Abstract
The emergence of COVID-19 disease due to SARS-CoV-2 at the end of 2019 was rapidly associated with the isolation of the strain from co-culture onto VERO cells. These isolations quickly made it possible to carry out the first tests for antiviral agents’ susceptibility and drug repurposing. However, it seems important to make an inventory of all the cells that can support the growth of this virus and evaluate possible differences between isolates. In the present work, we tested 4 strains of SARS-CoV-2 locally isolated on a panel of 34 cell lines present in our laboratory and commonly used for the isolation of human pathogenic microorganism. After inoculation, cells were observed for cytopathic effects and quantitative real-time polymerase reaction was used to measure the virus replication on the cells. We were able to obtain growth on 7 cell lines, 6 simian, and the human Caco-2. The cytopathogenic effects are variable, ranging from lysis of the cell monolayer in 48–72 h to no cytopathic effect in spite of intense multiplication, as in Caco-2 cells. Interestingly, effect and multiplication varied widely according to the strain tested. In this paper, we explored the species specificity and tissue tropism of SARS-CoV-2 in vitro on a panel of cells available in our laboratory and identified human and animal cell lines susceptible to support SARS-CoV-2 replication. Our work highlights the importance of testing multiple strains when testing antiviral molecules and performing patho-physiological analyzes.
Keywords: SARS-Cov2, Covid-19, Coronavirus, Culture, Cell lines
Introduction
The current outbreak of the novel severe acute respiratory syndrome (2019-nCov then Covid-19) due to severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) started in Wuhan, China, in late December 2019 and has spread to many other countries [1–4]. To date, more than 84,000 cases and more than 4600 deaths have been reported across China due to SARS-Cov2, mostly in the region of Hubei (WHO, [5]). SARS-Cov-2 has disseminated in 188 countries, with currently more than 52 million confirmed cases and 1 million deaths around the world.
Coronaviruses are enveloped, positive single-stranded large RNA viruses that infect also a wide range of animals. The first description of coronavirus was made in 1966 by Tirell and Bynoe, who cultivated the viruses from patients with colds [6]. They were named coronavirus because of their morphology, spherical virions with a core shell and surface resembling to a solar crown, in Latin corona. Coronaviruses are divided into 4 subfamilies alpha, beta, delta and gamma-coronaviruses. The first two originate from mammals, in particular bats, while the other two come from pigs and birds. The genome size of coronaviruses ranges from approximately 27 to 34 kilobases. Severe disease and fatalities are caused essentially by beta-coronaviruses, whereas alpha-coronaviruses cause asymptomatic or mildly symptomatic infections. SARS-CoV and Middle East Respiratory Syndrome coronavirus (MERS-CoV) belong to the beta-coronavirus cluster [7], as well as the SARS-CoV-2 [8].
In this crisis situation, isolation of causative virus is indispensable for developing and evaluating diagnostic tools and therapeutics assays. The first isolation of SARS-CoV-2 was performed on human airway epithelial cells in China [8]. Subsequently, like SARS-CoV and MERS-CoV [9, 10], SARS-CoV-2 was isolated on VERO cells, which are kidney epithelial cells extracted from African green monkey [11–13]. In this paper, we investigated the susceptibility of a number of cells lines available in our laboratory collection to SARS-CoV-2. These cells were derived from a variety of species and tissues routinely used for the culture of micro-organisms. After inoculation with SARS-CoV-2, cells were observed for cytopathic effects and quantitative real-time polymerase reaction was used to measure ongoing replication on the cells growing the virus.
Materials and methods
Virus routine propagation
SARS-CoV-2-IHUMI2, IHUMI3, IHUMI669, and IHUMI2123 strains were isolated from human nasopharyngeal swab as previously described [14] and used for all tests. The 4-passage strains were grown in VERO E6 (ATCC® CRL-1586™) before subculture in different cell lines in Minimum Essential Medium culture medium (Ref. 21090022, Thermo Fisher Scientific) with 4% fetal calf serum (Ref. 10270106, Thermo Fisher Scientific) and 1% L-glutamine (Ref. 25030024, Thermo Fisher Scientific), without antibiotics at 37 °C under 5% CO2.
Multiple cell line assays
The cell lines tested are listed in Table 1. These cells are either routinely or occasionally used for microorganism isolation or for various diverse research projects in our laboratory. Cell lines to be tested were inoculated in 96-well flat bottom microplates (Ref. 020035, Dutscher) at 2*105 cells/ml into their specific growth medium (Table 1), without antibiotics and incubated to reach sub-confluence. At this stage, cells were infected with SARS-CoV-2 strains at 10−1 and 10−4 dilutions of VERO E6 supernatant, after filtration of 0.2 μ. Each day, cells were observed for SARS-CoV-2 specific cytopathic effects (CPE) for 7 days using EVOS™ FL Digital Inverted Fluorescence Microscope (Invitrogen™) with × 10 objective. On day 0 and day 7 after infection, supernatants were collected for subsequent quantification using reverse transcription-polymerase chain reaction (RT-PCR) targeting E-gene as previously described [16]. The difference of Ct between day 0 and Day 7 correspond to the Delta Ct (Δ Ct). Each condition was performed in triplicate. For cells for which a CPE effect was observed, cells were incubated with 10-fold dilutions of different viruses and incubated for 7 days at 37 °C. Each condition was performed in quadruplicate. Median tissue culture infectious dose (TCID50) was determined by the Reed and Muench method [17]. All experiments involving SARS-CoV-2 cultures were carried out in a Biosafety level 3 laboratory and conducted under appropriate conditions.
Table 1.
Cell lines tested for their susceptibility to SARS-CoV-2
| Cell lines | Species of origin | Cell types | Characteristics | References | Culture medium | Culture conditions |
|---|---|---|---|---|---|---|
| Aa23 | Aedes albopictus | Larva cells | Morphology: epithelial, culture properties: adherent | ATCC® CCL-125™ | L15 Leibovitz + 10% FBS + 8% tryptose phosphate | 28 °C |
| C6/36 | Aedes albopictus | Larva cells | Morphology: epithelial-like, culture properties: adherent | ATCC® CRL-1660™ | L15 Leibovitz + 10% FBS + 8% tryptose phosphate | 28 °C |
| S2 | Drosophila melanogaster | Embryo cells | Morphology: macrophage-like, culture properties: semi-adherent and suspension | Thermo R69007 | Schneider medium + 10% FBS | 28 °C |
| ISE6 | Ixodes scapularis | Embryo cells | Morphology: neuron-like, culture properties: adherent | ATCC® CRL-11974 | L15B + 10% FBS | 28 °C |
| IPL-LD-65Y | Lymantria dispar | Larvae cells | Morphology: fibroblastic like, culture properties: semi-adherent and suspension | ACC 181 (DSMZ) | TC-100 + 101% FBS | 28 °C |
| BGM | Cercopithecus aethiops | Kidney cells | Morphology: epithelial-like, culture properties: adherent | ECACC 90092601 | MEM + 10% FBS | 37 °C 5% CO2 |
| VERO/hSLAM | Cercopithecus aethiops | Kidney cells | Morphology: fibroblastic, culture properties: adherent | 04091501-1VL | MEM + 5% FBS + | 37 °C 5% CO2 |
| MA104 | Cercopithecus aethiops | Kidney cells | Morphology: epithelial, culture properties: adherent | ATCC® CRL-2378.1™ | MEM + 10% FBS | 37 °C 5% CO2 |
| VERO 81 | Cercopithecus aethiops | Kidney cells | Morphology: epithelial, culture properties: adherent | ATCC® CCL-81™ | MEM + 4% FBS | 37 °C 5% CO2 |
| VERO E6 | Cercopithecus aethiops | Kidney cells | Morphology: epithelial, culture properties: adherent | ATCC® CRL-1586™ | MEM + 10% FBS | 37 °C 5% CO2 |
| LLC-MK2 | Macaca mulatta | Kidney cells | Morphology: epithelial, culture properties: adherent | ATCC® CCL-7™ | M199 + 1% FBS | 37 °C 5% CO2 |
| HT-29 | Homo sapiens | Cells from colorectal adenocarcinoma | Morphology: epithelial, culture properties: adherent | ATCC® HTB-38™ | DMEM/F12 + 10% FBS | 37 °C 5% CO2 |
| Caco-2 | Homo sapiens | Cells from Colorectal adenocarcinoma | Morphology: epithelial, culture properties: adherent | ATCC® HTB-37™ | DMEM + 10% FBS + 1%AA | 37 °C 5% CO2 |
| HELA | Homo sapiens | Cervix cells from adenocarcinoma | Morphology: epithelial, culture properties: adherent | ATCC® CCL-2™ | MEM + 10% FBS | 37 °C 5% CO2 |
| HCT-8 | Homo sapiens | Colon cells from ileocecal colorectal adenocarcinoma | Morphology: epithelial, culture properties: adherent | ATCC® CCL-244™ | RPMI + 10% FBS | 37 °C 5% CO2 |
| HEP-2 | Homo sapiens | HeLa derived cell line from laryngeal epidermoid carcinoma | Morphology: epithelial, culture properties: adherent | ATCC® CCL-23™ | MEM + 5% FBS + 1% AA | 37 °C 5% CO2 |
| ECV304 | Homo sapiens | Cells from human cord/urinary bladder carcinoma | Morphology: epithelial/endothelial, culture properties: adherent | ATCC® CRL-1998™ | RPMI + 10% FBS | 37 °C 5% CO2 |
| HL-60 | Homo sapiens | Promyeloblast cells from human peripherical blood from acute promyelocytic leukemia | Morphology: lymphoblast-like, culture properties: suspension | ATCC® CCL-240™ | RPMI + 10% FBS | 37 °C 5% CO2 |
| MRC5 | Homo sapiens | Cells from lung | Morphology: fibroblast, culture properties: adherent | ATCC® CCL-171™ | MEM + 10% FBS | 37 °C 5% CO2 |
| THP1 | Homo sapiens | Cells from peripheral blood from acute monolytic leukemia | Morphology: monocytes, culture properties: suspension | ATCC® TIB-202™ | RPMI + 10% FBS | 37 °C 5% CO2 |
| BHK21 | Mesocricetus auratus | Kidney cells | Morphology: fibroblast, culture properties: adherent | ATCC® CCL-10™ | MEM + 4% FBS | 37 °C 5% CO2 |
| McCoy | Mus musculus | Unknown | Morphology: fibroblast, culture properties: adherent | ATCC® CRL-1696™ | MEM + 4% FBS | 37 °C 5% CO2 |
| L929 | Mus musculus | Cells from subcutaneous areolar and adipose | Morphology: fibroblast, culture properties: adherent | ATCC® CCL1™ | MEM + 4% FBS | 37 °C 5% CO2 |
| P388D1 | Mus musculus | Cells from lymphoma | Morphology: lymphoblast, culture properties: suspension | ATCC® CCL-46™ | MEM + 10% FBS | 37 °C 5% CO2 |
| RAW 264.7 | Mus musculus | Cells from Abelson murine leukemia virus-induced tumor | Morphology: monocyte/macrophage, culture properties: adherent | ATCC® TIB-71™ | MEM + 10% FBS + AA | 37 °C 5% CO2 |
| BA 886 | Bos taurus | Cells from bovine aorta | Morphology: endothelial, culture properties: adherent | [15] | DMEM/F12 + 10% FBS | 37 °C 5% CO2 |
| MDCK | Canis familiaris | Cells from kidney | Morphology: epithelial, culture properties: adherent | ATCC® CCL-34™ | MEM + 10% FBS | 37 °C 5% CO2 |
| DH82 | Canis familiaris | Cells from malignant histiocytosis | Morphology: macrophage-like, culture properties: adherent | ATCC® CRL-10389™ | MEM + 10% FBS | 37 °C 5% CO2 |
| OA3.Ts | Ovis aries | Testis cells | Morphology: epithelial, culture properties: adherent | ATCC® CRL-6546™ | DMEM + 10% FBS | 37 °C 5% CO2 |
| MDOK | Ovis aries | Kidney cells | Morphology: epithelial, culture properties: adherent | ATCC® CRL-1633™ | MEM + 10% FBS + 1% AA + 1% pyruvate | 37 °C 5% CO2 |
| R05T | Rousettus aegyptiacus | Head of fetus | Morphology: epithelial, culture properties: adherent | Bei resources NR-49169 | DMEM/F12 + 10% FBS | 37 °C 5% CO2 |
| R06E | Rousettus aegyptiacus | Fetus cells | Morphology: epithelial, culture properties: adherent | Bei resources NR-49168 | DMEM/F12 + 10% FBS | 37 °C 5% CO2 |
| TB1 Lu | Tadarida brasiliensis | Lung cells | Morphology: epithelial, culture properties: adherent | ATCC® CCL-88™ | DMEM + 10% FBS | 37 °C 5% CO2 |
| XTC-2 | Xenopus laevis | Tadpole cells | Morphology: epithelial, culture properties: adherent | CellBank Riken® RCB0771 | L15 Leibovitz + 5% FBS + 8% tryptose phosphate | 28 °C |
FBS, fetal bovine serum; AA, nonessential amino-acids
Results
Table 1 presents the panel of 34 cell lines present in the laboratory and tested for their susceptibility to the SARS-Cov-2 virus. Among these cell lines, 7 are able to support SARS-CoV-2 multiplication and are presented in Table 2. For these seven cell lines that supported growth of the virus, the Δ Ct between day 0 and day 7 at dilution 10−1 varied between 3.85 and 9.76, as shown in Table 2. Besides VERO E6 in which the virus was isolated and propagated, 4 African green monkey kidney cell lines supported replication of SARS-CoV-2 (VERO 81, VERO SLAM, MA104, and BGM cells) and produced CPE 48 h after SARS-CoV-2 infection. All produced evident CPEs. One human cell line supported virus replication, an epithelial cell line from colorectal adenocarcinoma (Caco-2). Caco-2 showed only discrete modification as compared to control but no real CPE.
Table 2.
Tested cell lines permissive to SARS-CoV-2
| Cell lines | CPE | IHUMI2 | IHUMI3 | IHUMI669 | IHUMI2123 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Δ Ct d0–d7 Dil. 10−1 |
Δ Ct d0–d7 Dil. 10−4 |
TCID50/ml | Δ Ct d0–d7 Dil. 10−1 |
Δ Ct d0–d7 Dil. 10−4 |
TCID50/ml | Δ Ct d0–d7 Dil. 10−1 |
Δ Ct d0–d7 Dil. 10−4 |
TCID50/ml | Δ Ct d0–d7 Dil. 10−1 |
Δ Ct d0–d7 Dil. 10−4 |
TCID50/ml | ||
| BGM | 48H | 5.17 | 11.3 | 6,3E+07 | 7.33 | 15.65 | 9,3E+04 | 9.76 | 17.32 | 6,3E+05 | 7.29 | 16.52 | 9,3E+06 |
| VERO/hSLAM | 48H | 6.48 | 15.66 | 2,0E+07 | 3.85 | 16.57 | 6,3E+06 | 7.51 | 17.53 | 6,3E+04 | 6.8 | 16.06 | 1,2E+05 |
| MA104 | 48H | 5.6 | 16.17 | 6,3E+07 | 4.12 | 9.37 | 4,3E+04 | 5.67 | 10.58 | 1,2E+05 | 6.05 | ˂ 3 | 2,0E+05 |
| VERO 81 | 48H | 5.25 | 14.92 | 4,3E+07 | 6.09 | 14.95 | 9,3E+07 | 6.23 | 12.87 | 4,3E+06 | 6.39 | 15.81 | 6,3E+05 |
| VERO E6 | 48H | 5.1 | 12.9 | 6,3E+06 | 3.97 | 13.03 | 4,3E+05 | 5.41 | 11.57 | 2,0E+04 | 4.88 | 13.94 | 9,3E+04 |
| LLC-MK2 | NO * | 4.65 | 15.07 | ND | 4.27 | ˂ 3 | ND | 5.03 | ˂ 3 | ND | 3.88 | ˂ 3 | ND |
| Caco-2 | NO * | 6.28 | 17.26 | ND | 5.92 | ˂ 3 | ND | 6.69 | 16.44 | ND | 5.43 | ˂ 3 | ND |
Dil., SARS-Cov-2 virus dilution; ND, not determined; d, day
*Modifications were observed
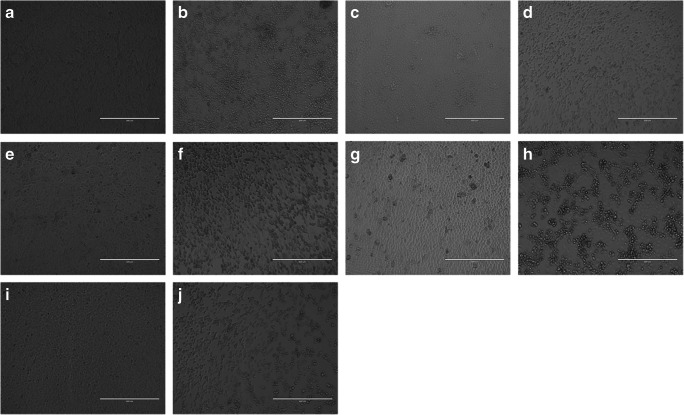
The morphological changes observed in the different cell lines are shown in Fig. 1.
Fig. 1.
Morphological changes observed in the different cell lines. a Noninfected VERO cells (× 10). b SARS-Cov-2 infected VERO cells at 48 h post-infection (× 10). c Non infected E6 VERO cells (× 10) d SARS-Cov-2 infected E6 VERO cells at 48 h post-infection (× 10). e Noninfected VERO/hSLAM cells (× 10). f SARS-Cov-2 infected VERO/hSLAM cells at 48 h post-infection (× 10). g Noninfected MA104 cells (× 10). h SARS-Cov-2 infected MA104 cells at 48 h post-infection (× 10). i Noninfected BGM cells (× 10). j SARS-Cov-2 infected BGM cells at 48 h post-infection (× 10)
LLC-MK2, a rhesus macaque epithelial kidney cell line, did not produce evident CPE.
For these seven cell lines that supported growth of the virus, the Δ Ct between day 0 and day 7 at dilution 10−4 varied between 9.37 and 17.32 as shown in Table 2. Viral multiplication was not associated with the intensity of CPE. Interestingly, according to the strain tested, major differences could be observed between permissivity, level of multiplication, and TCID50. In example, strain IHUMI2 as nearly always the highest TCID50 in spite its level of multiplication tested by RT-PCR can be lower. Twenty-seven other cell lines, derived from various species like insect, human, rodent, bovine, dog, sheep and bat cell lines, did not present any morphological changes or CPE and no difference of Δ Ct was observed.
Discussion
In the context of the SARS-CoV-2 epidemic, it was first important to develop rapid methods to isolate the virus. This was done easily using the common VERO E6 cell line, a highly virus permissive interferon deficient cell line [18]. In order to produce the virus in large quantities for vaccine research, to identify potential antiviral compounds, to understand intracellular trafficking and to develop innovative therapeutic approach, it is important to have other cell line, especially from human origin. In this paper, we explored the species specificity and tissue tropism of SARS-CoV-2 in vitro on a panel of cells available in our microbiology laboratory and identified human and animal cell lines susceptible to support SARS-CoV-2 multiplication.
Previous published reports showed that several monkey kidney cell lines are susceptible to SARS-CoV-2, specifically classical VERO cells, VERO E6 cells, VERO h/SLAM cells [8, 11–13, 19–22]. In this paper, we showed that all kidney cells derived from two species of monkey (African green monkey and rhesus macaque) support the growth of SARS-CoV-2, and all these cells, except for LLC-MK2 cell lines, presented CPE at 48 h post-infection. Unsurprisingly, MA104, BGM and LLC-MK2 already tested for SARS-CoV with very early CPE [23] and not previously tested with SARS-CoV-2, supported its growth.
In our first tests, HEP-2, an endothelial cell line suspected to be derived from laryngeal epidermoid carcinoma but in fact a clone derived from HELA cells [24], was first identified as susceptible to SARS-CoV-2 infection. SARS-CoV2 infection on our HEP-2 cells induced CPE after 120 h of infection with high virus multiplication. This result was unexpected, as previous studies on SARS-CoV showed that this virus did not infect HEP-2 cell lines, with no observable CPE or virus multiplication [23]. Based on PCR amplification of HPV18, a virus that chronically infects HEP-2 and HELA cells, we could detect that our HEP-2 cell line was misidentified. A new batch of cells ordered to the ATCC allowed to confirm that HEP-2 cells do not support growth of SARS-CoV-2.
A unique human cell line, Caco-2, epithelial cells from colorectal adenocarcinoma, was susceptible to SARS-CoV-2 with medium virus multiplication, but no specific CPE. Instead of CPE, we observed that the cell layer appears to be mottled more rapidly than in the control. This effect is rather seen in aging uninfected Caco-2. Previous studies showed that SARS-CoV and SARS-CoV-2 can infect Caco-2 cell lines [25, 26]. For SARS-CoV infections, CPE appeared on Caco-2 cell line 48 h post-infection [26], whereas, as observed, no obvious cell damage was found for SARS-CoV-2 infections [25]. This capability of SARS-CoV-2 to infect Caco-2 cells, could explain why patients infected with the virus present commonly gastrointestinal symptoms [27]. Moreover, SARS-CoV-2 RNA was detected in stools of patients infected with the virus, raising the question of viral gastrointestinal infection and fecal-oral transmission routes [28, 29]. However, to our knowledge, the virus could not be isolated from stools of infected patients.
We showed that 8 other human cells lines were not susceptible to SARS-CoV-2 (HEP-2, HT-29, HELA, HCT-8, ECV-304, HL-60, MRC5, and THP1 cell lines). In a recent paper of Chu et al. [25], SARS-CoV-2 was inoculated on 9 human cell lines. They showed that SARS-CoV-2 replicates also on Calu3 (lung adenocarcinoma), Huh7 (hepatocellular carcinoma), U251 (glioblastoma), and 293T (embryonic kidney) cell lines, whereas no growth was observed on A549 (lung adenocarcinoma), HFL (embryonic lung fibroblasts), and RD (rhabdomyosarcoma) cell lines. These data are consistent with the results observed in our study.
In this latter study, they evaluated the cell tropism profile of SARS-CoV-2 in non-human and non-primate cells originating from different animal species and showed that SARS-CoV-2 replicate in cat (feline kidney CRFK cells), rabbit (RK-13 rabbit kidney cells), and pig cells (PK-15 porcine kidney cells). In our study, we evaluated the susceptibility of SARS-CoV-2 in 19 animal cell lines. SARS-CoV-2 did not infect insect cells (Aa23, C6/36, S2, ISE6, and IPL-LD-65Y cells), rodent cells (BHK-21, McCoy, L929, P388 D1, and RAW 264.7 cells), bovine cells (BA886), bat cells (R05T, R06E, TB1 Lu cells), frog cells (XTC-2), dog cells (DH-2, MDCK cells), and sheep cells (OA3.Ts, MDOK cells).
Cellular entry of coronaviruses depends on the binding of the spike (S) protein to a specific cellular receptor and subsequent S protein priming by cellular proteases. Similarly to SARS-CoV [30, 31], SARS-CoV-2 seems to employ angiotensin-converting enzyme 2 (ACE2) as a receptor for cellular entry, and priming to be performed by the Transmembrane Serine Protease 2(TMPRSS2) [20, 32, 33]. This likely explains the specific permissivity of animal and kidney cell lines to the virus. ACE2 is expressed in various human tissues, such as heart, kidney, and testes, in addition to the lungs [34], indicating that SARS-CoV-2 may infect other tissues aside from the lungs. Moreover, Zhou et al. demonstrated that overexpressing ACE2 from different species in HeLa cells with human ACE2, pig ACE2, civet ACE2 (but not mouse ACE2) allowed SARS-CoV-2 infection and replication [20]. Hoffmann et al. reported similar findings for human and bat ACE-2 [35]. Additionally, Hoffmann et al. showed that treating VERO E6 cells, a monkey kidney cell line known to permit SARS-CoV replication, with an Anti-ACE-2 Antibody blocked the entry of vesicular stomatitis virus pseudotypes expressing the SARS-CoV-2 S protein [35]. A recent study conducted by Wang et al. reported that the existence of the novel SARS-CoV-2 (CD147-SP) route in host cells [36]. All these data suggest that SARS-CoV-2 is able to infect different tissues in human, but is also able to infect animals, and these information are concomitant with the variety of cell line that SARS-CoV-2 is able to infect. Finally, using 4 different isolates sampled at different time during the epidemic, we can observe that result of growth measured by RT-PCR and TCID50 can vary greatly and independently. This is of paramount importance because most studies testing drugs, survival on surfaces and efficiency of disinfectants are usually based on test performed on a unique strain. Our work could be an explanation of some discrepant results obtained i.e. about survival of SARS-CoV-2 on different supports [37–39]. Our results suggest all phenotypic tests should rather include a panel of strains.
Authors’ contributions
Wurtz Nathalie: writing–original draft preparation, analysis of results
Penant Gwilherm: methodology, investigation, writing–original draft preparation
Duclos Nathalie and Priscilla Jardot: methodology, investigation
Bernard La Scola: conceptualization, supervision, writing
Funding
This work has benefited from French state support, managed by the Agence Nationale de la Recherche (ANR), including the ‘Programme d’investissement d’avenir’ under the reference Méditerranée Infection 10-1AHU-03 and Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they no conflict of interest.
Ethics approval
Not applicable
Consent to participate
Not applicable
Code availability
Not applicable
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nathalie Wurtz and Gwilherm Penant contributed equally to this work.
References
- 1.Al-Tawfiq JA (2020) Asymptomatic coronavirus infection: MERS-CoV and SARS-CoV-2 (COVID-19). Travel Med Infect Dis. 10.1016/j.tmaid.2020 [DOI] [PMC free article] [PubMed]
- 2.Huang Y, Tu M, Wang S, Chen S, Zhou W, Chen D, et al. Clinical characteristics of laboratory confirmed positive cases of SARS-CoV-2 infection in Wuhan, China: a retrospective single center analysis. Travel Med Infect Dis. 2020;2020:101606. doi: 10.1016/j.tmaid.2020.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toit AD. Outbreak of a novel coronavirus. Nat Rev Microbiol. 2020;18:123. doi: 10.1038/s41579-020-0332-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO (2020) WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Accessed 27 May 2020
- 6.Tyrrell DAJ, Bynoe ML. Cultivation of viruses from a high proportion of patients with colds. Lancet. 1966;287:76–77. doi: 10.1016/s0140-6736(66)92364-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. New Engl J Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. New Engl J Med. 2003;348:1953–1966. doi: 10.1056/nejmoa030781. [DOI] [PubMed] [Google Scholar]
- 10.Park WB, Kwon N-J, Choe PG, Choi S-J, Oh HS, Lee SM, et al. Isolation of Middle East respiratory syndrome coronavirus from a patient of the 2015 Korean outbreak. J Korean Med Sci. 2016;31:315–320. doi: 10.3346/jkms.2016.31.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Nat Acad Sci. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J et al (2020) Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. Biorxiv. 10.1101/2020.03.02.972935
- 13.Park WB, Kwon N-J, Choi S-J, Kang CK, Choe PG, Kim JY, et al. Virus isolation from the first patient with SARS-CoV-2 in Korea. J Korean Med Sci. 2020;35:e84. doi: 10.3346/jkms.2020.35.e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al (2020) Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 1–3. doi: 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed]
- 15.Yunker C, Bryom B, Semu S. Cultivation of Cowdria ruminantium in bovine vascular endothelial cells. Kenya Vet. 1988;12:12–16. [Google Scholar]
- 16.Amrane S, Tissot-Dupont H, Doudier B, Eldin C, Hocquart M, Mailhe M, et al (2020) Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France, - January 31st to March 1st, 2020: a respiratory virus snapshot. Travel Med Infect Di 101632. doi: 10.1016/j.tmaid.2020.101632 [DOI] [PMC free article] [PubMed]
- 17.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 18.Desmyter J, Melnick JL, Rawls WE. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero) J Virol. 1968;2:955–961. doi: 10.1128/JVI.2.10.955-961.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, et al (2020) Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med 1–8. doi: 10.1038/s41591-020-0877-5 [DOI] [PubMed]
- 20.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caly L, Druce J, Roberts J, Bond K, Tran T, Kostecki R et al (2020) Isolation and rapid sharing of the 2019 novel coronavirus (SAR-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Australia. 10.5694/mja2.50569 [DOI] [PMC free article] [PubMed]
- 22.Kim J-M, Chung Y-S, Jo HJ, Lee N-J, Kim MS, Woo SH, et al. Identification of coronavirus isolated from a patient in Korea with COVID-19. Osong Public Heal Res Perspect. 2020;11:3–7. doi: 10.24171/j.phrp.2020.11.1.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye M, Druce J, Tran T, Kostecki R, Chibo D, Morris J, et al. SARS–associated coronavirus replication in cell lines. Emerg Infect Dis. 2006;12:128–133. doi: 10.3201/eid1201.050496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorphe P. A comprehensive review of Hep-2 cell line in translational research for laryngeal cancer. Am J Cancer Res. 2019;9:644–649. [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/s2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cinatl J, Hoever G, Morgenstern B, Preiser W, Vogel J-U, Hofmann W-K, et al. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell Mol Life Sci Cmls. 2004;61:2100–2112. doi: 10.1007/s00018-004-4222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tian Y, Rong L, Nian W, He Y. Review article: gastrointestinal features in COVID-19 and the possibility of faecal transmission. Aliment Pharm Ther. 2020;51:843–851. doi: 10.1111/apt.15731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First case of 2019 novel coronavirus in the United States. New Engl J Med. 2020;382:929–936. doi: 10.1056/nejmoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/jvi.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/s0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2000;275:33238–33243. doi: 10.1074/jbc.m002615200. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P et al (2020) SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. Biorxiv. 10.1101/2020.03.14.988345
- 37.Pastorino B, Touret F, Gilles M, de Lamballerie X, Charrel RN. Prolonged infectivity of SARS-CoV-2 in fomites. Emerg Infect Dis. 2020;26(9):2256–2257. doi: 10.3201/eid2609.201788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riddell S, Goldie S, Hill A, Eagles D, Drew TW. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol J. 2020;17(1):145. doi: 10.1186/s12985-020-01418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.