbial taxonomic composition or functional metabolic pathways. Importantly, this separation manifested at 4 months (timepoint B) and 6 months (timepoint C), but it was not present at 6 weeks (timepoint A) before treatment began.
Figure 3.
Cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T; red) and HEU infants not treated with cotrimoxazole (CTX-N; blue) significantly separate by resistance gene annotation profiles, but not by microbial taxonomic or functional pathway annotation profiles. Points are individual samples and labels are centers of gravity for CTX-T infants and CTX-N infants from canonical analysis of principal coordinates on dissimilarity matrices calculated by Bray-Curtis. Permutational multivariate analysis of variance was used to determine if group separations were significant, and P values are reported above each plot. Input data was generated by MetaPhlAn2 (microbial taxonomic profiles) for A–C, by HUMAnN2 (functional pathway annotations) for D–F, and by ShortBRED (resistance gene annotations) for G–I. Abbreviations: CAP1, Canonical Analysis of Principal coordinates; MDS1, multidimensional scaling.
CTX-T Infants Had Higher Resistance Gene Abundance Than CTX-N Infants Following Cotrimoxazole Treatment
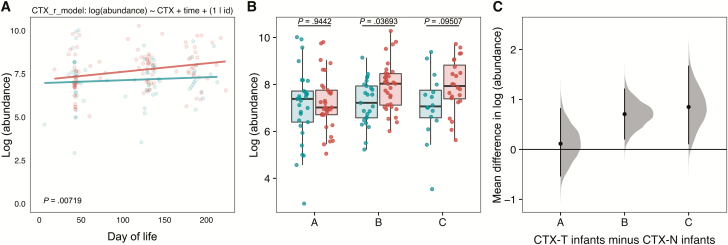
To understand drivers of group separation for resistance genes in timepoints B and C (Figure 3H and 3I), we first looked at resistance gene abundance. Using a linear mixed-effects model with resistance gene abundance as the response variable, we found that cotrimoxazole treatment was a significant predictor of increased resistance gene prevalence (Figure 4A; P = .00719). Additionally, comparisons of CTX-T infants and CTX-N infants binned by timepoint (Figure 4B and 4C) demonstrated increased resistance gene prevalence measured by log-normalized resistance gene abundance in timepoint B (mean, 0.71 [95% CI, .2–1.2]) and timepoint C (mean, 0.85 [95% CI, .1–1.7]) in the CTX-T infants. The timepoint C comparison was not significant by false discovery rate–corrected Wilcoxon testing, but it was significant when tested by bootstrapping to calculate a 95% CI.
Figure 4.
Total resistance gene abundance is higher in cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T) than in HEU infants not treated with cotrimoxazole (CTX-N). A, Points represent individual HEU infant samples colored by treatment group (red for CTX-T infants and blue for CTX-N infants), and lines represent predictions of simple linear regression models for the 2 groups. The x-axis for each plot is the day of life for each infant calculated from their day of birth, and the y-axis is log-transformed total resistance gene abundance. The formula for the linear mixed-effects model is reported above the plot. The model was compared to a null model without the cotrimoxazole treatment variable (CTX) included using a likelihood ratio test. The P value for this comparison is reported at bottom left. B, Points are individual HEU infant samples binned by treatment group (red for CTX-T and blue for CTX-N) and by time. The y-axis is log-transformed total resistance gene abundance. Unpaired Wilcoxon tests were used to compare the treated to untreated samples within collection times. C, Gray distributions show the difference at each collection time between the CTX-T infants and CTX-N infants log-transformed total resistance gene abundance for 5000 bootstrapped subsamples. The black points are the mean value for this distribution, and black lines are 95% confidence intervals.
Microbial Taxonomic and Resistance Gene α-Diversity Increased Significantly in CTX-T Infants Following Cotrimoxazole Exposure
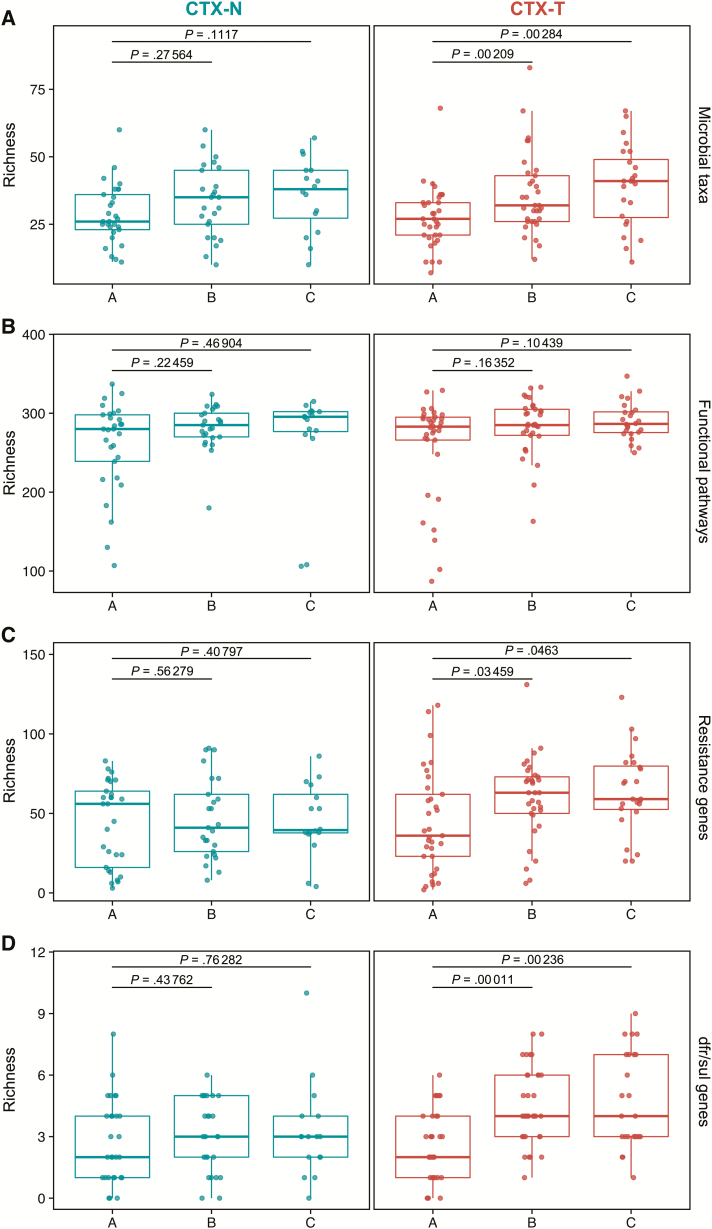
Next, we investigated infant gut microbial taxonomic, functional metabolic pathway, and resistance gene α-diversity longitudinally in CTX-T infants and CTX-N infants (Figure 5A–C). The α-diversity is diversity in an area independent of other areas (in this study, the area is within each infant’s gut). We consistently observed that microbial taxonomic, functional metabolic pathway, and resistance gene α-diversity did not significantly increase from timepoint A to timepoints B or C for CTX-N infants. In contrast, CTX-T infants showed significant increases in α-diversity over time measured by richness (number of unique values) for microbial taxonomic profiles (Figure 5A; timepoint A to timepoint B: P = .0021; timepoint A to timepoint C: P = .0028) and resistance genes (Figure 5C; timepoint A to timepoint B: P = .034; timepoint A to timepoint C: P = .046). Shannon index α-diversity measurements yielded similar results (Supplementary Notes A and Supplementary Figure 2). Importantly, maternal CD4 T-cell count and reported illnesses did not significantly alter HEU infant gut microbial taxa α-diversity (Supplementary Notes D and Supplementary Figures 3–5).
Figure 5.
Resistance richness (α-diversity) significantly increases over time in cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T) for total resistance genes and for dfr/sul resistance genes. Points represent richness values for individual samples, and boxes show median values (dark middle line) and first and third quartiles (lower and upper lines). The x-axis groups for each plot are the times of collection, and the y-axis for each plot is richness. Paired Wilcoxon tests (signed rank) were used to compare the latter 2 collections (timepoints B and C) to the first collection (timepoint A), and P values are reported above the graph with black lines depicting the comparisons. Graphs show HEU infants not treated with cotrimoxazole (CTX-N; blue) on the left and CTX-T infants (red) on the right. Richness was calculated for microbial taxa (A), functional pathways (B), resistance genes (C), and trimethoprim- and sulfonamide-resistance (dfr/sul) genes (D).
Clinical resistance to trimethoprim is often mediated by transferrable dfr genes, and clinical resistance to sulfamethoxazole is often mediated by transferrable sul genes [10]. We observed significant increases in α-diversity of cotrimoxazole-specific dfr and sul resistance genes for CTX-T infants relative to the preintervention timepoint (Figure 5D; timepoint A to timepoint B: P = .00011; timepoint A to timepoint C: P = .00236).
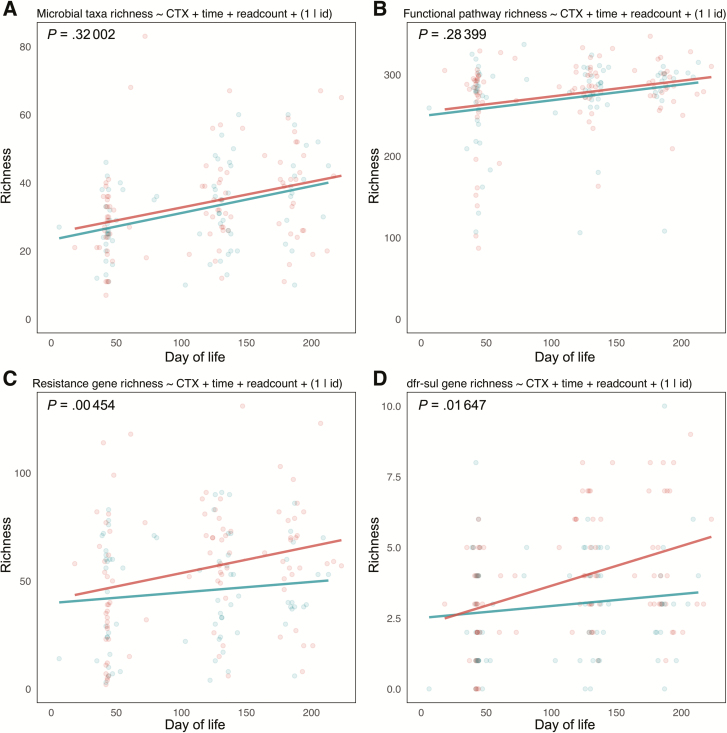
Cotrimoxazole Treatment Resulted in Significantly Higher Resistance Gene α-Diversity for CTX-T Infants Compared to CTX-N Infants
To determine if within treatment group differences were significant between treatment groups, we constructed linear mixed-effects models with α-diversity as the response variable and treatment, day of life, and sample read count as fixed-effects variables. We also included subject identity as a random-effects variable to account for repeated measures (Figure 6 and Supplementary Figure 6). When compared to null models where cotrimoxazole treatment was not included as a fixed effect, cotrimoxazole treatment significantly differentiated resistance gene richness (Figure 6C; P = .0045) and dfr/sul gene richness (Figure 6D; P = .016), but it did not have a significant effect on microbial taxa richness (Figure 6A) or functional pathway richness (Figure 6B).
Figure 6.
Resistance gene richness (α-diversity) is significantly higher in cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T) compared to HEU infants not treated with cotrimoxazole (CTX-N). Points represent individual patient samples colored by treatment group (red for CTX-T infants and blue for CTX-N infants), and lines represent predictions of simple linear regression models for the 2 groups. The x-axis for each plot is the day of life for each infant calculated from their day of birth, and the y-axis is richness. Models were made for microbial taxa (A), functional pathways (B), resistance genes (C), and trimethoprim- and sulfonamide-resistance (dfr/sul) genes (D). Formulas for each linear mixed-effects model are reported above the plots, and these models were compared using likelihood-ratio tests to null models made without the cotrimoxazole treatment variable (CTX) included. The P values for these comparisons of linear mixed-effects models are reported at top left of each graph.
Cotrimoxazole Treatment Resulted in Lower Microbial Taxonomic and Resistance Gene β-Diversity for CTX-T Infants Compared to CTX-N Infants
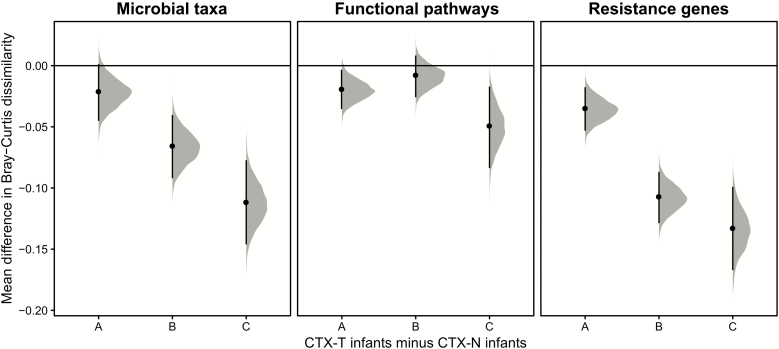
We next compared microbial taxa, functional pathway, and resistance gene β-diversities of CTX-T infants and CTX-N infants within our 3 collection timepoints (Figure 7 and Supplementary Figure 7). We calculated pairwise Bray-Curtis dissimilarities [45] for our samples’ microbial taxonomic profiles, functional metabolic pathways, and resistance genes. β-diversity is diversity between 2 samples, and higher diversity indicates that sample compositions are more different from each other.
Figure 7.
Microbial taxa, functional pathway, and resistance gene β-diversity are lower in cotrimoxazole-treated human immunodeficiency virus–exposed, uninfected (HEU) infants (CTX-T) than in HEU infants not treated with cotrimoxazole (CTX-N). Data are shown for β-diversity calculated from microbial taxonomic profiles, functional metabolic pathways, and resistance genes. Distributions show the difference at each collection timepoint between the CTX-T cohort’s Bray-Curtis dissimilarities and the CTX-N cohort’s Bray-Curtis dissimilarities for 5000 bootstrapped subsamples. The black points are the mean value for this distribution, and black lines are 95% confidence intervals.
Within-treatment group comparisons showed sustained decreases for CTX-T infants in microbial taxa, functional metabolic pathway, and resistance gene β-diversity from 6 weeks to 4 months and to 6 months (Supplementary Notes C and Supplementary Figure 8).
In all cases, CTX-T infants had lower microbial taxonomic, functional pathway, and resistance gene β-diversity than CTX-N infants (Figure 7 and Supplementary Figure 7), indicating that microbiome characteristics in CTX-T infants became more similar to one another after starting cotrimoxazole treatment. This dissimilarity was significantly lower in CTX-T infants compared to CTX-N infants for microbial taxa at timepoint B (mean, −0.067 [95% CI, −.092 to −.04]) and timepoint C (mean, −0.11 [95% CI, −.15 to −.077]). The magnitude of difference between CTX-T and CTX-N infants also increased with time. Interestingly, functional pathways were significantly different between CTX-T infants and CTX-N infants at timepoint A (mean, −0.020 [95% CI, −.035 to −.0033]) and timepoint C (mean, −0.050 [95% CI, −.084 to −.017]) but not at timepoint B (mean, −0.0088 [95% CI, −.026 to .0083]). Resistance gene β-diversity was significantly lower in timepoint A (mean, −0.036 [95% CI, −.053 to −.018]), timepoint B (mean, −0.11 [95% CI, −.13 to −.087]), and timepoint C (mean, −0.13 [95% CI, −.17 to −.099]) in CTX-T infants compared with CTX-N infants. Resistance gene differences between timepoint A and timepoints B and C were also significant, with 95% CIs not overlapping between early and later timepoints for the difference between CTX-T infants and CTX-N infants.
DISCUSSION
Here we investigate the effects of cotrimoxazole prophylaxis on developing HEU infant gut microbiomes, finding that cotrimoxazole treatment has little effect on taxonomic or functional metabolic pathway α-diversity, but increases resistance gene α-diversity. While α-diversity increases or remains the same following cotrimoxazole treatment, taxonomic, functional metabolic pathway, and resistance gene β-diversity decreases.
Studies have noted that early-life microbiota perturbations have long-term health consequences [23–25, 46]. Perturbations can result from many factors, including HIV exposure in utero [47, 48] and antibiotic treatment [18]. In contrast to studies showing that microbiota taxonomic α-diversity decreases following antibiotic treatment [49–52], we did not find significant decreases in CTX-T infants compared to CTX-N infants, corresponding with our results for functional metabolic pathways and keystone taxa abundance (Supplementary Notes B). Gibson et al [21] showed that early-life microbiome α-diversity changes in preterm infants vary by antibiotic, so this may be specific for cotrimoxazole action on developing HEU infant gut microbiomes. Since cotrimoxazole is broad-spectrum [53], it may evenly reduce gut bacterial load, making its effects undetectable by measuring α-diversity or relative abundance. Indeed, 2 studies of HIV-infected populations found that cotrimoxazole had insignificant effect on gut microbial diversity, but differences in study design qualify comparisons to our study (see Supplementary Notes F) [12, 54]. Alternatively, if cotrimoxazole reduces α-diversity, its effects on HEU infant gut microbiomes may be insignificant compared to perturbation from HIV exposure in utero [47, 55]. This possibility is supported by small but statistically significant increases in taxonomic α-diversity from timepoint A to timepoints B and C in CTX-T infants but not in CTX-N infants. This increase in microbial taxa α-diversity may result from initial suppression by cotrimoxazole followed by colonization with cotrimoxazole resistant or tolerant species. Finally, cotrimoxazole resistance may be prevalent enough in our cohort’s early colonizers that cotrimoxazole treatment does not perturb the HEU infant gut microbial community. Studies showing high resistance rates in low- and middle-income countries, including South Africa [56, 57], support this possibility [58]. Together, our taxonomic and functional microbiome data do not indicate large or consistent HEU infant gut microbiome dysbiosis in response to cotrimoxazole treatment.
In contrast with results for taxonomic composition and functional metabolic pathways, cotrimoxazole treatment significantly changed HEU infant antibiotic resistomes. Changes manifested as increases in resistance gene prevalence and resistance gene α-diversity. Importantly, increases are found for trimethoprim and sulfamethoxazole–specific resistance genes and for resistance genes as a whole. This is consistent with results by Powis et al [59] showing that Escherichia coli and Klebsiella pneumoniae commensal bacteria isolated from CTX-T infants had increased cotrimoxazole and amoxicillin resistance.
Microbiota β-diversity decreases over time for infants in this cohort, but in CTX-T infants, β-diversity reductions are swifter and more pronounced than in CTX-N infants. This could be from selective pressure of cotrimoxazole treatment on specific bacterial taxa. As functional metabolic pathways and resistance genes are tied to bacterial taxa, their β-diversity would also be sensitive to the selective pressure of antibiotics. Interestingly, sensitivity of microbial taxonomic profiles, functional metabolic pathways, and resistance genes to cotrimoxazole treatment differs. Resistance genes are most sensitive to antibiotic pressures and functional metabolic pathways are least sensitive. Functional metabolic pathways are likely conserved between multiple bacterial taxa, and this conservation would make them invariant to selective pressures [60–62].
Our study adds to growing work on HEU infants [13, 15, 47, 48, 59, 63] (see Supplementary Notes F). To our knowledge, this is the first randomized controlled trial comparing cotrimoxazole-treated and untreated HEU infant gut microbiomes. Despite our efforts, this study has limitations. Since our collection timepoints only extend approximately 6 months, further changes may manifest between CTX-T and CTX-N infants after our sampling. Alternatively, resistance gene prevalence and diversity differences may equalize between the 2 groups. Our collection times are staggered due to patient appointment availability, and this staggering could reduce power to detect microbiota differences. Second, we can only establish relative and not absolute abundance differences; thus, we cannot determine if CTX-T infants and CTX-N infants differ in total gut bacterial load. Additionally, we do not have maternal metagenomes, in utero data, or cytokine information. Prior research has shown maternal factors such as diet and antimicrobial use can impact infant microbiomes and inflammatory markers [64, 65]. Finally, adherence difficulties are mentioned in the WHO guidelines [9]. Given daily dosing regimens, we cannot verify treatment adherence beyond verbal confirmation from mothers that infants received cotrimoxazole and did not receive other antibiotics.
Our study sheds light on the effects of cotrimoxazole prophylaxis on HEU infant microbiomes and provides additional information for future treatment recommendations. Unlike some other studies on antibiotic use in infants [66–69], we did not observe significant microbiome dysbiosis. However, increases we observed in resistance gene prevalence and diversity are important for ongoing public health and antibiotic stewardship discussions [58]. WHO guidelines say that “understanding whether people living with HIV using cotrimoxazole affects community cotrimoxazole resistance and whether community cotrimoxazole resistance affects treatment failure for other infectious diseases is important for national efforts to combat antimicrobial resistance” [9]. Low- and middle-income countries, including South Africa, often have high background antibiotic resistance rates [58]. Unnecessary antibiotic prescriptions may contribute to resistance disparities, and there is mounting evidence that cotrimoxazole treatment of HEU infants in countries with low malaria prevalence is ineffective [13]. As South Africa has low malaria prevalence and high infant HIV exposure, WHO recommendations on prophylactic cotrimoxazole may unnecessarily contribute to increasing resistance; however, more research on treatment consequences of high background antibiotic resistance for HIV-exposed infants is needed to refine guidelines. Additionally, dried blood PCR testing takes time and money [70]; thus, cotrimoxazole treatment often begins before HIV status is known. Furthermore, HIV-exposed infants who are HIV-negative at birth may contract HIV during breastfeeding [71]. These factors must also be considered in WHO recommendations, and they highlight urgency to develop rapid and accessible technologies to determine HIV infection status.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. E. M.-G. and A. C. conceived the clinical study design. A. W. D., G. D., I. T., and B. B. conceived the metagenomic study design. A. W. D., under advisement of G. D., conducted the analysis and drafted the manuscript. E. M.-G. led sample collection and tabulated clinical metadata with help from B. D., and B. B., B. W., and X. S. extracted and sequenced the stool samples. T. K. helped with data processing and analysis. All authors critically reviewed the manuscript and provided feedback.
Acknowledgments. The authors thank the clinics for the allocated clinic space and their partnership in taking care of these mother–child pairs. The authors acknowledge all the mothers, their children, and the study team for their commitment to this investigation. The authors also thank members of the Dantas laboratory for helpful discussions of the results, and the Edison Family Center for Genome Sciences and Systems Biology staff, Eric Martin, Brian Koebbe, and Jessica Hoisington-López for technical support and sequencing expertise.
Disclaimer. The findings and conclusions of this study are those of the authors and do not reflect official positions of the funding agencies.
Financial support. This work is supported in part by awards to G. D. from the National Institute of General Medical Sciences (grant number R01 GM099538), the National Institute of Allergy and Infectious Diseases (grant number R01 AI123394), and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant number R01 HD092414) of the National Institutes of Health; the Edward Mallinckrodt Jr Foundation (Scholar Award); and the Bill & Melinda Gates Foundation Grand Challenges (GCE phase I award number OPP1161117). A. W. D. received support from the Institutional Program Unifying Population and Laboratory-Based Sciences Burroughs Wellcome Fund grant to Washington University. E. M.-G. received support from the International Atomic Energy Agency international training fellowship and the University of KwaZulu-Natal postdoctoral scholarship.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Slogrove A, Reikie B, Naidoo S, et al. . HIV-exposed uninfected infants are at increased risk for severe infections in the first year of life. J Trop Pediatr 2012; 58:505–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Slogrove AL, Goetghebuer T, Cotton MF, Singer J, Bettinger JA. Pattern of infectious morbidity in HIV-exposed uninfected infants and children. Front Immunol 2016; 7:164. doi:10.3389/fimmu.2016.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adler C, Haelterman E, Barlow P, Marchant A, Levy J, Goetghebuer T. Severe infections in HIV-exposed uninfected infants born in a European country. PLoS One 2015; 10:e0135375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thea DM, St Louis ME, Atido U, et al. . A prospective study of diarrhea and HIV-1 infection among 429 Zairian infants. N Engl J Med 1993; 329:1696–702. [DOI] [PubMed] [Google Scholar]
- 5. Newell ML, Coovadia H, Cortina-Borja M, Rollins N, Gaillard P, Dabis F; Ghent International AIDS Society (IAS) Working Group on HIV Infection in Women and Children Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet 2004; 364:1236–43. [DOI] [PubMed] [Google Scholar]
- 6. Brahmbhatt H, Bishai D, Wabwire-Mangen F, Kigozi G, Wawer M, Gray RH; Rakai Project Group Polygyny, maternal HIV status and child survival: Rakai, Uganda. Soc Sci Med 2002; 55:585–92. [DOI] [PubMed] [Google Scholar]
- 7. Newell ML, Brahmbhatt H, Ghys PD. Child mortality and HIV infection in Africa: a review. AIDS 2004; 18(Suppl 2):S27–34. [DOI] [PubMed] [Google Scholar]
- 8. Shapiro RL, Lockman S, Kim S, et al. . Infant morbidity, mortality, and breast milk immunologic profiles among breast-feeding HIV-infected and HIV-uninfected women in Botswana. J Infect Dis 2007; 196:562–9. [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization. Guidelines on post-exposure prophylaxis for HIV and the use of cotrimoxazole prophylaxis for HIV-related infections among adults, adolescents and children: recommendations for a public health approach: December 2014 supplement to the 2013 consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. 2014. Available at: https://www.ncbi.nlm.nih.gov/books/NBK298965/. Accessed 25 February 2019. [PubMed] [Google Scholar]
- 10. Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 2001; 32:1608–14. [DOI] [PubMed] [Google Scholar]
- 11. Bwakura-Dangarembizi M, Kendall L, Bakeera-Kitaka S, et al. . A randomized trial of prolonged co-trimoxazole in HIV-infected children in Africa. N Engl J Med 2014; 370:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bourke CD, Gough EK, Pimundu G, et al. . Cotrimoxazole reduces systemic inflammation in HIV infection by altering the gut microbiome and immune activation. Sci Transl Med 2019; 11. doi:10.1126/scitranslmed.aav0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lockman S, Hughes M, Powis K, et al. . Effect of co-trimoxazole on mortality in HIV-exposed but uninfected children in Botswana (the Mpepu Study): a double-blind, randomised, placebo-controlled trial. Lancet Glob Health 2017; 5:e491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis NL, Barnett EJ, Miller WC, et al. . Impact of daily cotrimoxazole on clinical malaria and asymptomatic parasitemias in HIV-exposed, uninfected infants. Clin Infect Dis 2015; 61:368–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis NL, Wiener J, Juliano JJ, et al. . Breastfeeding, Antiretrovirals and Nutrition (BAN) Study Team; Breastfeeding, Antiretrovirals and Nutrition (BAN) Study Team Co-trimoxazole prophylaxis, asymptomatic malaria parasitemia, and infectious morbidity in human immunodeficiency virus-exposed, uninfected infants in Malawi: the BAN study. Clin Infect Dis 2017; 65:575–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization. Guiding principles for complementary feeding of the breastfed child. Geneva, Switzerland: WHO, 2003. [Google Scholar]
- 17. Yatsunenko T, Rey FE, Manary MJ, et al. . Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015; 17:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robertson RC, Manges AR, Finlay BB, Prendergast AJ. The human microbiome and child growth—first 1000 days and beyond. Trends Microbiol 2019; 27:131–47. [DOI] [PubMed] [Google Scholar]
- 20. Cai X, Wardlaw T, Brown DW. Global trends in exclusive breastfeeding. Int Breastfeed J 2012; 7:12. doi:10.1186/1746-4358-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gibson MK, Wang B, Ahmadi S, et al. . Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol 2016; 1:16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science 2016; 352:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nobel YR, Cox LM, Kirigin FF, et al. . Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 2015; 6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Korpela K, Salonen A, Virta LJ, et al. . Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016; 7:10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korpela K, Salonen A, Virta LJ, Kekkonen RA, de Vos WM. Association of early-life antibiotic use and protective effects of breastfeeding: role of the intestinal microbiota. JAMA Pediatr 2016; 170:750–7. [DOI] [PubMed] [Google Scholar]
- 26. Coutsoudis A, Daniels B, Moodley-Govender E, et al. . Randomised controlled trial testing the effect of cotrimoxazole prophylaxis on morbidity and mortality outcomes in breastfed HIV-exposed uninfected infants: study protocol. BMJ Open 2016; 6:e010656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dan M, Richardson J, Miliotis MD, Koornhof HJ. Comparison of preservation media and freezing conditions for storage of specimens of faeces. J Med Microbiol 1989; 28:151–4. [DOI] [PubMed] [Google Scholar]
- 28. Agapova S, Stephenson K, Manary M, et al. . Detection of low-concentration host mRNA transcripts in Malawian children at risk for environmental enteropathy. J Pediatr Gastroenterol Nutr 2013; 56:66–71. [DOI] [PubMed] [Google Scholar]
- 29. Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 2015; 10:e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Truong DT, Franzosa EA, Tickle TL, et al. . MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 2015; 12:902–3. [DOI] [PubMed] [Google Scholar]
- 32. Franzosa EA, McIver LJ, Rahnavard G, et al. . Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 2018; 15:962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol 2015; 11:e1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia B, Raphenya AR, Alcock B, et al. . CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017; 45:D566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH; UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015; 31:926–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. [Google Scholar]
- 37. Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag, 2016. [Google Scholar]
- 38. Kassambara A. ggpubr: ‘ggplot2’ based publication ready plots. 2018. Available at: https://cran.r-project.org/package=ggpubr [Google Scholar]
- 39. Wilke CO. cowplot: streamlined plot theme and plot annotations for ‘ggplot2’. 2019. Available at: https://cran.r-project.org/package=cowplot [Google Scholar]
- 40. Oksanen J, Blanchet FG, Friendly M, et al. . vegan: community ecology package. 2019. Available at: https://cran.r-project.org/package=vegan [Google Scholar]
- 41. Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 2003; 84:511–25. [Google Scholar]
- 42. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015; 67:1–48. [Google Scholar]
- 43. Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nat Methods 2019; 16:565–6. [DOI] [PubMed] [Google Scholar]
- 44. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995; 57:289–300. [Google Scholar]
- 45. Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 1957; 27:325–49. [Google Scholar]
- 46. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med 2016; 22:713–22. [DOI] [PubMed] [Google Scholar]
- 47. Bender JM, Li F, Martelly S, et al. . Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med 2016; 8:349ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Claassen-Weitz S, Gardner-Lubbe S, Nicol P, et al. . HIV-exposure, early life feeding practices and delivery mode impacts on faecal bacterial profiles in a South African birth cohort. Sci Rep 2018; 8:5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 2007; 1:56–66. [DOI] [PubMed] [Google Scholar]
- 50. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A 2011; 108(Suppl 1):4554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal microbiota. Gut Microbes 2010; 1:279–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. National Center for Biotechnology Information. PubChem database. Cotrimoxazole, CID 358641. Bethesda, MD: NCBI, 2019. [Google Scholar]
- 54. Monaco CL, Gootenberg DB, Zhao G, et al. . Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 2016; 19:311–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oldenburg CE, Sie A, Coulibaly B, et al. . Effect of commonly used pediatric antibiotics on gut microbial diversity in preschool children in Burkina Faso: a randomized clinical trial. Open Forum Infect Dis 2018; 5:ofy289. doi:10.1093/ofid/ofy289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams PCM, Isaacs D, Berkley JA. Antimicrobial resistance among children in sub-Saharan Africa. Lancet Infect Dis 2018; 18:e33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tadesse BT, Ashley EA, Ongarello S, et al. . Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis 2017; 17:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Klein EY, Tseng KK, Pant S, Laxminarayan R. Tracking global trends in the effectiveness of antibiotic therapy using the drug resistance index. BMJ Global Health 2019; 4:e001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Powis KM, Souda S, Lockman S, et al. . Cotrimoxazole prophylaxis was associated with enteric commensal bacterial resistance among HIV-exposed infants in a randomized controlled trial, Botswana. J Int AIDS Soc 2017; 20. doi:10.1002/jia2.25021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Franzosa EA, Morgan XC, Segata N, et al. . Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 2014; 111:E2329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A 2011; 108:14288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van der Horst C, Chasela C, Ahmed Y, et al. . Breastfeeding, Antiretroviral, and Nutrition Study Team Modifications of a large HIV prevention clinical trial to fit changing realities: a case study of the breastfeeding, antiretroviral, and nutrition (BAN) protocol in Lilongwe, Malawi. Contemp Clin Trials 2009; 30:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pärnänen K, Karkman A, Hultman J, et al. . Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun 2018; 9:3891. doi:10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Baumann-Dudenhoeffer AM, D’Souza AW, Tarr PI, Warner BB, Dantas G. Infant diet and maternal gestational weight gain predict early metabolic maturation of gut microbiomes. Nat Med 2018; 24:1822–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fouhy F, Guinane CM, Hussey S, et al. . High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 2012; 56:5811–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ajslev TA, Andersen CS, Gamborg M, Sørensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond) 2011; 35:522–9. [DOI] [PubMed] [Google Scholar]
- 68. Koenig JE, Spor A, Scalfone N, et al. . Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011; 108(Suppl 1):4578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wei S, Mortensen MS, Stokholm J, et al. . Short- and long-term impacts of azithromycin treatment on the gut microbiota in children: a double-blind, randomized, placebo-controlled trial. EBioMedicine 2018; 38:265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cassol S, Salas T, Arella M, Neumann P, Schechter MT, O’Shaughnessy M. Use of dried blood spot specimens in the detection of human immunodeficiency virus type 1 by the polymerase chain reaction. J Clin Microbiol 1991; 29:667–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miotti PG, Taha TE, Kumwenda NI, et al. . HIV transmission through breastfeeding: a study in Malawi. JAMA 1999; 282:744–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Shotgun metagenomic reads (quality-checked and filtered for human reads) generated for this study were uploaded to the National Center for Biotechnology Information (NCBI) under the project code PRJNA549787 (see BioSample accession metadata file for individual sample accession codes). Metadata files for these shotgun metagenomic reads are also included in this submission: (1) SACTX_HEU_Infant_metadata.txt; (2) SACTX_DOL_metadata_mod.txt; (3) SACTX_HEU_Infant_bodyMeasurements.txt; (4) SACTX_HEU_Infant_Bloodwork.txt; (5) NCBI_BioSample_AccessionList.txt.