Abstract
Fanconi anemia (FA) is a rare genetic disorder, characterized by birth defects, progressive bone marrow failure, and a predisposition to cancer. This devastating disease is caused by germline mutations in any one of the 22 known FA genes, where the gene products are primarily responsible for the resolution of DNA interstrand cross-links (ICLs), a type of DNA damage generally formed by cytotoxic chemotherapeutic agents. However, the identity of endogenous mutagens that generate DNA ICLs remains largely elusive. In addition, whether DNA ICLs are indeed the primary cause behind FA phenotypes is still a matter of debate. Recent genetic studies suggest that naturally occurring reactive aldehydes are a primary source of DNA damage in hematopoietic stem cells (HSCs), implicating that they could play a role in genome instability and FA. In addition, emerging lines of evidence indicate that the FA pathway constitutes a general surveillance mechanism for the genome by protecting against a variety of DNA replication stresses. Therefore, understanding the DNA repair signaling that is regulated by the FA pathway, and the types of DNA lesions underlying the FA pathophysiology is crucial for the treatment of FA and FA-associated cancers. Here, we review recent advances in our understanding of the relationship between reactive aldehydes, bone marrow dysfunction, and FA biology in the context of signaling pathways triggered during FA-mediated DNA repair and maintenance of the genomic integrity.
Keywords: Fanconi anemia, bone marrow failure, reactive aldehydes, ALDH2, DNA-protein cross-link
1. INTRODUCTION
Fanconi anemia (FA) is a rare chromosome instability syndrome that affects one in every ~100,000 births. First described in 1927 by the Swiss pediatrician Guido Fanconi, FA is a genetic disease mainly seen in children and is characterized by progressive bone marrow failure (BMF), developmental abnormalities, and increased susceptibility to multiple malignancies early in life (Figure 1). Germline mutation in any one of the 22 identified FA genes (FANCA to FANCW) causes FA in an autosomal recessive manner, except for FANCB, which is instead X-linked. Although FA is a genetically and phenotypically heterogenous disease, typical symptoms include congenital abnormalities, such as skeletal deformities of the thumb and forearm, endocrine dysfunctions frequently leading to short stature, reduced fertility, hearing loss, and café-au-lait spots (Auerbach 2009; Kee and D’Andrea 2012; Tsui and Crismani 2019). Furthermore, patients are predisposed to a variety of cancers, including acute myeloid leukemia (AML) and squamous cell carcinoma of the head and neck (Kutler et al. 2003a; Kutler et al. 2003b; Kutler et al. 2016). Additionally, monoallelic mutations of certain FA subtypes, including BRCA2 (FANCD1), BRIP1 (FANCJ), PALB2 (FANCN), RAD51C (FANCO), or BRCA1 (FANCS) have been associated with familial predisposition to breast, ovarian, and pancreatic cancers (D’Andrea 2010; Kottemann and Smogorzewska 2013). However, the most frequently mutated FA genes are those corresponding to proteins functioning in the DNA damage signaling, upstream of the enzymatic DNA repair performed by the former gene products (see below for details). FA is an orphan disease for which there are no standard treatments yet available. Current treatments are confined to alleviation of symptoms, and the average lifespan of FA patients is between 20 and 30 years, with a few patients reaching their 40s and 50s. The most prevalent symptom of FA is hematological malignancies, including BMF, which affects 75% to 90% of FA patients within the first decade of their life (Butturini et al. 1994; Kutler et al. 2003b). There are two indirect therapies of BMF: androgens, which increase red blood cell and platelet counts while having some detrimental side effects when used long-term, and hematopoietic growth factors such as G-CSF or GM-CSF, which improve neutrophil counts (Alter et al. 2000; Velazquez and Alter 2004). Currently, the best treatment option to counteract BMF is hematopoietic stem cell (HSC) transplantation using a histocompatible, or ideally a sibling donor, before the onset of hematopoietic defects. Despite extending the life expectancy of FA patients, this procedure has long-term complications including organ toxicities, graft-versus-host disease, and endocrinopathies. Because FA is a genetic disease caused by defined FA gene mutations, another promising therapeutic strategy is gene therapy. Unfortunately, limited availability of HSCs in FA patients, low efficiency of viral transduction, and an inherent risk of leukemia due to the insertion of a transgene near a proto-oncogene are major barriers to be circumvented. This strategy has recently been rejuvenated with clinical trials in phase I and II (NCT03157804, NCT03814408, and NCT04069533), testing the potential of a modified lentiviral vector whose insertion sites in HSCs are analyzed, for treating BMF of patients with FA subtype A. Corrected HSCs eventually gave rise to long-term functional bone marrow stem and progenitor cells(Rio et al. 2019). In addition, for affected children who suffer from hypothyroidism or growth hormone deficiency, thyroid and growth hormone therapies have been employed to improve the endocrine functions, with reasonable success (Dupuis-Girod et al. 2001; Eyal et al. 2008).
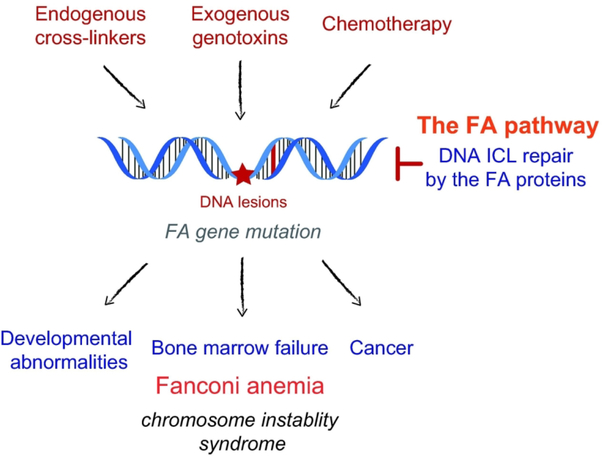
Figure 1. The causes and effects of Fanconi anemia (FA).
FA is a rare genetic disorder with onset of symptoms at a young age, primarily affecting bone marrow function in conjunction with developmental abnormalities. It is caused by germ-line mutations in the FA genes, which lead to deficiencies in coping with DNA damage, especially damage from DNA ICLs. The FA gene products constitutes the FA DNA repair pathway that resolves DNA ICLs and other lesions generated by endogenous cross-linkers, exogenous genotoxins, and cytotoxic chemotherapeutic agents such as platinum and nitrogen mustards. Due to their reduced ability to counteract genome instability, affected children are highly susceptible to a variety of cancers.
Early detection of FA symptoms and availability of better treatment options have allowed children with FA to become adults with FA. Unfortunately, these patients are at high risks of developing malignancies, including head and neck squamous cell carcinoma, cervical/gynecological cancers, and AML (Alter et al. 2003; Kutler et al. 2003a; Taniguchi and D’Andrea 2006). Due to the inherent hypersensitivity of FA patient cells to chemotherapy and radiotherapy, standard cancer treatment options are often inadequate for FA patients. Thus, it is critical to identify signs of tumorigenesis as early as possible, and adults with FA are encouraged to undergo routine dental evaluations, gynecologic exams, and bone marrow aspirates.
Mechanistically, FA is considered as a disease of chromosome instability, resulting from a defect in the repair of DNA damage, especially a failure to resolve DNA interstrand cross-links (ICLs). Accordingly, FA patient-derived cells are hypersensitive to ICL-inducing agents such as diepoxybutane (DEB) and mitomycin C (MMC), which cause high levels of chromosomal aberrations, including chromosomal breaks and quadriradial formation. In fact, this unique characteristic is often employed in the clinic to diagnose FA patients, using a DEB-induced chromosome breakage test. DNA ICLs form a covalent link between the Watson-Crick strands of DNA and prevent the separation of the DNA duplex, which has the potential to inhibit both DNA replication and transcription. DNA ICLs are thus extremely cytotoxic and effective in killing cancer cells when produced in large numbers by cytotoxic chemotherapy, such as platinum and nitrogen mustards, and have been shown to be particularly efficient for the treatment of leukemia (DeVita and Chu 2008).
Although defects in DNA ICL repair caused by mutations in FA genes are thought to underlie the pathogenesis of FA, the exact nature of the cellular lesions responsible for this chromosome instability syndrome remains ill-defined. The previously mentioned alkylating agents, those able to produce DNA ICLs, are often used as a standard-of-care for cancer treatment, however, they do not account for the etiology of FAas they are neither environmental chemicals nor metabolites of living organisms. Although DNA ICLs may be induced upon exposure to environmental mutagens, it is not sufficient to explain the profound deficiency in bone marrow function, together with many developmental abnormalities seen in FA. A plausible hypothesis explaining the pathogenesis of FA is the accumulation of unresolved DNA damage in the HSC compartment, occurring as early as in utero, and culminating in progressive loss of hematopoietic functions during childhood, together with an increased risk for hematological malignancies. This model is based upon the existence of endogenous DNA damage, presumably DNA ICLs that are specifically repaired by the FA gene products, and a high susceptibility of the HSC compartment to those genotoxins in FA patients. However, endogenous sources causing DNA ICLs in vivo have not been fully substantiated. Furthermore, direct evidence proving that defective DNA repair contributes to BMF has been lacking. Understanding the nature of these genotoxins and the underlying mechanisms by which they affect FA patients would have a significant impact on the treatment of FA and FA-associated malignancies. In this review, we will discuss our current understanding of the signaling associated with DNA ICL repair, as well as the molecular basis underlying BMF and genomic instability in FA patients. We will focus on recent advances in the study of naturally-derived aldehydes and DNA-protein adducts, and how this knowledge could be applied to developing future therapies for FA.
2. FA PATHWAY SIGNALING
2.1. Recognition of a DNA ICL and initiation of the FA pathway
At the molecular level, the 22 currently known FA gene products constitute a common DNA repair mechanism, called the FA pathway (Ceccaldi et al. 2016). The FA pathway coordinates multiple layers of DNA damage signaling and enzymatic DNA repair processes to primarily resolve DNA ICLs encountered during DNA replication (Kim and D’Andrea 2012). In a cell-free system derived from Xenopus egg extracts, a DNA ICL is recognized by the convergence of two replication forks (Zhang et al. 2015). In such an ICL-induced stalled fork, the CDC45/MCM2–7/GINS (CMG) helicase complex needs to be unloaded for the leading strand to approach the ICL. This process is promoted by the polyubiquitination of the MCM7 subunit by the RING-type ubiquitin E3 ligase TRAIP (TRAF-interacting protein, RNF206) and subsequent extraction from DNA by the AAA+ ATPase p97/VCP (Fu et al. 2011; Fullbright et al. 2016; Wu et al. 2019) (Figure 2A). The FANCM-FAAP24 complex recognizes the stalled fork structure and recruits RPA, thereby promoting ATR-CHK1 checkpoint activation (Collis et al. 2008; Huang et al. 2010). Two histone-fold-containing proteins, MHF1 (FAAP16) and MHF2 (FAAP10), form a complex with FANCM to stimulate FANCM association to chromatin, thereby contributing to the activation of FA signaling (Singh et al. 2010) (Figure 2B). Additionally, ubiquitin-like with PHD and RING finger domain 1 (UHRF1) is involved in DNA ICL sensing (Liang et al. 2015; Tian et al. 2015). FANCM also works together with proliferation cell nuclear antigen (PCNA) and the Bloom syndrome protein (BLM) complex to promote repair and replication traversal of DNA ICLs via FANCM’s translocase and DNA binding activities (Huang et al. 2013; Ling et al. 2016; Rohleder et al. 2016; Huang et al. 2019). Whether FANCM is a bona fide FANC gene is not clear, as initial FANCM association was not causative and several patients with biallelic nonsense FANCM mutations do not present clinical symptoms of FA, although hypersensitivity to chemotherapies and high cancer incidence have been noted (Singh et al. 2009; Bogliolo et al. 2018; Catucci et al. 2018).
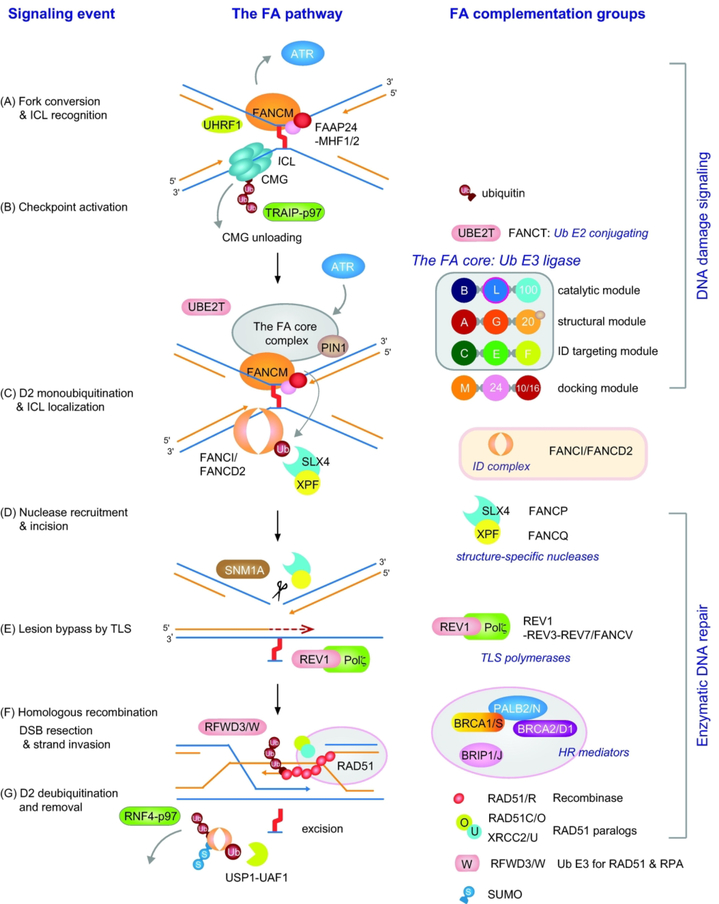
Figure 2. FA signaling in the DNA ICL repair pathway.
(A) A DNA ICL is recognized by the FANCM-FAAP24-MHF1/2 (FAAP16/FAAP10) complex and UHRF1 at stalled replication forks. CMG unloading is required for the replication fork to approach the ICL and is mediated by TRAIP-dependent MCM7 polyubiquitination and extraction from DNA by the AAA+ ATPase p97/VCP. (B) FANCM at stalled forks promotes the ATR checkpoint and activates the FA core complex, targeting it to DNA ICLs. The FA core complex is composed of three central modules that are responsible for catalysis, structural integrity, and substrate targeting. (C) UBE2T ubiquitin E2 conjugating enzyme and the FA core ubiquitin E3 ligase complex monoubiquitinate FANCD2 to target the ID complex to ICLs. (D) FANCD2-Ub functions as a platform to recruit the SLX4/FANCP-XPF/FANCQ nuclease complex to incise and unhook the DNA ICL. The SNM1A exonuclease may process the unhooked intermediate to facilitate downstream lesion bypass. (E) The lesion bypass by the REV1-polζ TLS polymerase complex restores the nascent strand and resumes replication. (F) The DNA double-strand break (DSB) ends are processed and repaired by HR, which is mediated by the recombinase RAD51/FANCR and its associated HR factors. Regulated turnover of RAD51 (and RPA) by RFWD3/FANCW is required for completion of the HR step. (G) FANCD2-Ub activity is downregulated by the USP1-UAF1 deubiquitinase complex and p97-dependent extraction of the ID complex from DNA lesions.
2.2. FANCD2 monoubiquitination by the FA core complex
The FANCM-FAAP24-MHF1/2 complex acts as a docking platform to recruit the FA core complex to DNA ICLs and initiate the FA pathway (Horejsi et al. 2009). The FA core complex is a multi-subunit ubiquitin E3 ligase, which is composed of at least eight FA proteins, namely FANCA/B/C/E/F/G/L/M, and the FA-associated proteins FAAP100 and FAAP20 (Walden and Deans 2014). The FA core complex, in conjunction with the ubiquitin E2 conjugating enzyme UBE2T/FANCT, is responsible for the monoubiquitination of FANCD2 at lysine 561 in the FANCI-FANCD2 (ID) heterodimer (Garcia-Higuera et al. 2001; Hira et al. 2015; Rickman et al. 2015) (Figure 2C). Multiple FANCI phosphorylations in the ID complex are required for efficient FANCD2 monoubiquitination (Ishiai et al. 2008). The monoubiquitinated ID complex accumulates at ICL lesions and recruits downstream factors to initiate the enzymatic processing of the ICL. The mechanism by which the ID complex is targeted to ICL lesions is not clear, but a recent cryogenic electron microscopy (cryo-EM) studyof the ID complex suggests that its recruitment to a stalled fork may precede the monoubiquitination event (Liang et al. 2016). Indeed, another structural analysis on the ID complex has revealed that the FANCD2 ubiquitination event remodels the ID complex to convert it into a sliding DNA clamp, which may work as a processivity factor to coordinate the downstream repair reactions, indicating that the role of monoubiquitin may be more complex than previously suggested (Wang et al. 2019b).
Individual subunits of the FA core complex are known to undergo various posttranslational modifications during cell cycle progression and after DNA damage, such as ATR-dependent phosphorylations, indicating that the activity of the FA core complex is regulated at multiple levels (Jo and Kim 2015). In vitro reconstitution studies have revealed that the FA core complex is composed of three distinct modules: FANCB-FANCL-FAAP100 (B-L-100), FANCA-FANCG-FAAP20 (A-G-20), and FANCC-FANCE-FANCF (C-E-F) (Huang et al. 2014; Rajendra et al. 2014). Among these, FANCL in the B-L-100 module is a RING-type ubiquitin E3 ligase that constitutes a minimal catalytic core sufficient for monoubiquitinating FANCD2 in vitro (Alpi et al. 2008). The C-E-F subcomplex connects the FA core complex to FANCM and FANCD2, while the A-G-20 trimer functions as a chromatin-targeting module and a scaffold for the FA core complex (Huang et al. 2014; Rajendra et al. 2014; van Twest et al. 2017). Intriguingly, a recent cryo-EM study has revealed that the FA core complex is composed of two B-L-100 heterotrimers around which the other subunits assemble to generate an extended asymmetric structure, suggesting that the RING domains of the two FANCLs may each have a distinct E3 ligase function toward the ID complex (Shakeel et al. 2019). This asymmetry may also be important for creating a proper ID binding site to DNA and remodeling the ID complex to access the ubiquitination site (Wang et al. 2019c). In addition, FAAP20 from the A-G-20 trimer plays a key role in preserving the integrity of the FA core complex by directly interacting with FANCA to prevent its proteasomal degradation and subsequent loss of FA core complex activity (Ali et al. 2012; Kim et al. 2012; Leung et al. 2012). Phosphorylation-dependent cis-trans isomerization catalyzed by the PIN1 isomerase and the SKP1-CUL1-F-box (SCF)FBW7 ubiquitin E3 ligase-mediated degradation was shown to modulate FAAP20 stability, underscoring the role of dynamic posttranslational modifications within the FA core complex in preserving its integrity and function (Kim et al. 2012; Wang et al. 2016; Wang et al. 2019a). Additionally, a missense mutation has been discovered in a breast cancer patient with FA-like phenotypes that disrupts the FAAP20-FANCA interaction, suggesting that maintaining this interaction is critical for regulating FA activity and limiting FA-associated tumorigenesis (Xie et al. 2015). As deletion of individual FA proteins in the FA core complex is sufficient for abrogating damage-inducible FANCD2 monoubiquitination in the cells, it is not surprising to note that most of the pathological FA mutations are concentrated in the subunits of the FA core complex required for FA pathway activation and that FANCA mutation is most prevalent among them(Levran et al. 2005).
2.3. Enzymatic processing of a DNA ICL
The primary role of FANCD2 monoubiquitin (FANCD2-Ub) at a DNA ICL is thought to recruit SLX4/FANCP and its associated 3′-flap structure-specific endonuclease XPF/FANCQ-ERCC1 heterodimer, which cooperates with SLX4 to generate nucleolytic incisions and unhook the DNA ICL (Knipscheer et al. 2009; Klein Douwel et al. 2014) (Figure 2D). SLX4 utilizes its ubiquitin-binding zinc finger 4 (UBZ4) motif to specifically recognize FANCD2-Ub (Yamamoto et al. 2011). Subsequently, XPF-ERCC1 cuts on 5′ of the ICL to generate an entry site for the 5′−3′ exonuclease SNM1A, which digests past the ICL to facilitate processing of the unhooked ICL intermediate (Abdullah et al. 2017; Buzon et al. 2018). This incision step generates a double-strand DNA break (DSB) at the stalled fork, and the opposite nascent leading strand is restored by translesion DNA synthesis (TLS), where bypass of the adducted base involves TLS polymerases REV1 and Pol ζ, a heterodimeric complex of REV3 and REV7/FANCV (Budzowska et al. 2015) (Figure 2E). It has been shown in Xenopus egg extracts that REV1-Pol ζ recruitment requires the FA core complex, indicating that the FA core complex controls both FANCD2 activation and TLS for remodeling of a DNA ICL (Budzowska et al. 2015).
Following incision and TLS, repair of replication-associated DSBs is mediated by homology-directed repair, using the restored nascent strand as a template for strand invasion and homologous recombination (HR) promoted by the RAD51/FANCR recombinase (Figure 2F). Heterozygous mutations in RAD51 found in FA patients act in a dominant-negative fashion to destabilize stalled forks, resulting in unrestricted nuclease activity (Ameziane et al. 2015; Wang et al. 2015; Zadorozhny et al. 2017). PALB2/FANCN binds directly to BRCA2/FANCD1 and BRCA1/FANCS to promote the formation of RAD51 nucleofilaments, with the help of the DNA helicase BRIP1/FANCJ, and the RAD51 paralogs RAD51C/FANCO and XRCC2/FANCU (Clauson et al. 2013; Kottemann and Smogorzewska 2013; Michl et al. 2016). Heterozygous mutations of several FA genes involved in HR are associated with increased susceptibility to breast and ovarian cancers, emphasizing the connection between the FA pathway and the BRCA HR repair network (D’Andrea 2010; Niraj et al. 2019). Intriguingly, recent studies have revealed that RFWD3/FANCW, a ubiquitin E3 ligase for RPA and RAD51, is mutated in FA patients, indicating that regulated degradation of RPA and RAD51 from sites of DNA ICL repair is critical for replication-associated HR processes and suppression of the FA phenotype (Feeney et al. 2017; Knies et al. 2017).
Lastly, FANCD2-Ub is downregulated by the USP1-UAF1 deubiquitinating enzyme complex, and the remaining adduct is removed via nucleotide excision repair (NER) to complete the ICL repair (Nijman et al. 2005; Cohn et al. 2007) (Figure 2G). In addition, timely extraction of FANCD2 from chromatin by the SUMO-targeted ubiquitin E3 ligase RNF4 and ATPase p97 plays an important role in modulating the balance of active FANCD2 levels at sites of DNA repair, which may be necessary for restricting nuclease activity and allowing selective responses of the ID complex during the multiple steps of DNA repair (Gibbs-Seymour et al. 2015).
Together, FANCD2 monoubiquitination by the FA core complex constitutes an essential gateway for DNA ICL repair by connecting upstream DNA damage signaling to downstream enzymatic DNA repair processes. Intriguingly, FANCD2 and FANCI also play additional roles in DNA replication fork protection, maintenance of common fragile sites, regulation of origin firing, histone chaperone activity, homology-directed repair during CRISPR-Cas9 genome editing, and resolution of DNA:RNA hybrids, or R-loops, indicating that the FA pathway has evolved to be a general genome maintenance mechanism to counteract various types of DNA replication stresses (Schlacher et al. 2012; Chen et al. 2015; Schwab et al. 2015; Madireddy et al. 2016; Higgs et al. 2018; Richardson et al. 2018).
3. UNDERLYING CAUSES OF BONE MARROW FAILURE IN FA
3.1. Stem cell defect and bone marrow failure in FA
FA is characterized by the progressive development of aplastic anemia due to failure of the bone marrow to produce blood cells. Interestingly, blood counts of FA patients are mostly normal at birth. Hematological defects generally begin to be detected by the age of seven, when platelet count drops first, followed by leukocytes, culminating in pancytopenia once all other blood cell types are found severely depleted. Since all blood cell lineages are affected, this points to a defect in the HSC of the bone marrow (Butturini et al. 1994; Kutler et al. 2003b). Studies using the CD34 marker to trace a HSC-enriched bone marrow fraction have shown that very young FA patients already exhibit significantly decreased HSC counts, even before the onset of pancytopenia, suggesting that the HSC defect starts before birth (Kelly et al. 2007; Ceccaldi et al. 2012). In fact, the FA pathway is considered particularly important for hematopoiesis in humans, as knocking-down FA genes in human embryonic stem cells impairs maturation of the hematopoietic lineage (Tulpule et al. 2010). This indicates that the FA pathway is not only essential in utero, but also critical for proper HSC function, presumably due to a high susceptibility of HSCs to DNA damage. Then, what is the nature of DNA damage in HSCs? It has been a difficult question to address due to limitations in studying hematopoiesis in a rare disease in humans, as well as in animal models. For instance, FA mouse models generally do not recapitulate the severity of the FA phenotype, and they do not spontaneously develop BMF, despite showing lower numbers of HSCs and an impaired ability to regenerate blood after transplantation (Haneline et al. 1999; Parmar et al. 2009; Zhang et al. 2010; Bakker et al. 2013). Recent Fancd2−/− mice generated by CRISPR-Cas9 gene editing have been engineered with a disruption site different from the previous ES-targeted knockout model and exhibit severe FA phenotypes, including progressive anemia. This different result further exemplifies how different mutations, even in the same FA gene, can produce radically different phenotypes, at the root of FA heterogeneity (Yang et al. 2019).
3.2. DNA damage as a driver for bone marrow failure in FA
Despite the discrepancy between FA patients and animal models, a few lines of evidence suggest that unresolved DNA damage is indeed responsible for HSC dysfunction and progression of BMF. First, it is widely known that bone marrow is the first organ system to fail upon total body irradiation, as it is one of the most radiosensitive tissues in the body. The huge load of DNA damage caused by irradiation produces defects in HSCs leading to their inability to maintain hematopoiesis (Niedernhofer 2008; Milyavsky et al. 2010). Second, gross chromosomal rearrangements, typically due to defective DNA repair, are observed in FA patients with hematological malignancies such as myelodysplastic syndrome (MDS) and AML. Such disorders are believed to originate from the stem and/or progenitor cell compartments, arguing for the notion that the HSC defect in FA is, at least in part, due to accumulated DNA damage (Butturini et al. 1994; Bonnet and Dick 1997; Alter et al. 2000; Nilsson et al. 2000; Nilsson et al. 2007; Chen et al. 2008; Welch et al. 2012). Third, defects in other DNA repair pathways and the DNA damage response (DDR) in mice induce a profound defect in HSCs, leading to spontaneous BMF as shown in the genetic backgrounds of DNA-PKcs3A/3A, LigIVY288C/Y288C, or Rad50S/S (Ruzankina et al. 2007; Niedernhofer 2008; Zhang et al. 2011).
A key cellular response upon genotoxic stress is to stimulate the p53-dependent checkpoint in G1, resulting in either cell cycle arrest or programmed cell death. Consequently, deleting p53 from FA-deficient cells partially alleviates their hypersensitivity to ICL-inducing agents, however it also increases genomic instability and tumor formation in Fancd2−/− or Fancc−/− mice (Freie et al. 2003; Houghtaling et al. 2005). Studies on primary bone marrow cells from FA patients and Fancd2−/− or Fancg−/− mice have demonstrated that accumulated DNA damage triggers the p53-p21 axis and cell cycle arrest, leading to HSC depletion and progressive BMF in FA (Ceccaldi et al. 2012). Accordingly, p53 inactivation rescues the defects of hematopoietic progenitors in both FA patients and FA mice (Niedernhofer 2008; Ceccaldi et al. 2012). Genetic deletion of p21, however, was not able to rescue the HSC defects of Fancd2−/− mice, suggesting that the DDR signaling mediated by p53 may be different between human and mouse (Zhang et al. 2013). Apoptosis has also been observed following cytokinesis failure in FA-deficient HSCs, raising the possibility that FA proteins may have independent functions in mitosis. However, this could also be a consequence of an increased number of ultrafine DNA bridges arising from unresolved DNA damage (Chan et al. 2009; Naim and Rosselli 2009; Vinciguerra et al. 2010). Together, attrition of HSC pools caused by DNA damage-dependent cell cycle arrest and apoptosis is an important mechanism underlying BMF in FA patients. Although p53 inactivation may prevent HSC depletion of FA patients, it is at the cost of inducing chromosome instability and tumor development.
4. REACTIVE ALDEHYDES AS AN ENDOGENOUS SOURCE OF DNA DAMAGE
4.1. Endogenous sources of DNA damage behind FA
Given that depletion of HSCs due to unresolved DNA damage causes FA, identifying the source of this endogenous DNA damage is critical for understanding FA pathogenesis. Reactive oxygen species (ROS) are recognized as one of the most pervasive types of reactive molecules in cells. They are generated as a by-product of the electron transport chain and lipid peroxidation, and can damage not only DNA, but also RNA and cellular proteins. Circumstantial lines of evidence support the hypothesis that oxidative stress sensitizes hematopoietic cells in FA by inducing DNA damage. Seminal studies have reported that FA-deficient cells produce increased levels of ROS and that they grow better under low oxygen tension, while exhibiting fewer chromosomal aberrations (Joenje et al. 1981; Schindler and Hoehn 1988; Korkina et al. 1992; Degan et al. 1995). Moreover, knocking out superoxide dismutase 1 (SOD1), one of the key enzymes protecting against ROS, in Fancc−/− mice leads to bone marrow hypocellularity, where erythroid, myeloid, and early-B lymphoid colonies show decreased proliferation and survival capabilities (Hadjur et al. 2001). However, the number of HSCs is similar to that in the wild-type mice, and there is no sign of the developmental defects or chromosomal aberrations that are commonly observed in FA patients.
Reactive aldehydes are known to generate a variety of DNA lesions including DNA ICLs. These highly reactive molecules, especially simple aldehydes such as acetaldehyde (CH3CHO) or formaldehyde (CH2O), are not only abundant in the environment, but also produced as common byproducts from various metabolic pathways. For instance, acetaldehyde is generated during carbohydrate catabolism and ethanol oxidation. Formaldehyde is naturally produced during histone demethylation at nucleosomes, one-carbon metabolism, and oxidative demethylation from DNA base damage (Trewick et al. 2002; Walport et al. 2012; Burgos-Barragan et al. 2017). Additionally, other reactive aldehydes such as 4-hydroxynonenal (4-HNE) and acrolein are produced via lipid peroxidation (Voulgaridou et al. 2011). Because of the presence of a carbonyl group, these molecules are highly reactive toward proteins and DNA, and have been shown to form DNA adducts in vitro and in vivo (McGhee and Von Hippel 1977; Wang et al. 2000; Cheng et al. 2003; Wang et al. 2009; Garcia et al. 2011). Formaldehyde has been shown to induce DNA ICLs by generating a methylene bridge between the exocyclic amino groups of adjacent DNA bases, whereas acetaldehyde has been shown to induce ICLs mostly by reacting with guanines (Chaw et al. 1980; Duxin and Walter 2015). Formaldehyde is also known to induce cross-links between DNA and proteins, and has been classified as carcinogenic as it is associated with a higher risk for developing nasopharyngeal cancers and leukemia (Cogliano et al. 2004).
4.2. Reactive aldehydes and FA
A series of extensive mouse genetic studies, led by the Patel laboratory, have provided invaluable insights into the relationship between aldehyde metabolism and the FA pathway. Mouse models of FA typically do not recapitulate the severity of the human disease in that they rarely display BMF, developmental abnormalities, or a predisposition to cancer (Parmar et al. 2009; Bakker et al. 2013). Mice, like humans, possess various detoxifying enzymes capable of eliminating toxic intracellular aldehydes, one of which is aldehyde dehydrogenase 2 (Aldh2/ALDH2), where its role in acetaldehyde catabolism is well established in humans (Vasiliou et al. 2004). Strikingly, Aldh2−/− Fancd2−/− double knockout (DKO) mice cannot be born unless their mother carries at least one wild-type allele of Aldh2 (Langevin et al. 2011). Although Fancd2−/− mice display reduced HSC content, growth retardation, and increased tumor incidence, Aldh2−/− mice do not have an obvious phenotype and are born at normal Mendelian ratios. Nevertheless, Aldh2−/− Fancd2−/− DKO embryos were reported to die between E9.5 and E13.5, supporting the notion that acetaldehyde catabolism is essential for fetal development in the absence of the FA pathway. Since acetaldehyde can passively diffuse across the placental membrane, maternal detoxification could compensate for the deficiency of a fetus and allow for its survival (Oberbeck et al. 2014).
Furthermore, when Aldh2−/− Fancd2−/− embryos were challenged with ethanol, their viability decreased due to developmental defects, and pups frequently displayed kinked tails or eye defects (Langevin et al. 2011). Between 3 to 6 months of age, most Aldh2−/− Fancd2−/− mice die from an illness equivalent to acute lymphoblastic leukemia (ALL), with neoplastic cells being positive for the pan-T-cell marker CD3. Additionally, challenging younger DKO mice with ethanol was shown to impair hematopoiesis, which leads to BMF, with bone marrow cells displaying increased levels of γH2AX, a marker of DSBs (Langevin et al. 2011).
A follow-up study has shown that old Aldh2−/− Fancd2−/− mice that did not succumb to leukemia spontaneously develop aplastic anemia (Garaycoechea et al. 2012). This profound hematopoietic defect is restricted to the stem and progenitor cell populations. Aldh2−/− Fanca−/− mice also exhibit severe developmental defects (when maternal aldehyde catabolism is defective) and attrition of the hematopoietic stem and progenitor cells, reinforcing the idea that genotoxicity from endogenous aldehydes causes depletion of HSCs, which leads to BMF in FA patients (Oberbeck et al. 2014) (Figure 3). Hence, HSCs appear to possess a two-tier protection mechanism whereby high levels of reactive aldehydes are counteracted by aldehyde catabolism, while unresolved DNA damage caused by aldehydes is processed by the FA pathway (Garaycoechea and Patel 2014). By doing so, the FA pathway prevents the formation of DSBs stemming from aldehyde-derived DNA lesions, thereby protecting stem cells from gross chromosome deletions and rearrangements, which may be predominantly generated by microhomology-mediated end-joining repair (Garaycoechea et al. 2018). It is interesting to note that the hematological phenotypes of human FA patients are only recapitulated in mice when ALDH2 activity is lost, which may be due to the fact that the two species may differ in their capacity to detoxify aldehydes. Also, the short lifespan of mice may not provide sufficient time for the complete exhaustion of the HSC pool, especially since mice are fed a standard chow diet in a controlled laboratory environment.
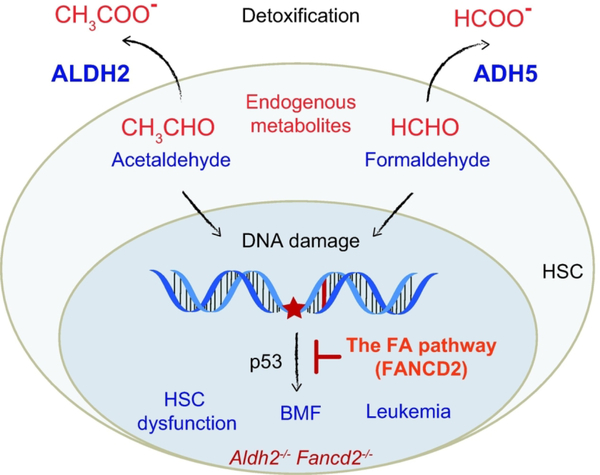
Figure 3. Mechanisms of reactive aldehyde detoxification cooperating to preserve HSC function.
The elevated ALDH2 and ADH5 activities in HSCs alleviate the genotoxic effect of endogenous reactive aldehydes, such as acetaldehyde and formaldehyde. DNA damage caused by reactive aldehydes is resolved by the FA pathway, which would otherwise result in exhaustion of the HSC pool via activation of p53. In Aldh2−/− Fancd2−/− DKO mice, the burden of accumulated reactive aldehydes in the absence of the FA pathway leads to HSC dysfunction, spontaneous BMF, and cellular transformation.
Intriguingly, more than half a billion people, mainly from Southeast Asia, carry a semidominant negative mutation in ALDH2 (ALDH2*2), which is responsible for the so-called Asian flushing syndrome. The E487K mutation (or E504K in the full-length protein) drastically reduces ALDH2 activity and predisposes carriers to squamous cell carcinoma of the esophagus from alcohol consumption (Brooks et al. 2009; Van Wassenhove et al. 2016). Accordingly, Japanese FA patients carrying this ALDH2 polymorphism are subject to a much earlier onset of BMF than those from other ethnicities, starting before the age of 7 months for homozygous carriers (Hira et al. 2013). These results further highlight the detrimental effect of reactive aldehydes in contributing to the pathogenesis of FA.
Ethanol is primarily metabolized by alcohol dehydrogenase (ADH) into acetaldehyde, which is then metabolized into acetate by ALDH2 (Crabb et al. 2004). The effect of alcohol exposure during fetal development has been observed, such as in the fetal alcohol syndrome (FAS), where maternal alcohol consumption causes congenital learning disability (Abel and Sokol 1991). It is therefore tempting to speculate that the DNA damage caused by acetaldehyde might be involved in the pathogenesis of FAS, as well. Additionally, epidemiological studies have correlated maternal alcohol consumption with an increased risk of acute childhood leukemia for their child, which is in line with the hypersensitivity of HSCs to acetaldehyde as demonstrated in the FA studies (MacArthur et al. 2008; Latino-Martel et al. 2010).
A similar synthetic lethal relationship has been observed between a formaldehyde-detoxifying enzyme, alcohol dehydrogenase 5 (ADH5), and the FA pathway. While knocking out both Fancc and Aldh2 in chicken DT40 B-cells does not render cells hypersensitive to acetaldehyde, inactivation of Fancc or Fancl in the Adh5−/− strain is synthetically lethal (Langevin et al. 2011; Rosado et al. 2011). This discrepancy may be explained by the higher abundance of endogenous formaldehyde compared to endogenous acetaldehyde, and/or greater genotoxicity of formaldehyde in chicken cells. Adh5 and Fancd2 are synthetically lethal in mice as well, unless crossed in hybrid backgrounds, where Adh5−/− Fancd2−/− DKO mice are born at sub-Mendelian ratios (Pontel et al. 2015). These DKO mice are smaller than wild-type littermates and suffer from a loss of HSCs, leading to rapid BMF 3 to 7 weeks after birth. Furthermore, they show karyomegaly in many organs, especially in the liver, which coincides with the activation of the DDR, possibly explaining their smaller size. Kidney function is also compromised, indicating that failure to detoxify endogenous formaldehyde may cause widespread DNA damage to two key detoxifying organs, the liver and the kidneys, in addition to the HSC compartment (Pontel et al. 2015). Together, these genetic models that recapitulate the FA phenotypes in the absence of aldehyde detoxification support the notion that aldehyde genotoxicity is primarily responsible for BMF in FA patients.
5. DNA-PROTEIN CROSS-LINK REPAIR
5.1. Reactive aldehydes and DNA-protein cross-links
Reactive aldehydes, especially formaldehyde, have been known to induce DNA-protein cross-links (DPCs) in addition to DNA cross-links (Klages-Mundt and Li 2017). The loss of formaldehyde detoxification has more severe and dramatic consequences than that of acetaldehyde detoxification, hinting that formaldehyde may well be one of the main sources of endogenous damage in FA (Rosado et al. 2011; Pontel et al. 2015). Formaldehyde produces DPCs by forming a methylene bridge between nucleophilic amino acid side chains and exocyclic amines of DNA bases (Conaway et al. 1996; Quievryn and Zhitkovich 2000; Duxin and Walter 2015). Moreover, most ICL-inducing drugs are also able to produce DPCs, suggesting that the endogenous lesions triggering the FA pathway may involve DPCs in addition to DNA ICLs (Chválová et al. 2007; Loeber et al. 2008; Loeber et al. 2009; Michaelson-Richie et al. 2010).
DPCs constitute a group of complex and diverse DNA lesions. They involve the proteins in close proximity to DNA that become covalently and irreversibly bound to it through a chemical reaction that engages an endogenous or exogenous cross-linker (Vaz et al. 2017). Notably, some DPCs are generated by enzymatic reactions associated with DNA metabolism, such as topoisomerase 1 and 2 cleavage complexes (TOP1cc and TOP2cc), DNA polymerase β, or poly (ADP-ribose) polymerase 1 (PARP1). DPCs are ubiquitous in cells, and like DNA ICLs, they are cytotoxic lesions that impede the progression of DNA replication and transcription machineries. They also block the DNA repair machineries and chromatin-remodeling factors from accessing DNA (Barker et al. 2005). If not removed from DNA, DPCs induce mutations, replication fork breakage, and chromosomal aberrations (Stingele and Jentsch 2015). In particular, formaldehyde and acetaldehyde have been reported to cause a variety of chromosomal aberrations in mammalian cells (Mechilli et al. 2008; Lorenti Garcia et al. 2009).
5.2. Mechanisms of DPC repair
Studies using Xenopus egg extracts and an artificial DPC formed by the bacterial methyltransferase, M. HpaII, cross-linked to DNA, have shown that DPC repair is coupled to DNA replication and initiated when the CMG helicase collides with the DPC (Duxin et al. 2014) (Figure 4). In contrast to DNA ICL repair, DPC repair does not require CMG unloading or nucleolytic incisions of the parental strands. Instead, replication fork collision with the DPC triggers its proteolysis, followed by leading strand extension via TLS. Intriguingly, the DNA helicase RTEL1 was shown to facilitate CMG bypass of the DPC by unwinding DNA beyond the DPC, and this bypass is necessary for efficient DPC proteolysis, revealing a mechanism that prevents an accidental destruction of CMG by DPC proteases (Sparks et al. 2019).
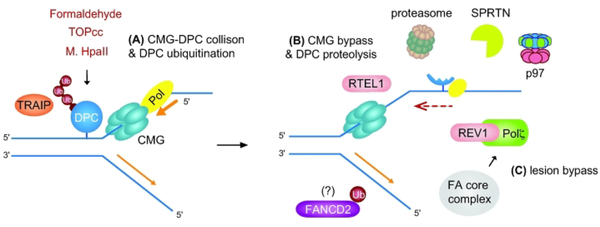
Figure 4. DNA protein cross-link (DPC) repair pathway.
(A) DPCs are generated by physiological processes (e.g. formaldehyde from histone demethylation), nucleic acids metabolism (e.g. TOPcc intermediate), or experimentally in vitro (e.g. M. HpaII adduct introduced into a plasmid). In Xenopus egg extracts, when the CMG helicase collides a DPC lesion, the ubiquitin E3 ligase TRAIP stimulates DPC ubiquitination and proteasome targeting. (B) CMG bypasses an intact DPC, which is facilitated by the DNA helicase RTEL1. CMG bypass is required for efficient DPC proteolysis, which occurs either by ubiquitin-dependent proteasome activity or by the metalloprotease SPRTN with the help of the ATP-driven p97 segregase in a post-replicative manner. Unlike DNA ICL repair, this process does not involve CMG unloading or incision of a stalled fork. (C) Leading strand extension resumes via TLS, which is mediated by the REV1-Pol ζ (FANCV) TLS polymerase complex. The role of FANCD2-Ub in DPC repair is currently not clear, and whether the FA pathway directly regulates DPC repair has yet to be determined. The FA core complex may promote the recruitment of TLS polymerases independently of FANCD2-Ub.
Several proteolytic mechanisms and players have been implicated in replication-associated DPC repair. The metalloprotease Wss1 (weak suppressor of smt3) in S. cerevisiae targets DPCs and is required for cellular survival against formaldehyde or camptothecin-induced lesions (Stingele et al. 2014). Several independent studies in mammalian cells have demonstrated that the SPRTN metalloprotease (also known as DVC1 or C1orf124) is a protease that resolves DPCs during DNA replication (Lopez-Mosqueda et al. 2016; Stingele et al. 2016; Vaz et al. 2016; Mórocz et al. 2017). SPRTN is associated with the replisome machinery, where it may monitor replication-blocking lesions during replication fork progression. By recruiting p97 to DNA lesions, SPRTN promotes the extraction of proteins from chromatin for degradation, indicating that p97 may assist the proteolysis and clearance of DPC intermediates. A conserved HEXXH motif is present in both Wss1 and SPRTN to mediate DPC processing independently of proteasome activity (Fielden et al. 2018). The proteolytic activity of SPRTN requires its interaction with single-stranded DNA, suggesting that the DPC needs to be in close proximity to the traveling replication fork (Li et al. 2019). SPRTN-deficient cells exhibit elevated levels of TOPcc in unstressed conditions and are hypersensitive to topoisomerase inhibitors, arguing for the physiological role of SPRTN in protecting cells from endogenously generated DPCs. Moreover, Sprtn hypomorphic mice suffer from spontaneous liver cancer, suggesting that SPRTN may be particularly important for counteracting formaldehyde-induced DPCs that are predominantly produced in the liver (Maskey et al. 2017).
Besides SPRTN-dependent proteolysis, replication-coupled DPC removal requires ubiquitin-mediated DPC degradation, which is facilitated by the ubiquitin E3 ligase TRAIP. An in vitro study using Xenopus egg extracts has demonstrated that TRAIP is necessary for DPC polyubiquitination and proteasome recruitment (Larsen et al. 2019). TRAIP also plays a role in replication-coupled DNA ICL repair, where TRAIP- and p97-dependent unloading of the CMG helicase complex promotes remodeling and processing of a DNA ICL lesion by the FA pathway (Fullbright et al. 2016; Wu et al. 2019). Notably, the ubiquitin chain length controlled by TRAIP determines the subtypes of DNA ICL repair in vitro, highlighting the central role of TRAIP in the choice of replication-coupled DNA ICL repair pathways (Wu et al. 2019).
It may also be possible that known DNA repair pathways cooperate with SPRTN- and/or TRAIP-dependent DPC proteolysis. Previous studies in yeast and mammalian cells have demonstrated that NER and HR pathways contribute to cellular resistance toward formaldehyde-induced DPCs, while showing some discrepancies (de Graaf et al. 2009; Lorenti Garcia et al. 2009; Rosado et al. 2011; Klages-Mundt and Li 2017). Conflicting results obtained for NER suggest that this pathway might be able to excise only DPCs with specific structural characteristics, such as a small size, while studies in bacteria have shown that HR could deal with DPC regardless of their size (Nakano et al. 2007).
5.3. The FA pathway and DPC repair
While the role of the FA pathway in DNA ICL repair is well established, its role in DPC repair remains unclear. Given that the genotoxicity of both DNA ICLs and DPCs are pronounced during DNA replication and that multiple mammalian DNA repair mutants show shared sensitivities to both lesions, the components of the FA pathway may directly or indirectly contribute to the resolution of DPCs as well. Interestingly, at least in Xenopus egg extracts, FANCD2 is not required for the repair of M. HpaII-linked DPC, and the Rev1-Pol ζ TLS polymerase complex is able to bypass such DPC without DNA incision (Duxin et al. 2014). However, the FA core complex may contribute to the recruitment of TLS polymerases independently of the ID complex, as previously described (Kim et al. 2012; Budzowska et al. 2015). ALDH2 deficiency exerts a more severe effect on Fanca−/− mice than on Fancd2−/− mice, indicating that the FA core complex, or FANCA alone, may have additional functions, independent of promoting FANCD2 monoubiquitination (Oberbeck et al. 2014). The chemically-induced DPC lesions that are generated in vitro may have, however, a different chemistry and signaling compared to the DPC lesions produced by formaldehyde inside the cells. It is not currently known whether SPRTN cooperates with the FA pathway to resolve DNA ICLs or DPCs. C. elegans lacking both dvc-1 (SPRTN) and fcd-2 (FANCD2) are more sensitive to chronic exposure to cisplatin in comparison to single mutants, raising the possibility that cisplatin-induced ICLs may be repaired by FANCD2, while cisplatin-induced DPCs are repaired by SPRTN (Stingele et al. 2016). Moreover, Fancd2−/− mouse embryonic fibroblasts (MEFs) are proficient in DPC repair after formaldehyde exposure (Stingele et al. 2016). Similarly, FANCD2 depletion in HeLa cells has no effect on DPC levels detected at the chromatin (Vaz et al. 2016).
In contrast, several other studies have implicated the role of FA proteins in DPC repair. FA-deficient chicken and mammalian cell lines were reported to be hypersensitive to formaldehyde and DNA methyltransferase 1 (DNMT1) DPCs generated by the cytosine analog 5-aza-2′-deoxycytidine (5-aza-dC), respectively (Ridpath et al. 2007; Rosado et al. 2011; Orta et al. 2013). FancG-deficient Chinese hamster ovary (CHO) cells exhibit increased chromatid breaks and radial fusion upon induction of DNMT1 DPCs due to their failure to engage HR (Orta et al. 2013). FANCD2-depleted cells were shown to be hypersensitive to formaldehyde in several studies (Karanja et al. 2014; Vaz et al. 2016). Interestingly, it was shown that treatment of human cells with exogenous acetaldehyde is able to trigger FANCD2 monoubiquitination (Marietta et al. 2009; Langevin et al. 2011). Since DPCs constitute a diverse group of structurally distinct DNA lesions, it is possible that a subset of DPCs, in a certain context, may require FANCD2-dependent nucleolytic incision of the DNA-protein adduct or rely on a noncanonical role of the FA core complex that does not involve FANCD2 monoubiquitination. The development of various types of chemically-defined DPCs and their induction at different phases of the cell cycle may increase our understanding of the role of the FA pathway in DPC repair.
6. CONCLUSIONS AND FUTURE DIRECTIONS
Here, we have overviewed the core signaling of DNA ICL repair mediated by the FA pathway and discussed the key role of naturally-derived aldehydes as an endogenous source of DNA damage that contributes to the pathogenesis of FA. A series of genetic studies over the last decade have revealed that detoxification of aldehydes constitutes an essential barrier to limit genotoxicity against HSC function, thus preventing the manifestation of BMF in FA patients. Given that reactive aldehydes exacerbate FA phenotypes, this discovery reinforces the causal relationship between DNA repair defects in HSCs and BMF in FA. The FA gene products form a common genome maintenance pathway that resolves DNA lesions encountered during DNA replication, mostly DNA ICLs, while it may also be involved in resolving DPC adducts that are produced by formaldehyde or various DNA repair-associated enzymatic reactions. Besides unresolved DNA ICLs and DPCs, DNA replication stress aggravated due to the loss of general genome surveillance mechanisms in the absence of the FA pathway may also contribute to the FA phenotypes. Currently, it is not completely understood whether reactive aldehydes are bona fide endogenous genotoxins that are sufficient to cause FA alone. A direct approach to modulate aldehyde levels in vivo may be necessary to determine whether physiological levels of reactive aldehydes are indeed correlated with the severity of the disease. In this sense, using ALDH2 agonists, such as Alda-1 that boosts the detoxifying ALDH2 enzyme activity, may help answer this question and ultimately provide a new therapeutic strategy to alleviate the symptoms of FA (Gross et al. 2015). Alda-1 acts as an allosteric regulator that is able to correct the structural defect of the E487K ALDH2*2 mutant, which may have broad clinical implications to the diseases in which ALDH2 activation is beneficial (Perez-Miller et al. 2010). Intriguingly, a recent study has revealed that metformin (N,N-dimethylbiguanide) works as an aldehyde scavenger that improves hematopoiesis and delays tumor formation in a Fancd2−/− mouse model, suggesting that metformin may directly target the source of DNA damage responsible for FA (Zhang et al. 2016). Since metformin has been widely used for decades in the clinic as a first-line treatment for type II diabetes, it could have a significant benefit to FA patients, once its optimal dose, timing of delivery, and biological effects are established in FA (Cicero et al. 2012). A pilot clinical trial of metformin tailored to FA patients is addressing some of these issues (NCT03398824). Together, a better understanding of the nature of genotoxins associated with FA etiology will have a greater impact toward other BMF disorders, cancer, aging, and FAS, especially given the importance of maternal aldehyde detoxification to normal fetal development.
ACKNOWLEDGMENTS
We thank members in the Kim laboratory, Alexandra Weinheimer, Jennifer Park, and Arafat Kahn, for helpful suggestions and editing. This work was supported by the National Institutes of Health (R01 CA218132) and American Cancer Society to H.K. (132235-RSG-18-037-DMC).
Abbreviations:
- AML
acute myeloid leukemia
- BMF
bone marrow failure
- CMG
CDC45/MCM2–7/GINS
- DDR
DNA damage response
- DEB
diepoxybutane
- DNA ICL
DNA interstrand cross-link
- DPC
DNA-protein cross-link
- DSB
double-strand DNA break
- FA
Fanconi anemia
- FANCD2-Ub
FANCD2 monoubiquitin
- FAS
fetal alcohol syndrome
- HR
homologous recombination
- HSCs
hematopoietic stem cells
- ID
FANCI-FANCD2 complex
- MMC
mitomycin C
- TLS
translesion DNA synthesis
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- Abdullah UB, McGouran JF, Brolih S, Ptchelkine D, El-Sagheer AH, Brown T, McHugh PJ. 2017. RPA activates the XPF-ERCC1 endonuclease to initiate processing of DNA interstrand crosslinks. EMBO J 36(14):2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel EL, Sokol RJ. 1991. A Revised Conservative Estimate of the Incidence of FAS and its Economic Impact. Alcoholism: Clinical and Experimental Research 15(3):514–524. [DOI] [PubMed] [Google Scholar]
- Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, Grassman E, Auerbach AD, Pang Q, Meetei AR. 2012. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood 119(14):3285–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi AF, Pace PE, Babu MM, Patel KJ. 2008. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell 32(6):767–777. [DOI] [PubMed] [Google Scholar]
- Alter BP, Caruso JP, Drachtman RA, Uchida T, Velagaleti GVN, Elghetany MT. 2000. Fanconi Anemia: Myelodysplasia as a Predictor of Outcome. Cancer Genetics and Cytogenetics 117(2):125–131. [DOI] [PubMed] [Google Scholar]
- Alter BP, Greene MH, Velazquez I, Rosenberg PS. 2003. Cancer in Fanconi anemia. Blood 101(5):2072–2072. [DOI] [PubMed] [Google Scholar]
- Ameziane N, May P, Haitjema A, van de Vrugt HJ, van Rossum-Fikkert SE, Ristic D, Williams GJ, Balk J, Rockx D, Li H, Rooimans MA, Oostra AB, Velleuer E, Dietrich R, Bleijerveld OB, Maarten Altelaar AF, Meijers-Heijboer H, Joenje H, Glusman G, Roach J, Hood L, Galas D, Wyman C, Balling R, den Dunnen J, de Winter JP, Kanaar R, Gelinas R, Dorsman JC. 2015. A novel Fanconi anaemia subtype associated with a dominant-negative mutation in RAD51. Nat Commun 6:8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. 2009. Fanconi anemia and its diagnosis. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 668(1):4–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ST, de Winter JP, Riele Ht. 2013. Learning from a paradox: recent insights into Fanconi anaemia through studying mouse models. Disease Models & Mechanisms 6(1):40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker S, Weinfeld M, Murray D. 2005. DNA–protein crosslinks: their induction, repair, and biological consequences. Mutation Research/Reviews in Mutation Research 589(2):111–135. [DOI] [PubMed] [Google Scholar]
- Bogliolo M, Bluteau D, Lespinasse J, Pujol R, Vasquez N, d’Enghien CD, Stoppa-Lyonnet D, Leblanc T, Soulier J, Surralles J. 2018. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet Med 20(4):458–463. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nature Medicine 3(7):730–737. [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Enoch M-A, Goldman D, Li T-K, Yokoyama A. 2009. The Alcohol Flushing Response: An Unrecognized Risk Factor for Esophageal Cancer from Alcohol Consumption. PLOS Medicine 6(3):e1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budzowska M, Graham TG, Sobeck A, Waga S, Walter JC. 2015. Regulation of the Rev1-pol zeta complex during bypass of a DNA interstrand cross-link. EMBO J 34(14):1971–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Barragan G, Wit N, Meiser J, Dingler FA, Pietzke M, Mulderrig L, Pontel LB, Rosado IV, Brewer TF, Cordell RL, Monks PS, Chang CJ, Vazquez A, Patel KJ. 2017. Mammals divert endogenous genotoxic formaldehyde into one-carbon metabolism. Nature 548(7669):549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. 1994. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study [see comments]. Blood 84(5):1650–1655. [PubMed] [Google Scholar]
- Buzon B, Grainger R, Huang S, Rzadki C, Junop MS. 2018. Structure-specific endonuclease activity of SNM1A enables processing of a DNA interstrand crosslink. Nucleic Acids Res 46(17):9057–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catucci I, Osorio A, Arver B, Neidhardt G, Bogliolo M, Zanardi F, Riboni M, Minardi S, Pujol R, Azzollini J, Peissel B, Manoukian S, De Vecchi G, Casola S, Hauke J, Richters L, Rhiem K, Schmutzler RK, Wallander K, Torngren T, Borg A, Radice P, Surralles J, Hahnen E, Ehrencrona H, Kvist A, Benitez J, Peterlongo P. 2018. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med 20(4):452–457. [DOI] [PubMed] [Google Scholar]
- Ceccaldi R, Parmar K, Mouly E, Delord M, Kim Jung M, Regairaz M, Pla M, Vasquez N, Zhang Q-S, Pondarre C, Peffault de Latour R, Gluckman E, Cavazzana-Calvo M, Leblanc T, Larghero J, Grompe M, Socié G, D’Andrea Alan D, Soulier J. 2012. Bone Marrow Failure in Fanconi Anemia Is Triggered by an Exacerbated p53/p21 DNA Damage Response that Impairs Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 11(1):36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R, Sarangi P, D’Andrea AD. 2016. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol 17(6):337–349. [DOI] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nature Cell Biology 11(6):753–760. [DOI] [PubMed] [Google Scholar]
- Chaw YFM, Crane LE, Lange P, Shapiro R. 1980. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry 19(24):5525–5531. [DOI] [PubMed] [Google Scholar]
- Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, Lund EA, Sam TN, Kersey JH. 2008. Malignant Transformation Initiated by Mll-AF9: Gene Dosage and Critical Target Cells. Cancer Cell 13(5):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Jones MJ, Yin Y, Crist SB, Colnaghi L, Sims RJ 3rd, Rothenberg E, Jallepalli PV, Huang TT. 2015. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58(2):323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng G, Shi Y, Sturla SJ, Jalas JR, McIntee EJ, Villalta PW, Wang M, Hecht SS. 2003. Reactions of Formaldehyde Plus Acetaldehyde with Deoxyguanosine and DNA: Formation of Cyclic Deoxyguanosine Adducts and Formaldehyde Cross-Links. Chemical Research in Toxicology 16(2):145–152. [DOI] [PubMed] [Google Scholar]
- Chválová K, Brabec V, Kašpárková J. 2007. Mechanism of the formation of DNA–protein cross-links by antitumor cisplatin. Nucleic Acids Research 35(6):1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero AF, Tartagni E, Ertek S. 2012. Metformin and its clinical use: new insights for an old drug in clinical practice. Arch Med Sci 8(5):907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauson C, Scharer OD, Niedernhofer L. 2013. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb Perspect Med 3(10):a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliano V, Grosse Y, Baan R, Straif K, Secretan B, Ghissassi FE. 2004. Advice on formaldehyde and glycol ethers. The Lancet Oncology 5(9):528. [DOI] [PubMed] [Google Scholar]
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D’Andrea AD. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 28(5):786–797. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. 2008. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell 32(3):313–324. [DOI] [PubMed] [Google Scholar]
- Conaway CC, Whysner J, Verna LK, Williams GM. 1996. Formaldehyde mechanistic data and risk assessment: Endogenous protection from DNA adduct formation. Pharmacology & Therapeutics 71(1):29–55. [DOI] [PubMed] [Google Scholar]
- Crabb DW, Matsumoto M, Chang D, You M. 2004. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proceedings of the Nutrition Society 63(1):49–63. [DOI] [PubMed] [Google Scholar]
- D’Andrea AD. 2010. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med 362(20):1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf B, Clore A, McCullough AK. 2009. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair 8(10):1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degan P, Bonassi S, Caterina MD, Korkina LG, Pinto L, Scopacasa F, Zatterale A, Calzone R, Pagano G. 1995. In vivo accumulation of 8-hydroxy-2’-deoxyguanosine in DNA correlates with release of reactive oxygen species in Fanconi’s anaemia families. Carcinogenesis 16(4):735–742. [DOI] [PubMed] [Google Scholar]
- DeVita VT, Chu E. 2008. A History of Cancer Chemotherapy. Cancer Research 68(21):8643–8653. [DOI] [PubMed] [Google Scholar]
- Dupuis-Girod S, Gluckman E, Souberbielle J-C, Brauner R. 2001. Growth hormone deficiency caused by pituitary stalk interruption in Fanconi’s anemia. The Journal of Pediatrics 138(1):129–133. [DOI] [PubMed] [Google Scholar]
- Duxin Julien P, Dewar James M, Yardimci H, Walter Johannes C. 2014. Repair of a DNA-Protein Crosslink by Replication-Coupled Proteolysis. Cell 159(2):346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Walter JC. 2015. What is the DNA repair defect underlying Fanconi anemia? Current Opinion in Cell Biology 37:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal O, Blum S, Mueller R, Smith FO, Rose SR. 2008. Improved growth velocity during thyroid hormone therapy in children with Fanconi anemia and borderline thyroid function. Pediatric Blood & Cancer 51(5):652–656. [DOI] [PubMed] [Google Scholar]
- Feeney L, Munoz IM, Lachaud C, Toth R, Appleton PL, Schindler D, Rouse J. 2017. RPA-Mediated Recruitment of the E3 Ligase RFWD3 Is Vital for Interstrand Crosslink Repair and Human Health. Mol Cell 66(5):610–621 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielden J, Ruggiano A, Popović M, Ramadan K. 2018. DNA protein crosslink proteolysis repair: From yeast to premature ageing and cancer in humans. DNA Repair 71:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freie B, Li X, Ciccone SLM, Nawa K, Cooper S, Vogelweid C, Schantz L, Haneline LS, Orazi A, Broxmeyer HE, Lee S-H, Clapp DW. 2003. Fanconi anemia type C and p53 cooperate in apoptosis and tumorigenesis. Blood 102(12):4146–4152. [DOI] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. 2011. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146(6):931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullbright G, Rycenga HB, Gruber JD, Long DT. 2016. p97 Promotes a Conserved Mechanism of Helicase Unloading during DNA Cross-Link Repair. Mol Cell Biol 36(23):2983–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. 2012. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489:571. [DOI] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Mulderrig L, Louzada S, Yang F, Guilbaud G, Park N, Roerink S, Nik-Zainal S, Stratton MR, Patel KJ. 2018. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 553:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Patel KJ. 2014. Why does the bone marrow fail in Fanconi anemia? Blood 123(1):26–34. [DOI] [PubMed] [Google Scholar]
- Garcia CCM, Angeli JPF, Freitas FP, Gomes OF, de Oliveira TF, Loureiro APM, Di Mascio P, Medeiros MHG. 2011. [13C2]- Acetaldehyde Promotes Unequivocal Formation of 1,N2-Propano-2 ′ -deoxyguanosine in Human Cells. Journal of the American Chemical Society 133(24):9140–9143. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7(2):249–262. [DOI] [PubMed] [Google Scholar]
- Gibbs-Seymour I, Oka Y, Rajendra E, Weinert BT, Passmore LA, Patel KJ, Olsen JV, Choudhary C, Bekker-Jensen S, Mailand N. 2015. Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol Cell 57(1):150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross ER, Zambelli VO, Small BA, Ferreira JC, Chen CH, Mochly-Rosen D. 2015. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu Rev Pharmacol Toxicol 55:107–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Ung K, Wadsworth L, Dimmick J, Rajcan-Separovic E, Scott RW, Buchwald M, Jirik FR. 2001. Defective hematopoiesis and hepatic steatosis in mice with combined deficiencies of the genes encoding Fancc and Cu/Zn superoxide dismutase. Blood 98(4):1003–1011. [DOI] [PubMed] [Google Scholar]
- Haneline LS, Gobbett TA, Ramani R, Carreau M, Buchwald M, Yoder MC, Clapp DW. 1999. Loss of FancC Function Results in Decreased Hematopoietic Stem Cell Repopulating Ability. Blood 94(1):1–8. [PubMed] [Google Scholar]
- Higgs MR, Sato K, Reynolds JJ, Begum S, Bayley R, Goula A, Vernet A, Paquin KL, Skalnik DG, Kobayashi W, Takata M, Howlett NG, Kurumizaka H, Kimura H, Stewart GS. 2018. Histone Methylation by SETD1A Protects Nascent DNA through the Nucleosome Chaperone Activity of FANCD2. Mol Cell 71(1):25–41 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, Ogawa S, Matsuo K, Takata M, Yabe M. 2013. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood 122(18):3206–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira A, Yoshida K, Sato K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, Ito E, Kojima S, Kurumizaka H, Ogawa S, Takata M, Yabe H, Yabe M. 2015. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am J Hum Genet 96(6):1001–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horejsi Z, Collis SJ, Boulton SJ. 2009. FANCM-FAAP24 and HCLK2: roles in ATR signalling and the Fanconi anemia pathway. Cell Cycle 8(8):1133–1137. [DOI] [PubMed] [Google Scholar]
- Houghtaling S, Granville L, Akkari Y, Torimaru Y, Olson S, Finegold M, Grompe M. 2005. Heterozygosity for p53 (Trp53+/−) accelerates epithelial tumor formation in fanconi anemia complementation group D2 (Fancd2) knockout mice. Cancer Research 65(1):85. [PubMed] [Google Scholar]
- Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. 2013. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol Cell 52(3):434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhang J, Bellani MA, Pokharel D, Gichimu J, James RC, Gali H, Ling C, Yan Z, Xu D, Chen J, Meetei AR, Li L, Wang W, Seidman MM. 2019. Remodeling of Interstrand Crosslink Proximal Replisomes Is Dependent on ATR, FANCM, and FANCD2. Cell Rep 27(6):1794–1808 e1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Kim JM, Shiotani B, Yang K, Zou L, D’Andrea AD. 2010. The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response. Mol Cell 39(2):259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Leung JW, Lowery M, Matsushita N, Wang Y, Shen X, Huong D, Takata M, Chen J, Li L. 2014. Modularized functions of the Fanconi anemia core complex. Cell Rep 7(6):1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, Saberi A, Kinoshita E, Kinoshita-Kikuta E, Koike T, Tashiro S, Elledge SJ, Takata M. 2008. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol 15(11):1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo U, Kim H. 2015. Exploiting the Fanconi Anemia Pathway for Targeted Anti-Cancer Therapy. Mol Cells 38(8):669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joenje H, Arwert F, Eriksson AW, de Koning H, Oostra AB. 1981. Oxygen-dependence of chromosomal aberrations in Fanconi’s anaemia. Nature 290(5802):142–143. [DOI] [PubMed] [Google Scholar]
- Karanja KK, Lee EH, Hendrickson EA, Campbell JL. 2014. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle 13(10):1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D’Andrea AD. 2012. Molecular pathogenesis and clinical management of Fanconi anemia. The Journal of Clinical Investigation 122(11):3799–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PF, Radtke S, Kalle Cv, Balcik B, Bohn K, Mueller R, Schuesler T, Haren M, Reeves L, Cancelas JA, Leemhuis T, Harris R, Auerbach AD, Smith FO, Davies SM, Williams DA. 2007. Stem Cell Collection and Gene Transfer in Fanconi Anemia. Molecular Therapy 15(1):211–219. [DOI] [PubMed] [Google Scholar]
- Kim H, D’Andrea AD. 2012. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26(13):1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Yang K, Dejsuphong D, D’Andrea AD. 2012. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol 19(2):164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klages-Mundt NL, Li L. 2017. Formation and repair of DNA-protein crosslink damage. Science China Life Sciences 60(10):1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. 2014. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54(3):460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies K, Inano S, Ramirez MJ, Ishiai M, Surralles J, Takata M, Schindler D. 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J Clin Invest 127(8):3013–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326(5960):1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkina LG, Samochatova EV, Maschan AA, Suslova TB, Cheremisina ZP, Afanas’ev IB. 1992. Release of active oxygen radicals by leukocytes of Fanconi anemia patients. Journal of Leukocyte Biology 52(3):357–362. [DOI] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493(7432):356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP, Singh B. 2003a. High Incidence of Head and Neck Squamous Cell Carcinoma in Patients With Fanconi Anemia. JAMA Otolaryngology–Head & Neck Surgery 129(1):106–112. [DOI] [PubMed] [Google Scholar]
- Kutler DI, Patel KR, Auerbach AD, Kennedy J, Lach FP, Sanborn E, Cohen MA, Kuhel WI, Smogorzewska A. 2016. Natural history and management of Fanconi anemia patients with head and neck cancer: A 10-year follow-up. Laryngoscope 126(4):870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD. 2003b. A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101(4):1249–1256. [DOI] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475:53. [DOI] [PubMed] [Google Scholar]
- Larsen NB, Gao AO, Sparks JL, Gallina I, Wu RA, Mann M, Räschle M, Walter JC, Duxin JP. 2019. Replication-Coupled DNA-Protein Crosslink Repair by SPRTN and the Proteasome in Xenopus Egg Extracts. Molecular Cell 73(3):574–588.e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latino-Martel P, Chan DSM, Druesne-Pecollo N, Barrandon E, Hercberg S, Norat T. 2010. Maternal Alcohol Consumption during Pregnancy and Risk of Childhood Leukemia: Systematic Review and Meta-analysis. Cancer Epidemiology Biomarkers & Prevention 19(5):1238. [DOI] [PubMed] [Google Scholar]
- Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. 2012. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci U S A 109(12):4491–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Diotti R, Pujara K, Batish SD, Hanenberg H, Auerbach AD. 2005. Spectrum of sequence variations in the FANCA gene: an International Fanconi Anemia Registry (IFAR) study. Hum Mutat 25(2):142–149. [DOI] [PubMed] [Google Scholar]
- Li F, Raczynska JE, Chen Z, Yu H. 2019. Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan. Cell Reports 26(12):3336–3346.e3334. [DOI] [PubMed] [Google Scholar]
- Liang CC, Li Z, Lopez-Martinez D, Nicholson WV, Venien-Bryan C, Cohn MA. 2016. The FANCD2-FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat Commun 7:12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Zhan B, Yoshikawa Y, Haas W, Gygi SP, Cohn MA. 2015. UHRF1 is a sensor for DNA interstrand crosslinks and recruits FANCD2 to initiate the Fanconi anemia pathway. Cell Rep 10(12):1947–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling C, Huang J, Yan Z, Li Y, Ohzeki M, Ishiai M, Xu D, Takata M, Seidman M, Wang W. 2016. Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discov 2:16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber R, Michaelson E, Fang Q, Campbell C, Pegg AE, Tretyakova N. 2008. Cross-Linking of the DNA Repair Protein O6-Alkylguanine DNA Alkyltransferase to DNA in the Presence of Antitumor Nitrogen Mustards. Chemical Research in Toxicology 21(4):787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. 2009. Proteomic Analysis of DNA−Protein Cross-Linking by Antitumor Nitrogen Mustards. Chemical Research in Toxicology 22(6):1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maddi K, Prgomet S, Kalayil S, Marinovic-Terzic I, Terzic J, Dikic I. 2016. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife 5:e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenti Garcia C, Mechilli M, Proietti De Santis L, Schinoppi A, Katarzyna K, Palitti F. 2009. Relationship between DNA lesions, DNA repair and chromosomal damage induced by acetaldehyde. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 662(1):3–9. [DOI] [PubMed] [Google Scholar]
- MacArthur AC, McBride ML, Spinelli JJ, Tamaro S, Gallagher RP, Theriault G. 2008. Risk of childhood leukemia associated with parental smoking and alcohol consumption prior to conception and during pregnancy: the cross-Canada childhood leukemia study. Cancer Causes & Control 19(3):283–295. [DOI] [PubMed] [Google Scholar]
- Madireddy A, Kosiyatrakul ST, Boisvert RA, Herrera-Moyano E, Garcia-Rubio ML, Gerhardt J, Vuono EA, Owen N, Yan Z, Olson S, Aguilera A, Howlett NG, Schildkraut CL. 2016. FANCD2 Facilitates Replication through Common Fragile Sites. Mol Cell 64(2):388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marietta C, Thompson LH, Lamerdin JE, Brooks PJ. 2009. Acetaldehyde stimulates FANCD2 monoubiquitination, H2AX phosphorylation, and BRCA1 phosphorylation in human cells in vitro: Implications for alcohol-related carcinogenesis. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 664(1):77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskey RS, Flatten KS, Sieben CJ, Peterson KL, Baker DJ, Nam H-J, Kim MS, Smyrk TC, Kojima Y, Machida Y, Santiago A, van Deursen JM, Kaufmann SH, Machida YJ. 2017. Spartan deficiency causes accumulation of Topoisomerase 1 cleavage complexes and tumorigenesis. Nucleic Acids Research 45(8):4564–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Von Hippel PH. 1977. Formaldehyde as a probe of DNA structure. 3. Equilibrium denaturation of DNA and synthetic polynucleotides. Biochemistry 16(15):3267–3276. [DOI] [PubMed] [Google Scholar]
- Mechilli M, Schinoppi A, Kobos K, Natarajan AT, Palitti F. 2008. DNA repair deficiency and acetaldehyde-induced chromosomal alterations in CHO cells. Mutagenesis 23(1):51–56. [DOI] [PubMed] [Google Scholar]
- Michaelson-Richie ED, Loeber RL, Codreanu SG, Ming X, Liebler DC, Campbell C, Tretyakova NY. 2010. DNA−Protein Cross-Linking by 1,2,3,4-Diepoxybutane. Journal of Proteome Research 9(9):4356–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl J, Zimmer J, Tarsounas M. 2016. Interplay between Fanconi anemia and homologous recombination pathways in genome integrity. EMBO J 35(9):909–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, Lechman E, Hermans KG, Eppert K, Konovalova Z, Ornatsky O, Domany E, Meyn MS, Dick JE. 2010. A Distinctive DNA Damage Response in Human Hematopoietic Stem Cells Reveals an Apoptosis-Independent Role for p53 in Self-Renewal. Cell Stem Cell 7(2):186–197. [DOI] [PubMed] [Google Scholar]
- Mórocz M, Zsigmond E, Tóth R, Enyedi MZ, Pintér L, Haracska L. 2017. DNA-dependent protease activity of human Spartan facilitates replication of DNA–protein crosslink-containing DNA. Nucleic Acids Research 45(6):3172–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim V, Rosselli F. 2009. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nature Cell Biology 11(6):761–768. [DOI] [PubMed] [Google Scholar]
- Nakano T, Morishita S, Katafuchi A, Matsubara M, Horikawa Y, Terato H, Salem AMH, Izumi S, Pack SP, Makino K, Ide H. 2007. Nucleotide Excision Repair and Homologous Recombination Systems Commit Differentially to the Repair of DNA-Protein Crosslinks. Molecular Cell 28(1):147–158. [DOI] [PubMed] [Google Scholar]
- Niedernhofer LJ. 2008. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA repair 7(3):523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D’Andrea AD, Bernards R. 2005. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 17(3):331–339. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Åstrand-Grundstr m I, Arvidsson I, Jacobsson Br, Hellstr m-Lindberg E, Hast R, Jacobsen SEW. 2000. Isolation and characterization of hematopoietic progenitor/stem cells in 5q-deleted myelodysplastic syndromes: evidence for involvement at the hematopoietic stem cell level. Blood 96(6):2012–2021. [PubMed] [Google Scholar]
- Nilsson L, Edén P, Olsson E, Månsson R, Åstrand-Grundström I, Strömbeck B, Theilgaard-Mönch K, Anderson K, Hast R, Hellström-Lindberg E, Samuelsson J, Bergh G, Nerlov C, Johansson B, Sigvardsson M, Borg Å, Jacobsen SEW. 2007. The molecular signature of MDS stem cells supports a stem-cell origin of 5q− myelodysplastic syndromes. Blood 110(8):3005–3014. [DOI] [PubMed] [Google Scholar]
- Niraj J, Färkkilä A, D’Andrea AD. 2019. The Fanconi Anemia Pathway in Cancer. Annual Review of Cancer Biology 3(1):457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbeck N, Langevin F, King G, de Wind N, Crossan Gerry P, Patel Ketan J. 2014. Maternal Aldehyde Elimination during Pregnancy Preserves the Fetal Genome. Molecular Cell 55(6):807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orta ML, Calderón-Montaño JM, Domínguez I, Pastor N, Burgos-Morón E, López-Lázaro M, Cortés F, Mateos S, Helleday T. 2013. 5-Aza-2′-deoxycytidine causes replication lesions that require Fanconi anemia-dependent homologous recombination for repair. Nucleic Acids Research 41(11):5827–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K, D’Andrea A, Niedernhofer LJ. 2009. Mouse models of Fanconi anemia. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 668(1):133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Miller S, Younus H, Vanam R, Chen CH, Mochly-Rosen D, Hurley TD. 2010. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol 17(2):159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel Lucas B, Rosado Ivan V, Burgos-Barragan G, Garaycoechea Juan I, Yu R, Arends Mark J, Chandrasekaran G, Broecker V, Wei W, Liu L, Swenberg James A, Crossan Gerry P, Patel Ketan J. 2015. Endogenous Formaldehyde Is a Hematopoietic Stem Cell Genotoxin and Metabolic Carcinogen. Molecular Cell 60(1):177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quievryn G, Zhitkovich A. 2000. Loss of DNA–protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis 21(8):1573–1580. [PubMed] [Google Scholar]
- Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, Passmore LA. 2014. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell 54(5):858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Kazane KR, Feng SJ, Zelin E, Bray NL, Schafer AJ, Floor SN, Corn JE. 2018. CRISPR-Cas9 genome editing in human cells occurs via the Fanconi anemia pathway. Nat Genet 50(8):1132–1139. [DOI] [PubMed] [Google Scholar]
- Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O, Chandrasekharappa SC, Schindler D, Auerbach AD, Smogorzewska A. 2015. Deficiency of UBE2T, the E2 Ubiquitin Ligase Necessary for FANCD2 and FANCI Ubiquitination, Causes FA-T Subtype of Fanconi Anemia. Cell Rep 12(1):35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, Buerstedde J-M, Gillespie DAF, Sale JE, Yamazoe M, Bishop DK, Takata M, Takeda S, Watanabe M, Swenberg JA, Nakamura J. 2007. Cells Deficient in the FANC/BRCA Pathway Are Hypersensitive to Plasma Levels of Formaldehyde. Cancer Research 67(23):11117. [DOI] [PubMed] [Google Scholar]
- Rio P, Navarro S, Wang W, Sanchez-Dominguez R, Pujol RM, Segovia JC, Bogliolo M, Merino E, Wu N, Salgado R, Lamana ML, Yanez RM, Casado JA, Gimenez Y, Roman-Rodriguez FJ, Alvarez L, Alberquilla O, Raimbault A, Guenechea G, Lozano ML, Cerrato L, Hernando M, Galvez E, Hladun R, Giralt I, Barquinero J, Galy A, Garcia de Andoin N, Lopez R, Catala A, Schwartz JD, Surralles J, Soulier J, Schmidt M, Diaz de Heredia C, Sevilla J, Bueren JA. 2019. Successful engraftment of gene-corrected hematopoietic stem cells in non-conditioned patients with Fanconi anemia. Nat Med 25(9):1396–1401. [DOI] [PubMed] [Google Scholar]
- Rohleder F, Huang J, Xue Y, Kuper J, Round A, Seidman M, Wang W, Kisker C. 2016. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucleic Acids Res 44(7):3219–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. 2011. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nature Structural &Amp; Molecular Biology 18:1432. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. 2007. Deletion of the Developmentally Essential Gene ATR in Adult Mice Leads to Age-Related Phenotypes and Stem Cell Loss. Cell Stem Cell 1(1):113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler D, Hoehn H. 1988. Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. American journal of human genetics 43(4):429–435. [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22(1):106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, Cohn MA, Gibbons RJ, Deans AJ, Niedzwiedz W. 2015. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol Cell 60(3):351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakeel S, Rajendra E, Alcon P, O’Reilly F, Chorev DS, Maslen S, Degliesposti G, Russo CJ, He S, Hill CH, Skehel JM, Scheres SHW, Patel KJ, Rappsilber J, Robinson CV, Passmore LA. 2019. Structure of the Fanconi anaemia monoubiquitin ligase complex. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, Wahengbam K, Steltenpool J, Andreassen PR, Williams DA, Joenje H, de Winter JP, Meetei AR. 2009. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 114(1):174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, Sung P, Meetei AR. 2010. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 37(6):879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Chistol G, Gao AO, Raschle M, Larsen NB, Mann M, Duxin JP, Walter JC. 2019. The CMG Helicase Bypasses DNA-Protein Cross-Links to Facilitate Their Repair. Cell 176(1–2):167–181 e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Alte F, Hewitt G, Sarek G, Maslen Sarah L, Tsutakawa Susan E, Borg A, Kjær S, Tainer John A, Skehel JM, Groll M, Boulton Simon J. 2016. Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease SPRTN. Molecular Cell 64(4):688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele J, Jentsch S. 2015. DNA–protein crosslink repair. Nature Reviews Molecular Cell Biology 16:455. [DOI] [PubMed] [Google Scholar]
- Stingele J, Schwarz Michael S, Bloemeke N, Wolf Peter G, Jentsch S. 2014. A DNA-Dependent Protease Involved in DNA-Protein Crosslink Repair. Cell 158(2):327–338. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, D’Andrea AD. 2006. Molecular pathogenesis of Fanconi anemia: recent progress. Blood 107(11):4223–4233. [DOI] [PubMed] [Google Scholar]
- Tian Y, Paramasivam M, Ghosal G, Chen D, Shen X, Huang Y, Akhter S, Legerski R, Chen J, Seidman MM, Qin J, Li L. 2015. UHRF1 contributes to DNA damage repair as a lesion recognition factor and nuclease scaffold. Cell Rep 10(12):1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419(6903):174–178. [DOI] [PubMed] [Google Scholar]
- Tsui V, Crismani W. 2019. The Fanconi Anemia Pathway and Fertility. Trends in Genetics 35(3):199–214. [DOI] [PubMed] [Google Scholar]
- Tulpule A, Lensch MW, Miller JD, Austin K, D’Andrea A, Schlaeger TM, Shimamura A, Daley GQ. 2010. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood 115(17):3453–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Twest S, Murphy VJ, Hodson C, Tan W, Swuec P, O’Rourke JJ, Heierhorst J, Crismani W, Deans AJ. 2017. Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway. Mol Cell 65(2):247–259. [DOI] [PubMed] [Google Scholar]
- Van Wassenhove LD, Mochly-Rosen D, Weinberg KI. 2016. Aldehyde dehydrogenase 2 in aplastic anemia, Fanconi anemia and hematopoietic stem cells. Molecular Genetics and Metabolism 119(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliou V, Pappa A, Estey T. 2004. Role of Human Aldehyde Dehydrogenases in Endobiotic and Xenobiotic Metabolism. Drug Metabolism Reviews 36(2):279–299. [DOI] [PubMed] [Google Scholar]
- Vaz B, Popovic M, Newman JA, Fielden J, Aitkenhead H, Halder S, Singh AN, Vendrell I, Fischer R, Torrecilla I, Drobnitzky N, Freire R, Amor DJ, Lockhart PJ, Kessler BM, McKenna GW, Gileadi O, Ramadan K. 2016. Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Molecular cell 64(4):704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz B, Popovic M, Ramadan K. 2017. DNA-Protein Crosslink Proteolysis Repair. Trends in Biochemical Sciences 42(6):483–495. [DOI] [PubMed] [Google Scholar]
- Velazquez I, Alter BP. 2004. Androgens and liver tumors: Fanconi’s anemia and non-Fanconi’s conditions. American Journal of Hematology 77(3):257–267. [DOI] [PubMed] [Google Scholar]
- Vinciguerra P, Godinho SA, Parmar K, Pellman D, D’Andrea AD. 2010. Cytokinesis failure occurs in Fanconi anemia pathway–deficient murine and human bone marrow hematopoietic cells. The Journal of Clinical Investigation 120(11):3834–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulgaridou G-P, Anestopoulos I, Franco R, Panayiotidis MI, Pappa A. 2011. DNA damage induced by endogenous aldehydes: Current state of knowledge. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 711(1):13–27. [DOI] [PubMed] [Google Scholar]
- Walden H, Deans AJ. 2014. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 43:257–278. [DOI] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Schofield CJ. 2012. Mechanisms of human histone and nucleic acid demethylases. Current Opinion in Chemical Biology 16(5):525–534. [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, Abhyankar A, Sougnez C, Gabriel SB, Auerbach AD, Kowalczykowski SC, Smogorzewska A. 2015. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell 59(3):478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chan B, Tong M, Paung Y, Jo U, Martin D, Seeliger M, Haley J, Kim H. 2019a. Prolyl isomerization of FAAP20 catalyzed by PIN1 regulates the Fanconi anemia pathway. PLoS Genet 15(2):e1007983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jo U, Joo SY, Kim H. 2016. FBW7 regulates DNA interstrand cross-link repair by modulating FAAP20 degradation. Oncotarget 7(24):35724–35740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Cheng G, Balbo S, Carmella SG, Villalta PW, Hecht SS. 2009. Clear Differences in Levels of a Formaldehyde-DNA Adduct in Leukocytes of Smokers and Nonsmokers. Cancer Research 69(18):7170–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. 2000. Identification of DNA Adducts of Acetaldehyde. Chemical Research in Toxicology 13(11):1149–1157. [DOI] [PubMed] [Google Scholar]
- Wang R, Wang S, Dhar A, Peralta C, Pavletich NP. 2019b. DNA clamp function of the monoubiquitinated Fanconi Anemia FANCI-FANCD2 complex. bioRxiv:854133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang R, Peralta C, Yaseen A, Pavletich NP. 2019c. Structure of the Fanconi Anemia Core–UBE2T complex poised to ubiquitinate bound FANCI–FANCD2. bioRxiv:854158. [Google Scholar]
- Welch John S, Ley Timothy J, Link Daniel C, Miller Christopher A, Larson David E, Koboldt Daniel C, Wartman Lukas D, Lamprecht Tamara L, Liu F, Xia J, Kandoth C, Fulton Robert S, McLellan Michael D, Dooling David J, Wallis John W, Chen K, Harris Christopher C, Schmidt Heather K, Kalicki-Veizer Joelle M, Lu C, Zhang Q, Lin L, O’Laughlin Michelle D, McMichael Joshua F, Delehaunty Kim D, Fulton Lucinda A, Magrini Vincent J, McGrath Sean D, Demeter Ryan T, Vickery Tammi L, Hundal J, Cook Lisa L, Swift Gary W, Reed Jerry P, Alldredge Patricia A, Wylie Todd N, Walker Jason R, Watson Mark A, Heath Sharon E, Shannon William D, Varghese N, Nagarajan R, Payton Jacqueline E, Baty Jack D, Kulkarni S, Klco Jeffery M, Tomasson Michael H, Westervelt P, Walter Matthew J, Graubert Timothy A, DiPersio John F, Ding L, Mardis Elaine R, Wilson Richard K. 2012. The Origin and Evolution of Mutations in Acute Myeloid Leukemia. Cell 150(2):264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RA, Semlow DR, Kamimae-Lanning AN, Kochenova OV, Chistol G, Hodskinson MR, Amunugama R, Sparks JL, Wang M, Deng L, Mimoso CA, Low E, Patel KJ, Walter JC. 2019. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Kim H, Moreau LA, Puhalla S, Garber J, Al Abo M, Takeda S, D’Andrea AD. 2015. RNF4-mediated polyubiquitination regulates the Fanconi anemia/BRCA pathway. J Clin Invest 125(4):1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, Jiricny J, Takeda S, Hirota K. 2011. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A 108(16):6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Xie H, Zhong Y, Li D, Ke X, Ying H, Yu B, Zhang T. 2019. Severe Fanconi Anemia phenotypes in Fancd2 depletion mice. Biochem Biophys Res Commun 514(3):713–719. [DOI] [PubMed] [Google Scholar]
- Zadorozhny K, Sannino V, Belan O, Mlcouskova J, Spirek M, Costanzo V, Krejci L. 2017. Fanconi-Anemia-Associated Mutations Destabilize RAD51 Filaments and Impair Replication Fork Protection. Cell Rep 21(2):333–340. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dewar JM, Budzowska M, Motnenko A, Cohn MA, Walter JC. 2015. DNA interstrand cross-link repair requires replication-fork convergence. Nat Struct Mol Biol 22(3):242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-S, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, Watanabe-Smith K, Rathbun RK, Fleming WH, Bagby GC, Grompe M. 2010. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood 116(24):5140–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q-S, Watanabe-Smith K, Schubert K, Major A, Sheehan AM, Marquez-Loza L, Newell AEH, Benedetti E, Joseph E, Olson S, Grompe M. 2013. Fancd2 and p21 function independently in maintaining the size of hematopoietic stem and progenitor cell pool in mice. Stem Cell Research 11(2):687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QS, Tang W, Deater M, Phan N, Marcogliese AN, Li H, Al-Dhalimy M, Major A, Olson S, Monnat RJ Jr., Grompe M. 2016. Metformin improves defective hematopoiesis and delays tumor formation in Fanconi anemia mice. Blood 128(24):2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Yajima H, Huynh H, Zheng J, Callen E, Chen H-T, Wong N, Bunting S, Lin Y-F, Li M, Lee K-J, Story M, Gapud E, Sleckman BP, Nussenzweig A, Zhang CC, Chen DJ, Chen BPC. 2011. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. The Journal of Cell Biology 193(2):295. [DOI] [PMC free article] [PubMed] [Google Scholar]