Abstract
Mitochondrial-derived peptides (MDPs) are a novel class of bioactive microproteins that modify cell metabolism. The the eight MDPs that been characterized (e.g., humanin, MOTS-c, SHLPs1-6) attenuate disease pathology including Alzheimer’s disease, prostate cancer, macular degeneration, cardiovascular disease, and diabetes. The association between disease and human genetic variation in MDPs is underexplored, although two polymorphisms in humanin and MOTS-c associate with cognitive decline and diabetes, respectively, suggesting a precise role for MDPs in disease-modification. There could be hundreds of additional MDPs that have yet to be discovered. Altogether, MDPs could explain unanswered biological and metabolic questions and are part of a growing field of novel microproteins encoded by small open reading frames. In this review, the current state of MDPs are summarized with an emphasis on biological and therapeutic implications.
Keywords: Mitochondrial-derived peptides, Microproteins, Small open reading frames
1. Discovery of mitochondrial-derived peptides
In order to reduce the high number of non-coding open reading frames (ORFs), ORFs greater than approximately 100 codons were exclusively used to annotate the human genome, as the likelihood that an ORF encodes a bona fide protein increases with its codon length [1]. However, high-throughput DNA sequencing, enrichment-based mass spectrometry, and immunological peptide detection have revealed over a thousand bioactive eukaryotic microproteins encoded by nuclear small open reading frames (sORFs) and several microproteins encoded by mitochondrial sORFs [2]. Moreover, there are millions of uncharacterized putative nuclear-encoded and hundreds of mitochondrial sORFs, but the detection and biology of these microproteins have yet to be corroborated [3]. Novel microproteins could answer crucial biological questions (e.g., mitochondrial-nuclear communication, metabolic dysfunction, etc.) that have mystified the field for decades. Microproteins serve as therapeutic candidates both in the form of the microprotein itself or sORF gene target. In particular, mitochondrial microproteins – originally termed mitochondrial-derived peptides (MDPs) – have could explain how the mitochondrion communicates intracellularly and intercellularly in disease-specific contexts.
1.1. Humanin
The first MDP discovered was from a sORF encoded within the mitochondrial 16S rRNA and named humanin [4]. The detection of the 24-amino acid MDP humanin represented a paradigm shift in mitochondria biology and genetics because it was the first mitochondrial sORF-encoded microprotein found to have biological activity. Humanin was discovered by three independent laboratories in the early 2000s. First, Hashimoto et al. found humanin during a screen for genes that protected against amyloid beta (Aβ) toxicity [4,5]. These experiments revealed a cDNA fragment that mapped back to the mitochondrial 16S rRNA. Ultimately, this mitochondrial-derived cDNA was named humanin because it was found to encode for a microprotein that displayed profound protection against AD-related neurotoxicity, an effect that the original authors though potentially could restore the “humanity” of patients suffering from dementia. Second, Ikonen et al. found that humanin bound IGFBP3 using an unbiased yeast two hybridization system and enhanced the protective effects of IGFBP3 against Aβ toxicity [6]. Third, Guo et al. revealed humanin bounded the apoptotic protein BAX and mitigated cell apoptosis [7]. Since the discovery of humanin, hundreds of additional peer-reviewed reports have described humanin in the context of general biology, AD pathology attenuation, and chronic disease modification [8–21]. Humanin’s precise mechanism of action includes signaling through a trimeric receptor consisting of WSX-1, GP130 and CNTFR as well as a separate FRPL1 receptor [22,23] (Fig. 1). Downstream of humanin receptor activation are STAT3 and ERK1/2, which modified mitochondrial biology, cell proliferation, and cell survival [24]. In essence, humanin helped initiate the novel field of mitochondrial sORF-encoded microproteins (see Fig. 2).
Fig. 1. Humanin signaling cascade.
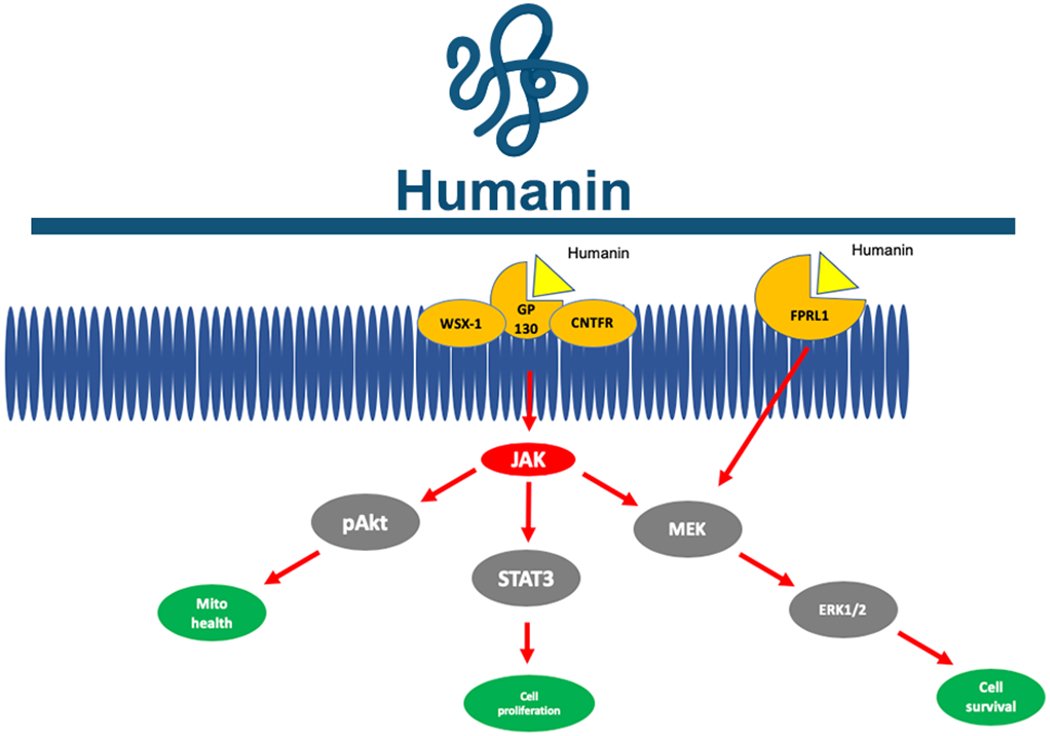
Humanin binds the trimeric WSX-1, GP130, and CNTFR receptor and the FPRL1 receptor to initiate intracellular cascades leading to mitochondrial maintenance and cell survival.
Fig. 2. Biological effects of MOTS-c.
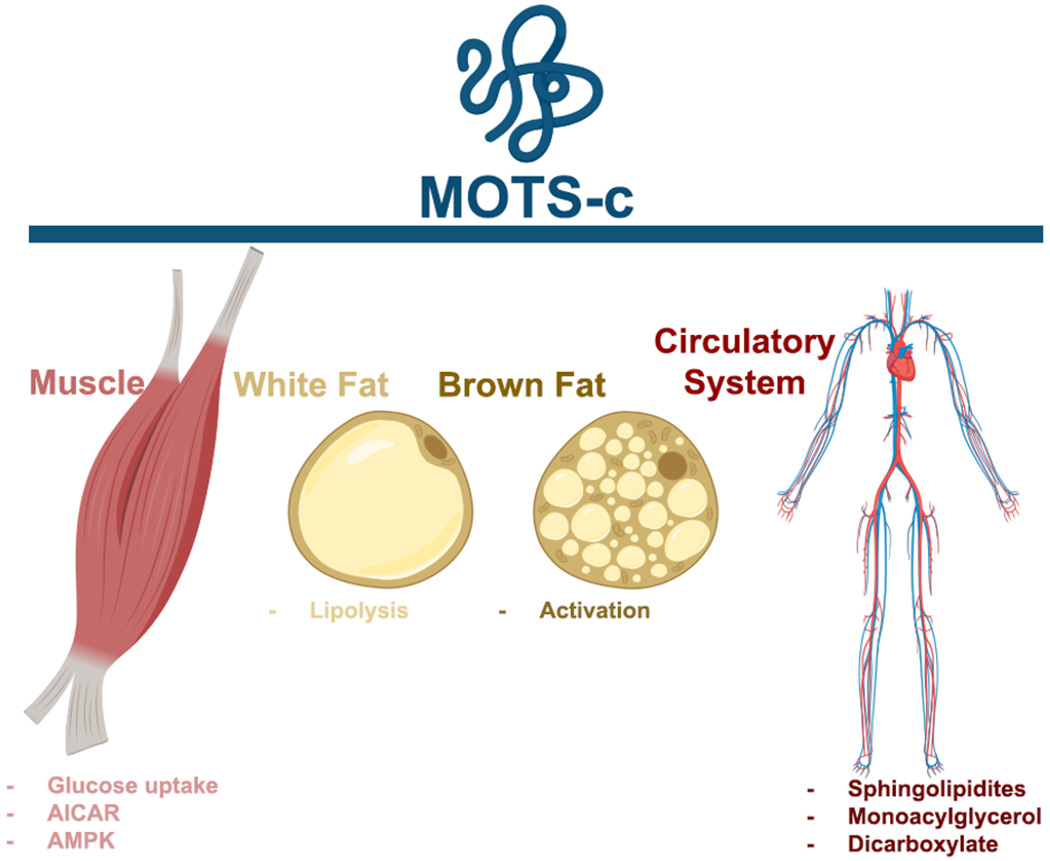
MOTS-c has been shown to act on muscle, white fat, brown fat, and circulate in the periphery.
1.2. MOTS-c and SHLPs
In addition to humanin, seven more MDPs have been characterized: MOTS-c and six humanin-like peptides (SHLPs 1–6). MOTS-c was the second MDP discovered and is a 51 bp sORF within the 12S rRNA that is highly conserved and translated in the cytosol [25]. The effects of the 16 amino-acid MOTS-c are vast but mediated through AMPK activity. MOTS-c has been reported to act profoundly on mouse skeletal muscle and has also been detected in skeletal muscle [26]. Administering MOTS-c to mice on a high-fat diet improved insulin resistance, attenuated high-fat diet induced weight gain, and minimized fat accumulation in the liver [27]. Lu et al. demonstrated that MOTS-c enhanced lipid catabolism in white adipose tissue, activated brown adipose tissue, and prevented ovariectomy-induced obesity and insulin resistance through AMPK [28]. An unbiased metabolomics approach demonstrated that MOTS-c administration to high-fat diet fed mice regulated sphingolipid metabolism, monoacylglycerol metabolism, and dicarboxylate metabolism [29]. MOTS-c has been described as an “exercise mimetic” that systematically regulates glucose and insulin action [25].
SHLPs are encoded by sORFs within the 16S rRNA and have many similar effects as humanin [9]. Five of the six SHLPs (SHLPs1-4 and 6) that were initially reported by Cobb et al. were detected at varying levels across tissues, and detection of these MDPs was validated by comparing levels in cells void of mitochondrial DNA (i.e., ρ0 cells) that produced no detectable SHLPs. The tissue specificity of certain SHLPs suggest precise mechanisms of action. Of the six SHLPs Cobb et al. characterized, SHLP2, SHLP3, and SHLP6 had the most profound effects on cellular biology. Specifically, SHLP2 and SHLP3 increased mitochondrial oxygen consumption rate and ATP while reducing reactive oxygen species. In the same report, SHLP2 protected cells against nervous system insults such as Aβ toxicity and macular degeneration. The original authors found SHLP2 in mouse and also in prostate at lower levels, whereas SHLP4 was detected in prostate at high levels. Although SHLP2 levels are lower in prostate tissue, follow-up studies on SHLP2 levels have predicted prostate cancer, an observation that hints at potential communication among MDPs such as SHLPs [30]. SHLPs have been characterized in a cytoprotective context, but SHLP6 has been reported to primarily induce apoptosis and is found in much lower quantity across all tissues [9]. The field is just beginning to understand the tissue specificity of MDPs.
1.3. Human genetic variation in MDPs
The degree of human genetic variation in MDPs is unclear. Most large-scale genome-wide association study (GWAS) analytic plans do not include the mitochondrial genome due to its haploid nature. If mitochondrial DNA variants are included in the GWAS analytic plan, potentially meaningful variants fail to be detected due to the conservative false-discovery Bonferroni correction typically used for nuclear DNA variants. Nevertheless, there have been reports on the associative effects of mitochondrial genetic variation on disease, although few interpret findings with careful consideration to the mitochondrial sORFome [31–36].
Two studies have examined human MDP genetic variation in large-scale population cohorts. First, Yen et al. found that the mitochondrial SNP rs2854128 associated with lower circulating humanin levels and cognitive age in the Health and Retirement Study (n = ~16,000) [37]. Specifically, humanin levels were 15% lower in reference allele carriers and especially low in those of African-American ancestry. rs2854128 represented the first known SNP to associate with a circulating, detectable MDP. Second, the mitochondrial SNP rs111033358 was found to increase risk of type 2 diabetes (T2D) in males of Asian ancestry in a comprehensive meta-analysis of over 27,000 individuals enrolled in the J-MICC, MEC, and TMM cohorts [38]. rslll033358 changes the 14th amino acid of MOTS-c from lysine to glutamine (K14Q), and administration of the mutant MOTS-c K14Q peptide failed to attenuate weight gain and improve insulin sensitivity compared to MOTS-c wild type in vivo [38]. Since human mitochondrial genetic variation varies substantially by ethnicity, it is plausible that ethnic-specific mitochondrial DNA mutations affect MDPs and, as a result, modify disease risk. For instance, mitochondrial genetic variation, captured by data reduction techniques, predicted height within and between self-reported ethnicities in the Health and Retirement study [39].
2. Classification of the putative mitochondrial sorfome
The 16,569 bp mitochondrial genome encodes for 13 large proteins involved in oxidative phosphorylation: ATP6, ATP8, CO1, CO2, CO3, CYB, ND1, ND2, ND3, ND4L, ND4, ND5, and ND6 [40]. Re-annotating the mitochondrial genome to include sORFs between 9 and 40 amino acids revealed hundreds putative MDP sORFs that can be separated into four classification categories based on the strand of origin (heavy or light) and genetic code (i.e., standard or mitochondrial-specific genetic code). There are several differences between the mitochondrial-specific genetic code and standard genetic code. The standard genetic code uses AGA and AGG codons for arginine, whereas the mitochondrial genetic code uses these codons for termination [41]. The UGA codon is used for termination in the standard genetic code, but the mitochondrial genetic code uses UGA for tryptophan [42]. Mitochondrial-specific genetic code also uses two alternative initiation codons (ATA and ATT) in addition to the standard initiation codon of ATG [43].
The largest class of MDP sORFs is derived from the mitochondrial-specific genetic code light-strand class (Fig. 3). The median amino-acid length of the mitochondrial-specific codon light-strand class is 17, and the distribution of these putative MDPs are skewed towards lengths around 10 amino acids. No mitochondrial-specific codon light-strand MDPs have been discovered yet. The mitochondrial-specific heavy-strand class encodes for putative MDPs with a larger median amino-acid length of 20. No MDPs from this class have been identified, either. MDP amino acid length distribution of standard genetic code class is not as positively skewed as the mitochondrial-specific class, and the median amino-acid lengths of the standard genetic code ORFome are 19 and 20 on the heavy and light strand, respectively. As the microprotein field continues to grow, it is possible that MDPs from these classes will be validated.
Fig. 3. Classes of MDPs.

Putative MDPs can be classified into four categories based on genetic code and strand specificity. The distribution graph on the far right illustrates putative MDP length (i.e., number of amino acids).
The eight MDPs that have been discovered are encoded by the standard genetic code class. The MDPs humanin, MOTS-c, and SHLP6 are encoded by the standard genetic code heavy-strand class, and the MDPs SHLP2-5 are encoded by the standard genetic code light-strand class. MOTS-c can only be translated in the cytosol because its sORF has tandem start and stop codons that are specific to the mitochondrial genetic code. The MOTS-c transcript, therefore, must be exported out of the mitochondrion into the cytosol. It has indeed been reported that mitochondrial RNA is exported out of the mitochondrion to the cytosol via unclear mechanisms [44–46]. Of the eight MDPs that have been characterized, humanin, MOTS-c, SHLP2, and SHLP6 have notable associations with several human diseases and lifespan.
3. Associative and experimental effects of MDP levels and diseases
Immunological detection methods for humanin, MOTS-c, SHLP2, and SHLP6 can be used on a variety of biological samples (e.g., blood, cell extracts, and tissue extracts). These techniques (e.g., ELISA, Western blot, etc.) detect low levels of both endogenous and synthetic MDPs and can be easily applied to measure MDP levels in diseased patients, various animal models, cellular experiments, and pharmacokinetic studies [26]. Several reports have quantified MDP levels in biological tissues in cross-sectional human association studies, in vitro experiments, and in vivo paradigms.
3.1. Humanin levels and cardiovascular function, cognition, and lifespan
Humanin has been measured in several biological contexts. Widmer et al. reported that humanin levels were lower in human patients with poor endothelial function versus individuals with healthy endothelial function [47]. Yen et al. observed that cognitive age and ethnicity associated with circulating levels of humanin [37]. Lee et al. observed that humanin positively correlated with lifespan and modified by the GH/IGF-1 axis in mice. In Lee et al. studies, transgenic GH mice displayed nearly 75% less circulating humanin compared to wild type mice. They also showed that humanin levels were over 30% higher in the Ames mouse model (i.e., a mouse model in which GH is deficient). Deleting IGF1 in mice also increased humanin levels over 15% and, conversely, knocking down IGFBP3 reduced humanin by 70% [48].
The precise pharmacokinetics and tissue distribution of humanin and analogues has been characterized using a humanin ELISA by Chen et al. [49]. The half-life of humanin is relatively fast, as about 80% of baseline humanin injected intraperitoneal (IP) is cleared from the circulatory system in 30 min. Notably, in the same report, Chen et al. reported that IGFBP3 knockout mice displayed a remarkably higher half-life of IP-injected humanin. Rather than the 80% of IP humanin clearance rate observed in wild type mice, IGFBP3 knockout only cleared 25% of IP-injected humanin after 30 min. In these wild type mice injected IP with humanin, circulating levels of IGFBP3 dropped by 20% after 2 h, suggesting a tightly regulated relationship between IGFBP3. Moreover, humanin levels were substantially higher (measured by ELISA) in a novel mouse model that overexpresses humanin via humanin sORF transgene [50].
3.2. MOTS-c levels and cardiovascular function and senescence
MOTS-c has been measured in tissues from mice, rats, and humans. Lee et al. reported that circulating MOTS-c levels were similar across species. In humans with coronary endothelial dysfunction, circulating MOTS-c was nearly 20% lower in diseased patients, which was similar to the decrease in circulating humanin in the same diseased patients [25]. In these same patients, improving coronary blood flow and epicardial function led to increased levels of MOTS-c. Levels of MOTS-c can also be detected in cellular extracts by using ELISA [51]. In fact, when cells undergo replicative senescence, MOTS-c levels dropped by ~20% [52]. It is expected that many biological conditions will present differences in MOTS-c levels that will be studied in the future.
3.3. SHLP2 and SHLP6 levels in prostate cancer and senescence
SHLP2 levels have been quantified in young and old male and female wild type mice. For example, all older mice regardless of gender displayed lower circulating SHLP2 levels, and female mice additionally displayed slightly lower levels of SHLP2 [9]. In addition, circulating SHLP2 levels have been used as a biomarker for prostate cancer [30]. That is, SHLP2 levels were nearly halved in prostate cancer patients, and SHLP2 levels were lower in black individuals compared to white individuals without prostate cancer, suggesting an ethnic-specific regulation that needs more follow-up. In the same report, adding SHLP2 levels to a standard predictive model for prostate cancer improved accuracy by nearly 30%. While the utility of the SHLP2 ELISA has been demonstrated, no observable differences in SHLP6 levels have been reported, although the SHLP6 ELISA has been used in senescence contexts [52]. Altogether, humanin, MOTS-c, SHLP2, and SHLP6 levels have potential to explain ethnic health disparities and disease pathology.
4. Conclusions and future research
Technological advancements in DNA sequencing, peptide detection, and computational power will undoubtedly reveal functions of current and soon-to-be discovered microproteins. In regards to MDPs, several unanswered questions will be answered as technology continues to evolve.
4.1. MDP translation
How MDPs are translated remain a crucial question. It is traditionally thought that the small size of the mitochondria encodes for 13 proteins, 22 transfer RNAs, and 2 ribosomal RNAs, but transcriptomics analyses suggests the mitochondria genome is more complex. Hallmark work from Mercer et al. revealed that the mitochondrial transcriptome included potentially 80 canonical and non-canonical cleavage sites [40]. That the mitochondria transcriptome possibly contains 80 cleavage sites is curious, and even more intriguing is the observation that the mitochondria also contains independently regulated small transcripts around the size of 15–30 nucleotides with unclear functions. Perhaps these small mitochondrial RNA fragments behave to modify translation of mitochondrial transcripts, but more work is needed to determine their function. Mercer et al. also reported that mitochondrial DNA contains numerous binding motifs, and RNaseA and PARE data suggest protein footprints are associated with sites of transcript termination and cleavage downstream of the light strand promoter. In addition, whether MDPs are translated in the cytosol or mitochondrion – or in both cytoplasmic and mitochondrial ribosomes – remains unclear. The ability to sequence nucleotides embedded within ribosomes might reveal the main site of translation. Mitochondrial RNA processing is complex and future work will elucidate the precise mechanisms of MDP translation.
4.2. Complete MDP human genetic variation
The degree of human mitochondrial genetic variation within mitochondrial sORFs and its associated effects on disease are still dramatically underexplored due to several reasons. One limitation of mitochondrial genome wide association studies (MiWAS) is the varying number of mitochondrial single nucleotide polymorphisms (SNPs) across sequencing chips. Large population genotyping data that used the Illumina HumanHap550 Genotyping BeadChip and Affymetrix Genome-Wide SNP Array 6.0 do not even contain mitochondrial SNPs. It is estimated that only 64 SNPs are necessary to tag mitochondrial SNPs with a frequency greater than 1% in European populations with an r-squared value of 0.8 [53]. In the Health and Retirement Study, Miller et al. showed that genetic principal components derived from nuclear or mitochondrial genotyping arrays explained a similar amount of the variance in Whites, Blacks, and Hispanics [39]. While a limited number of SNPs are necessary to tag certain populations confidently, the ideal analytic plan includes whole mitochondrial DNA sequencing, which will reveal rare mitochondrial DNA variants and the degree of heteroplasmy in certain mitochondrial genetic regions that contain sORFs. Large human population cohorts are beginning to conduct whole genome sequencing. These data sets will reflect comprehensive mitochondrial genetic architecture that haplogroup or mtSNP tagging analyses attempt to recapitulate. Whole mitochondrial DNA data could unravel the effects of certain mitochondrial genetic variants, many of which might change the function of MDPs.
4.3. Nuclear-encoded sORF microproteins that act on mitochondria
It is expected that ongoing research will reveal more microproteins not only derived from mitochondrial DNA but also nuclear DNA. For instance, Chu et al. characterized a novel microprotein derived from nuclear DNA, called PIGBOS, that localized to the mitochondrion and regulated uncoupling protein responses [54]. The same laboratory found a separate microprotein called MIEF1 that regulated mitochondrial translation [55]. Moreover, three independent, separate laboratories discovered the same microprotein encoded by a lncRNA that has been called mitoregulin, MPM, and MOXI. This novel microprotein supported super complexes and modified mitochondrial respiratory efficiency [56]. Separate reports noted that MOXI (mitoregulin or MPM) modified muscle and fat metabolism by acting on the mitochondria [57,58]. Since these newly discovered microproteins act on the mitochondria, future studies might focus on the interaction between nuclear-encoded microproteins and MDPs (e.g., effect of MIEF1 on MDP expression). Vice versa, how MDPs regulate nuclear-encoded microproteins, such as the mitochondrial MIEF1, could explain unclear biological processes. MOTS-c, for example, has been shown to regulate expression of NRF2 target genes by translocating to the nucleus [59]. Although NRF2 is not a microprotein, that MOTS-c regulates target nuclear genes suggests that MOTS-c and perhaps additional MDPs have the capacity to regulate nuclear-encoded genes. Whether MDPs modify expression of nuclear-encoded peptides is unclear, but future analyses might consider the effect of MDPs on nuclear-encoded peptides via RNASeq, ribosome profiling, and peptidomics.
4.4. Future directions
While technological improvements at the nucleotide and protein level have reveled several bioactive microproteins, there is still room for improvement. In particular, enhancing mass-spec enrichment analysis could yield several more MDPs and complement MDP ELISAs. Additionally, the advent of long-read sequencing could provide greater depth into MDP transcripts. Finally, the degree to which MDP genetic variation associates with human disease could become clear with whole mitochondrial genome sequencing data derived from large-scale human populations. Over the last two decades, eight MDPs have been characterized and their biology will continue to be explored, but it is expected that additional MDPs will have biological and therapeutic relevance.
Acknowledgments
Funding
This research was supported by NIH grants to Pinchas Cohen (R01AG061834, R56AG062693, P01AG034906, R01EY027363, U54CA233465) and NIA T32 AG000037 to Brendan Miller.
Footnotes
Declaration of competing interest
Pinchas Cohen is a consultant and stockholder of CohBar Inc.
References
- [1].Basrai MA, Hieter P, Boeke JD, Small open reading frames: beautiful needles in the haystack, Genome Res. 7 (1997) 768–771. [DOI] [PubMed] [Google Scholar]
- [2].Saghatelian A, Couso JP, Discovery and characterization of smORF-encoded bioactive polypeptides, Nat. Chem. Biol 11 (2015) 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Couso JP, Patraquim P, Classification and function of small open reading frames, Nat. Rev. Mol. Cell Biol 18 (2017) 575–589. [DOI] [PubMed] [Google Scholar]
- [4].Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y, Kita Y, Kawasumi M, Kouyama K, Doyu M, Sobue G, Koide T, Tsuji S, Lang J, Kurokawa K, Nishimoto I, A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hashimoto Y, Ito Y, Niikura T, Shao Z, Hata M, Oyama F, Nishimoto I, Mechanisms of neuroprotection by a novel rescue factor humanin from Swedish mutant amyloid precursor protein, Biochem. Biophys. Res. Commun 283 (2001) 460–468. [DOI] [PubMed] [Google Scholar]
- [6].Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T, Nishimoto I, Cohen P, Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC, Humanin peptide suppresses apoptosis by interfering with Bax activation, Nature 423 (2003) 456–461. [DOI] [PubMed] [Google Scholar]
- [8].Bachar AR, Scheffer L, Schroeder AS, Nakamura HK, Cobb LJ, Oh YK, Lerman LO, Pagano RE, Cohen P, Lerman A, Humanin is expressed in human vascular walls and has a cytoprotective effect against oxidized LDL-induced oxidative stress, Cardiovasc. Res 88 (2010) 360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, Cohen P, Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers, Aging 8 (2016) 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gong Z, Tas E, Muzumdar R, Humanin and age-related diseases: a new link? Front. Endocrinol 5 (2014) 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Muzumdar RH, Huffman DM, Atzmon G, Buettner C, Cobb LJ, Fishman S, Budagov T, Cui L, Einstein FH, Poduval A, Hwang D, Barzilai N, Cohen P, Humanin: a novel central regulator of peripheral insulin action, PloS One 4 (2009) e6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Muzumdar RH, Huffman DM, Calvert JW, Jha S, Weinberg Y, Cui L, Nemkal A, Atzmon G, Klein L, Gundewar S, Ji SY, Lavu M, Predmore BL, Lefer DJ, Acute humanin therapy attenuates myocardial ischemia and reperfusion injury in mice, Arterioscler. Thromb. Vasc. Biol 30 (2010) 1940–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yen K, Lee C, Mehta H, Cohen P, The emerging role of the mitochondrial-derived peptide humanin in stress resistance, J. Mol. Endocrinol 50 (2013) R11–R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yen K, Wan J, Mehta HH, Miller B, Christensen A, Levine ME, Salomon MP, Brandhorst S, Xiao J, Kim S-J, Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans, Sci. Rep 8 (2018) 14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yuan L, Liu XJ, Han WN, Li QS, Wang ZJ, Wu MN, Yang W, Qi JS, [Gly 14]-Humanin protects against amyloid beta peptide-induced impairment of spatial learning and memory in rats, Neurosci Bull 32 (2016) 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang W, Du Y, Bai M, Xi Y, Li Z, Miao J, S14G-humanin inhibits Abeta 1–42 fibril formation, disaggregates preformed fibrils, and protects against Abeta-induced cytotoxicity in vitro, J. Pept. Sci 19 (2013) 159–165. [DOI] [PubMed] [Google Scholar]
- [17].Zhang W, Zhang W, Li Z, Hao J, Zhang Z, Liu L, Mao N, Miao J, Zhang L, S14G-humanin improves cognitive deficits and reduces amyloid pathology in the middle-aged APPswe/PS1dE9 mice, Pharmacol. Biochem. Behav 100 (2012) 361–369. [DOI] [PubMed] [Google Scholar]
- [18].Zhang X, Urbieta-Caceres VH, Eirin A, Bell CC, Crane JA, Tang H, Jordan KL, Oh YK, Zhu XY, Korsmo MJ, Bachar AR, Cohen P, Lerman A, Lerman LO, Humanin prevents intra-renal microvascular remodeling and inflammation in hypercholesterolemic ApoE deficient mice, Life Sci. 91 (2012) 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mehta HH, Xiao J, Ramirez R, Miller B, Kim SJ, Cohen P, Yen K, Metabolomic profile of diet-induced obesity mice in response to humanin and small humanin-like peptide 2 treatment, Metabolomics 15 (2019) 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Oh YK, Bachar AR, Zacharias DG, Kim SG, Wan J, Cobb LJ, Lerman LO, Cohen P, Lerman A, Humanin preserves endothelial function and prevents atherosclerotic plaque progression in hypercholesterolemic ApoE deficient mice, Atherosclerosis 219 (2011) 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sreekumar PG, Ishikawa K, Spee C, Mehta HH, Wan J, Yen K, Cohen P, Kannan R, Hinton DR, The mitochondrial-derived peptide humanin protects RPE cells from oxidative stress, senescence, and mitochondrial dysfunction, Invest. Ophthalmol. Vis. Sci 57 (2016) 1238–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hashimoto Y, Kurita M, Aiso S, Nishimoto I, Matsuoka M, Humanin inhibits neuronal cell death by interacting with a cytokine receptor complex or complexes involving CNTF receptor alpha/WSX-1/gp130, Mol. Biol. Cell 20 (2009) 2864–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hashimoto Y, Kurita M, Matsuoka M, Identification of soluble WSX-1 not as a dominant-negative but as an alternative functional subunit of a receptor for an anti-Alzheimer’s disease rescue factor Humanin, Biochem. Biophys. Res. Commun 389 (2009) 95–99. [DOI] [PubMed] [Google Scholar]
- [24].Kim SJ, Guerrero N, Wassef G, Xiao J, Mehta HH, Cohen P, Yen K, The mitochondrial-derived peptide humanin activates the ERK1/2, AKT, and STAT3 signaling pathways and has age-dependent signaling differences in the hippocampus, Oncotarget 7 (2016) 46899–46912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J, Kim SJ, Mehta H, Hevener AL, de Cabo R, Cohen P, The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance, Cell Metabol. 21 (2015) 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kim SJ, Xiao J, Wan J, Cohen P, Yen K, Mitochondrially derived peptides as novel regulators of metabolism, J. Physiol 595 (2017) 6613–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee C, Kim KH, Cohen P, MOTS-c: a novel mitochondrial-derived peptide regulating muscle and fat metabolism, Free Radic. Biol. Med 100 (2016) 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lu H, Wei M, Zhai Y, Li Q, Ye Z, Wang L, Luo W, Chen J, Lu Z, MOTS-c peptide regulates adipose homeostasis to prevent ovariectomy-induced metabolic dysfunction, J. Mol. Med. (Berl.) 97 (2019) 473–485. [DOI] [PubMed] [Google Scholar]
- [29].Kim SJ, Miller B, Mehta HH, Xiao J, Wan J, Arpawong TE, Yen K, Cohen P, The mitochondrial-derived peptide MOTS-c is a regulator of plasma metabolites and enhances insulin sensitivity, Phys. Rep 7 (2019) e14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xiao J, Howard L, Wan J, Wiggins E, Vidal A, Cohen P, Freedland SJ, Low circulating levels of the mitochondrial-peptide hormone SHLP2: novel biomarker for prostate cancer risk, Oncotarget 8 (2017) 94900–94909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hudson G, Nalls M, Evans JR, Breen DP, Winder-Rhodes S, Morrison KE, Morris HR, Williams-Gray CH, Barker RA, Singleton AB, Hardy J, Wood NE, Burn DJ, Chinnery PF, Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease, Neurology 80 (2013) 2042–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kraja AT, Liu C, Fetterman JL, Graff M, Have CT, Gu C, Yanek LR, Feitosa MF, Arking DE, Chasman DI, Young K, Ligthart S, Hill WD, Weiss S, Luan J, Giulianini F, Li-Gao R, Hartwig FP, Lin SJ, Wang L, Richardson TG, Yao J, Fernandez EP, Ghanbari M, Wojczynski MK, Lee WJ, Argos M, Armasu SM, Barve RA, Ryan KA, An P, Baranski TJ, Bielinski SJ, Bowden DW, Broeckel U, Christensen K, Chu AY, Corley J, Cox SR, Uitterlinden AG, Rivadeneira F, Cropp CD, Daw EW, van Heemst D, de Las Fuentes L, Gao H, Tzoulaki I, Ahluwalia TS, de Mutsert R, Emery LS, Erzurumluoglu AM, Perry JA, Fu M, Forouhi NG, Gu Z, Hai Y, Harris SE, Hemani G, Hunt SC, Irvin MR, Jonsson AE, Justice AE, Kerrison ND, Larson NB, Lin KH, Love-Gregory LD, Mathias RA, Lee JH, Nauck M, Noordam R, Ong KK, Pankow J, Patki A, Pattie A, Petersmann A, Qi Q, Ribel-Madsen R, Rohde R, Sandow K, Schnurr TM, Sofer T, Starr JM, Taylor AM, Teumer A, Timpson NJ, de Haan HG, Wang Y, Weeke PE, Williams C, Wu H, Yang W, Zeng D, Witte DR, Weir BS, Wareham NJ, Vestergaard H, Turner ST, Torp-Pedersen C, Stergiakouli E, Sheu WH, Rosendaal FR, Ikram MA, Franco OH, Ridker PM, Perls TT, Pedersen O, Nohr EA, Newman AB, Linneberg A, Langenberg C, Kilpelainen TO, Kardia SLR, Jorgensen ME, Jorgensen T, Sorensen TIA, Homuth G, Hansen T, Goodarzi MO, Deary IJ, Christensen C, Chen YI, Chakravarti A, Brandslund I, Bonnelykke K, Taylor KD, Wilson JG, Rodriguez S, Davies G, Horta BL, Thyagarajan B, Rao DC, Grarup N, Davila-Roman VG, Hudson G, Guo X, Arnett DK, Hayward C, Vaidya D, Mook-Kanamori DO, Tiwari HK, Levy D, Loos RJF, Dehghan A, Elliott P, Malik AN, Scott RA, Becker DM, de Andrade M, Province MA, Meigs JB, Rotter JI, North KE, Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits, Am. J. Hum. Genet 104 (2019) 112–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu C, Yang Q, Hwang SJ, Sun F, Johnson AD, Shirihai OS, Vasan RS, Levy D, Schwartz F, Association of genetic variation in the mitochondrial genome with blood pressure and metabolic traits, Hypertension 60 (2012) 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tranah GJ, Santaniello A, Caillier SJ, D’Alfonso S, Martinelli Boneschi F, Hauser SL, Oksenberg JR, Mitochondrial DNA sequence variation in multiple sclerosis, Neurology 85 (2015) 325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van Oven M, Kayser M, Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation, Hum. Mutat 30 (2009) E386–E394. [DOI] [PubMed] [Google Scholar]
- [36].Yang TL, Guo Y, Shen H, Lei SF, Liu YJ, Li J, Liu YZ, Yu N, Chen J, Xu T, Cheng Y, Tian Q, Yu P, Papasian CJ, Deng HW, Genetic association study of common mitochondrial variants on body fat mass, PloS One 6 (2011) e21595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yen K, Wan J, Mehta HH, Miller B, Christensen A, Levine ME, Salomon MP, Brandhorst S, Xiao J, Kim SJ, Navarrete G, Campo D, Harry GJ, Longo V, Pike CJ, Mack WJ, Hodis HN, Crimmins EM, Cohen P, Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans, Sci. Rep 8 (2018) 14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zempo H, Kim S-J, Fuku N, Nishida Y, Higaki Y, Wan J, Yen K, Miller B, Vicinanza R, Miyamoto-Mikami E, Kumagai H, Naito H, Xiao J, Mehta HH, Lee C, Hara M, Patel YM, Setiawan VW, Moore TM, Hevener AL, Sutoh Y, Shimizu A, Kojima K, Kinoshita K, Tanaka K, Cohen P, A Pro-diabetogenic mtDNA Polymorphism in the Mitochondrial-Derived Peptide, MOTS-c, bioRxiv, 2019, p. 695585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Miller B, Arpawong TE, Jiao H, Kim SJ, Yen K, Mehta HH, Wan J, Carpten JC, Cohen P, Comparing the utility of mitochondrial and nuclear DNA to adjust for genetic ancestry in association studies, Cells 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS, The human mitochondrial transcriptome, Cell 146 (2011) 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bezerra AR, Guimaraes AR, Santos MA, Non-standard genetic codes define new concepts for protein engineering, Life 5 (2015) 1610–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Swart EC, Serra V, Petroni G, Nowacki M, Genetic codes with No dedicated stop codon: context-dependent translation termination, Cell 166 (2016) 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Osawa S, Ohama T, Jukes TH, Watanabe K, Evolution of the mitochondrial genetic code. I. Origin of AGR serine and stop codons in metazoan mitochondria, J. Mol. Evol 29 (1989) 202–207. [DOI] [PubMed] [Google Scholar]
- [44].Amikura R, Kashikawa M, Nakamura A, Kobayashi S, Presence of mitochondriatype ribosomes outside mitochondria in germ plasm of Drosophila embryos, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 9133–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jady BE, Ketele A, Kiss T, Dynamic association of human mRNP proteins with mitochondrial tRNAs in the cytosol, RNA 24 (2018) 1706–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Maniataki E, Mourelatos Z, Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein, RNA 11 (2005) 849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Widmer RJ, Flammer AJ, Herrmann J, Rodriguez-Porcel M, Wan J, Cohen P, Lerman LO, Lerman A, Circulating humanin levels are associated with preserved coronary endothelial function, Am. J. Physiol. Heart Circ. Physiol 304 (2013) H393–H397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee C, Wan J, Miyazaki B, Fang Y, Guevara-Aguirre J, Yen K, Longo V, Bartke A, Cohen P, IGF-I regulates the age-dependent signaling peptide humanin, Aging Cell 13 (2014) 958–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Chin YP, Keni J, Wan J, Mehta H, Anene F, Jia Y, Lue YH, Swerdloff R, Cobb LJ, Wang C, Cohen P, Pharmacokinetics and tissue distribution of humanin and its analogues in male rodents, Endocrinology 154 (2013) 3739–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zaman F, Zhao Y, Celvin B, Mehta HH, Wan J, Chrysis D, Ohlsson C, Fadeel B, Cohen P, Savendahl L, Humanin is a novel regulator of Hedgehog signaling and prevents glucocorticoid-induced bone growth impairment, Faseb. J 33 (2019) 4962–4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Qin Q, Delrio S, Wan J, Jay Widmer R, Cohen P, Lerman LO, Lerman A, Downregulation of circulating MOTS-c levels in patients with coronary endothelial dysfunction, Int. J. Cardiol 254 (2018) 23–27. [DOI] [PubMed] [Google Scholar]
- [52].Kim SJ, Mehta HH, Wan J, Kuehnemann C, Chen J, Hu JF, Hoffman AR, Cohen P, Mitochondrial peptides modulate mitochondrial function during cellular senescence, Aging 10 (2018) 1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].McRae AF, Byrne EM, Zhao ZZ, Montgomery GW, Visscher PM, Power and SNP tagging in whole mitochondrial genome association studies, Genome Res. 18 (2008) 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chu Q, Martinez TF, Novak SW, Donaldson CJ, Tan D, Vaughan JM, Chang T, Diedrich JK, Andrade L, Kim A, Zhang T, Manor U, Saghatelian A, Regulation of the ER stress response by a mitochondrial microprotein, Nat. Commun 10 (2019) 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rathore A, Chu Q, Tan D, Martinez TF, Donaldson CJ, Diedrich JK, Yates JR 3rd, Saghatelian A, MIEF1 microprotein regulates mitochondrial translation, Biochemistry 57 (2018) 5564–5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, Boudreau RL, Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency, Cell Rep. 23 (2018) 3710–3720 e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lin YF, Xiao MH, Chen HX, Meng Y, Zhao N, Yang L, Tang H, Wang JL, Liu X, Zhu Y, Zhuang SM, A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation, Cell Death Dis 10 (2019) 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Makarewich CA, Baskin KK, Munir AZ, Bezprozvannaya S, Sharma G, Khemtong C, Shah AM, McAnally JR, Malloy CR, Szweda LI, Bassel-Duby R, Olson EN, MOXI is a mitochondrial micropeptide that enhances fatty acid beta-oxidation, Cell Rep. 23 (2018) 3701–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim KH, Son JM, Benayoun BA, Lee C, The mitochondrial-encoded peptide MOTS-c translocates to the nucleus to regulate nuclear gene expression in response to metabolic stress, Cell Metabol 28 (2018) 516–524 e517. [DOI] [PMC free article] [PubMed] [Google Scholar]


