Abstract
Purpose
Recent studies have established the ability of centromere protein-A (CENP-A) to perform as an oncogene, regulating tumor progression. The aim of this research was to explore the relationship between CENP-A expression and clinical significance in gastric cancer (GC) patients.
Materials and Methods
Experiments with a microarray were conducted using the Affymetrix U133 plus 2.0 GeneChip Array. Upregulated differentially expressed genes (DEGs) were identified via the GEO2R and intersected using a Venn diagram. Bioinformatic databases Omcomine, GEPIA, and Ualcan were applied to investigate the expression level of CENP-A in GC. The real-time quantitative RT-PCR (qRT-PCR) was used to validate the level of CENP-A mRNA in GC. Immunohistochemistry (IHC) was employed to verify the protein levels of CENP-A, while the relationship between CENP-A expression and patients’ clinical parameters in GC was explored through the use of IHC. Kaplan-Meier analysis was conducted to evaluate the prognostic significance of CENP-A. Additionally, the Kaplan-Meier plotter database (KM plotter) was used to verify the prognostic function of CENP-A in GC patients.
Results
The results indicated that CENP-A was significantly overexpressed, both in protein and mRNA levels of GC tissues, compared to adjacent noncancerous tissues (P<0.05). Furthermore, we observed that CENP-A expression was positively associated with TNM stage, tumor classification, lymph node metastasis, distant metastasis, and Lauren type (P<0.05). Kaplan-Meier analysis showed that patients with an overexpression of CENP-A had significantly poorer overall survival (OS) times (P<0.05). Multivariate analysis suggested CENP-A may serve as an independent predicting factor for the poor outcome of GC patients.
Conclusion
Our results show that CENP-A upregulation is significantly correlated with advanced tumor progression and poor prognosis. CENP-A may function as a novel potential biomarker for predicting the clinical outcomes of GC patients.
Keywords: CENP-A, gastric cancer, survival analysis, poor prognosis
Introduction
Gastric cancer (GC) is one of the most aggressive diseases, with remarkably high lethality rates. It is the fifth most common malignancy and ranks in the top three major global causes of cancer-related deaths.1,2 Surgical resection is still the recommended therapy for GC, but despite undergoing this treatment, the 5-year survival rate remains low due to recurrence and metastasis.3 Despite advances in surgery, chemotherapy and radiotherapy, most GC patients still face a poor prognosis.4 Thus, discovery of the specific molecular mechanisms of GC tumorigenesis is becoming crucially important, in order to improve the therapeutic approaches for GC. Furthermore, new molecular markers that can predict prognosis or response to specific therapies, particularly in the era of targeted, personalized treatment, are urgently required.
In recent years, Centromere Protein (CENP) family members have been reported to be closely correlated with tumorigenesis, as well as tumor progression. The study of CENP and cancer is gaining more and more attention. As a member of the CENP family, CENP-A, also known as CenH3, is a centromere-specific 17-kDa protein, located in the chromaticity 2p23.3, with whole length genomic DNA of 8534bp, including three introns and five exons.5 It was identified as one of the earliest kinetochore components found in humans.6 It transpires to be a unique histone H3-like protein, only found in active centromeres. It replaces ordinary histone H3 in the centromeric chromatin, and plays an important role in specifying the location of centromeres on each chromosome.7,8 CENPA is needed for the assembly of the kinetochore and the establishment of cell viability, centromere activity and chromosome stability.9,10 Failure in any one of these events may lead to the development of malignances, mental illness, infertility and degenerative diseases.11,12 CENP-A upregulation is reported as possibly contributing to the spread of centromere heterochromatin along the arms of the chromosome, which may lead to anchoring deficiencies in microtubule-kinetochore along with genomic instability, ultimately causing carcinogenesis.13,14
Several studies indicate that CENP-A is widely expressed in solid tumors, including colorectal cancer, breast cancer, and lung cancer.15–17 The clinical value of CENP-A in GC is, however, still unknown, bringing new insight into the gastric cancer field.
In the present research, we firstly aim to seek and assess the clinical and pathological significance of CENP-A in GC, and its role in GC development.
Materials and Methods
Patients and Tissue Samples
Tissue specimens used for the subsequent experiments were collected from 120 GC patients who had received curative gastrectomy in the Department of Gastrointestinal Pancreatic Surgery, Zhejiang provincial people’s hospital (Hangzhou, Zhejiang, China), between 2011 and 2014, with ages ranging from 26 to 88 years (mean age = 58.8 years). None of GC patients received any other treatment prior to operation, such as radiation therapy and/or chemotherapy.
This study conforms to the modified Helsinki Declaration. Approval of the research protocol was obtained from the Ethics Committee of Zhejiang provincial people’s hospital. We confirm that all patients provided informed consent for their tissues to be used in this study.
Microarray Construction and Analysis
Total RNA was extracted from 20 pairs of gastric cancer and adjacent normal mucosa tissues, as instructed by the manufacturer (TRIzol TM, CA, Invitrogen, USA). Gene expression profiles were performed using the Affymetrix Gene Chip HG-U133 Plus 2.0 array. The array hybridization occurred in a Hybridization Oven 645 (Affymetrix Inc, Santa Clara, CA, USA) at 45 °C for 15 h. The arrays were then washed and stained in a Fluidics Station 450 (Affymetrix Inc, Santa Clara, CA, USA) according to the manufacturer’s instructions, and were eventually scanned by a Gene Chip scanner 3000 and Command Console Software 3.1, using default settings. A gene was regarded as differential if the value of T vs N signal log ratio changed as ≤0.5 fold or ≥2 fold.
Identification of Upregulated DEGs
Five gene expression profile datasets (GSE13911, GSE118916, GSE81948, GSE 29,998 and GSE26899) from the Gene Expression Omnibus (GEO) were applied for analysis. Identification of upregulated DEGs between GC and normal samples was obtained by GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/). Genes that met the cutoff criteria, logFC≥1.0 and adjusted P<0.05, were deemed to be upregulated DEGs. Next, a Venn diagram was utilized in order to obtain the intersection of the upregulated DEGs.
Bioinformatics Analysis
In this study, bioinformatic databases including Oncomine, GEPIA and Ualcan were applied to investigate the expression level of CENP-A. GEPIA is an interactive web-based tool, designed by Chinese scholars (Zefang Tang, Chenwei Li, etc.) for assessing mRNA expression data. It is based on 9736 tumors and 8587 noncancerous samples in the Cancer Genome Atlas (TCGA) and Genotype-tissue Expression dataset projects (GTEx).18
Next, Oncomine, a cancer microarray database,19 was employed to assess the CENP-A mRNA expression in GC. The screening conditions were as follows: gene name, CENP-A; analysis type, cancer vs normal analysis; type of data, mRNA. We selected a 1.5 fold change and top 10% gene rank as the threshold. A P-value ≤0.01 was considered significant.
In addition, we utilized the Ualcan database (http://ualcan.path.uab.edu.),20 which is a public online database providing estimates on the expression level of a target gene, along with its correlation with clinicopathologic features on patient survival, in order to measure the mRNA expression level of CENP-A in GC.
Quantitative Real-Time PCR (qRT-PCR)
We applied qRT-PCR to validate the level of CENP-A mRNA expression. Total RNA was isolated by a Trizol Reagent, following the manufacturer’s guidance. The mRNA was converted to cDNA through the application of SuperQuick RT MasterMix (CWBIO, Beijing, China), as instructed by the manufacturer. GAPDH acted as an endogenous control for CENP-A. The average was calculated, and the level of CENP-A expression was measured by 2–ΔΔCt, where ΔCt = Ct (CENP-A) – Ct (GAPDH). The thermocycling conditions were: 95 ° C for 5 min, amplification for 40 cycles, consisting of denaturation at 95 °C for 15 s, annealing at 60°C for 35 s, and extension at 60 °C for 30 s. The whole qRT-PCRs were executed in triplicate.
The CENP-A and GAPDH primers were listed as follows:
5′-CATCTCTTTGAGGACGCCTATCT-3′, (CENP-A, forward);
and 5′-TCCAGACAGCATCGCAGAATC-3′ (CENP-A, reverse);
5′-AGAAGGCTGGGGCTCATTTG-3′ (GAPDH, forward);
5′-AGGGGCCATCCACAGTCTTC-3′ (GAPDH, reverse).
Immunohistochemistry
Immunohistochemistry was applied to evaluate CENP-A protein expression levels. The paraffin-embedded, formalin-fixed, 4μm thick tissue sections were all dewaxed with xylene and dehydrated with alcohol, and then rinsed in PBS followed by antigen retrieval via high-pressure steam treatment in a citrate buffer (pH 6.0) for 20 mins. After incubation, with the primary rabbit anti-CENP-A monoclonal antibody at 1:400 dilution (Abcam, Cambridge, UK) held at 4 °C overnight, we washed the sections before incubating with a second antibody (SP9000; goat anti-mouse IgG; OriGene Technologies, Inc., Beijing, China) and stained with DAB (OriGene Technologies, Inc., Beijing, China). The nuclei were stained with a Mayer hematoxylin.
Two experienced pathologists performed blind analysis on all the slides. The rating method for CENP-A expression was based on staining strength and the proportion of positively stained tumor cells. In cases where the two pathologists gave differing scores, the final decision was made following discussion by two observers.
Staining strength and intensity was measured as follows: 0, negative; 1, weakly positive (+); 2, moderately positive (+ +); and 3, strongly positive (+ + +). Scores of 0 and 1+ were considered as negative for CENP-A upregulation, while scores of 2+ and 3+ were considered positive for CENP-A upregulation.
Statistical Analysis
All data were analyzed using SPSS19.0 software. The chi-square test was performed to evaluate the association between CENP-A expression and clinicopathologic parameters. The Kaplan-Meier method was employed to plot the survival curves, and the differences between subgroups were analyzed by the Log-rank test. A Cox proportional hazard model for multivariable analysis was used for factors with significance in univariable analysis. Two-tailed P <0.05 were deemed statistically significant.
Results
Identification of Upregulated DEGs
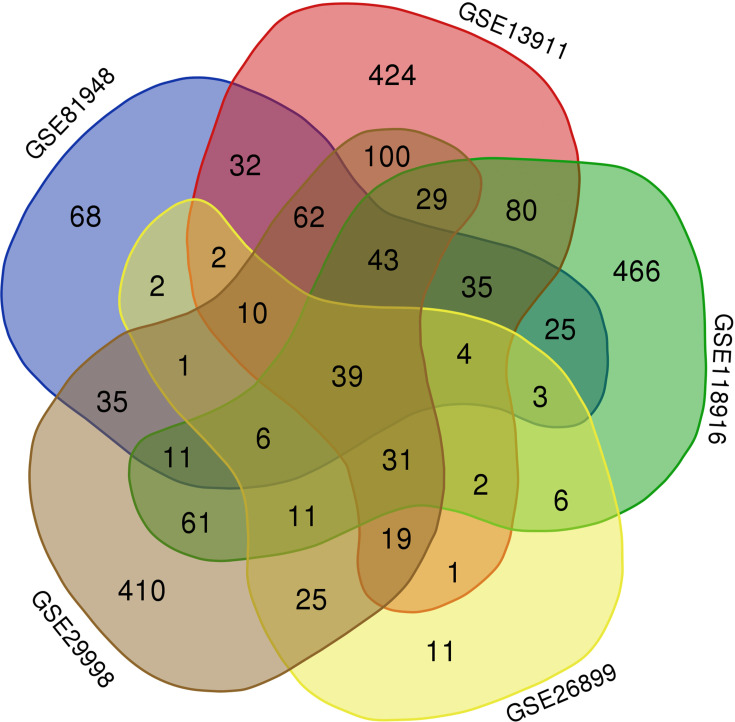
Among the five groups obtained from the Gene Expression Omnibus (GEO), a total of 39 DEGs were significantly upregulated, with the intersection presented in the form of a Venn diagram (Figure 1). Among the 39 upregulated DEGs, CENP-A, which has not been reported in GC, was selected for further study.
Figure 1.
Analysis of up-regulated differentially expressed genes (DEGs) in gastric cancer samples by Venn diagram. A total of 39 up-regulated DEGs were identified.
Expression of CENP-A mRNA in GC Determined by Bioinformatic Databases
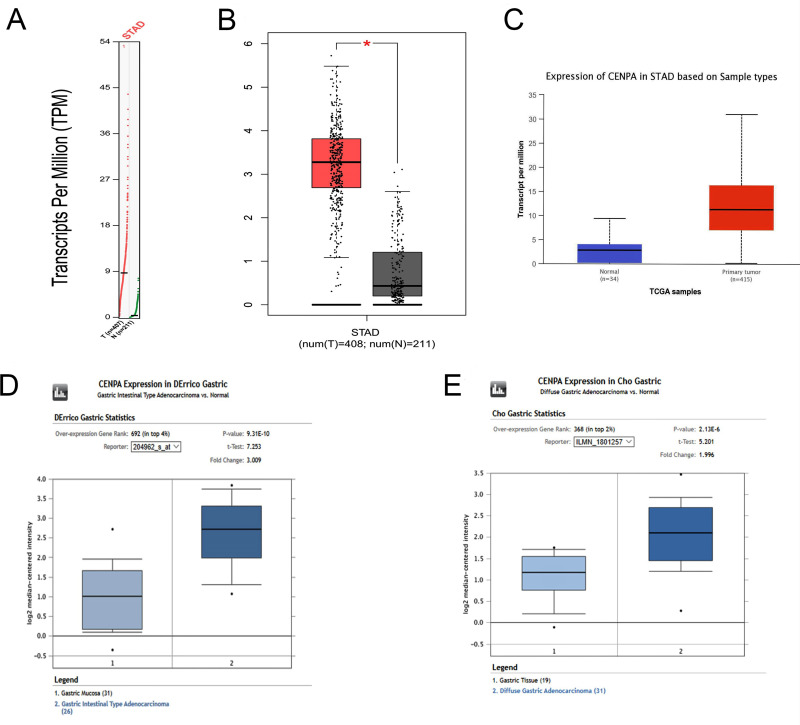
According to the GEPIA database, which is established on TCGA normal and GTEx data of 408 GC patients and 211 normal individuals, CENPA gene expression is significantly higher in GC patients when compared with normal individuals; the dot plot and boxplot were drawn (Figure 2A and B, P<0.05).
Figure 2.
Expression of CENP-A mRNA in gastric cancer from different bioinformatic databases. (A) Transcripts per million (TPM) of CENP-A in patients with gastric cancer and normal controls (STAD, stomach adenocarcinoma; T, tumor; N, normal tissue). (B) Box-whisker plot represented expression of CENP-A in gastric cancer compared with normal controls determined by the GEPIA database (*significant difference). (C) The expression level of CENP-A in gastric cancer versus normal controls, determined by the Ualcan database. (D, E) CENP-A was significantly upregulated in GC in two datasets (DErrico Gastric and Cho Gastric).
Moreover, we assessed CENP-A expression in the Ualcan database, consistent with GEPIA and Oncomine. The result demonstrated that CENPA expression was significantly higher in GC patients than normal (Figure 2C, P= 1.6E-12).
The mRNA expression level of CENP-A was additionally validated from two datasets (DErrico Gastric and Cho Gastric) by Oncomine.21,22 As shown in Figure 2D and E, we discovered significant upregulation of CENP-A in GC, compared with normal samples in both of the two datasets.
Upregulation of CENP-A Expression on the Microarray
Microarray profiles of primary GC patients were examined using human U133 Plus 2.0 GeneChip(r). Gene expression microarray signal quality achieved all Affymetrix data quality standards. SAM was used to scrutinize differences in gene expression. Following detection and statistical analysis of 20 pairs of GC microarray expression profiles, along with their corresponding noncancerous mucosa, and discovered that CENP-A overexpressed at least a threefold difference between cancer and normal tissues; CENP-A was significantly overexpressed in gastric cancer tissues (p<0.05) (Table 1).
Table 1.
Part of the Differentially Up-Regulated Genes
| Probe_Set_ID | P values | Fold Change (T vs N) | Gene_Symbol | Entrez Gene | Chromosome |
|---|---|---|---|---|---|
| 204849_at | 0.008 | 2.45 | TCFL5 | 10,732 | chr20q13.3-qter |
| 204855_at | 0.026 | 2.51 | SERPINB5 | 5268 | chr18q21.3 |
| 204886_at | 0.021 | 2.04 | PLK4 | 10,733 | chr4q28 |
| 204924_at | 0.008 | 2.15 | TLR2 | 7097 | chr4q32 |
| 204944_at | 0.001 | 2.07 | PTPRG | 5793 | chr3p21-p14 |
| 204959_at | 0.005 | 2.04 | MNDA | 4332 | chr1q22 |
| 204962_s_at | 0.007 | 3.75 | CENPA | 1058 | chr2p24-p21 |
| 204971_at | 0.006 | 0.41 | CSTA | 1475 | chr3q21 |
| 204992_s_at | 0.001 | 2.61 | PFN2 | 5217 | chr3q25.1 |
| 205009_at | 0.011 | 0.23 | TFF1 | 7031 | chr21q22.3 |
| 205042_at | 0.012 | 3.31 | GNE | 10,020 | chr9p13.3 |
| 205051_s_at | 0.002 | 0.39 | KIT | 3815 | chr4q11-q12 |
| 205052_at | 0.000 | 0.49 | AUH | 549 | chr9q22.31 |
| 205081_at | 0.008 | 2.18 | CRIP1 | 1396 | chr14q32.33 |
Note: P <0.05 were deemed statistically significant.
Abbreviations: T, tumor tissue; N, adjacent non-tumor tissue.
Expression of CENP-A mRNA in GC by RT-qPCR
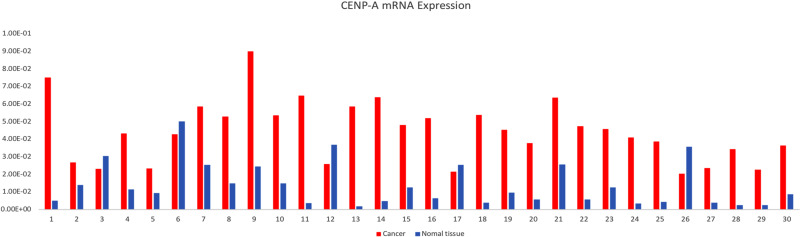
To further validate our gene array data, we analyzed the levels of CENP-A expression in 30 pairs of GC and corresponding noncancerous tissues at the mRNA level, by RT-qPCR. The results showed that 25 of the 30 patients (83.3%) possessed a higher expression level of CENP-A mRNA in GC tissues than in adjacent normal tissues. CENP-A mRNA levels in the GC tissues were significantly up-regulated (0.0445±0.0173 vs 0.0138±0.0124, P<0.05) (Figure 3). Moreover, CENP-A mRNA levels in GC patients with late stage (stage III/IV) were significantly higher than GC patients with early stage (stage I/II) (0.0506±0.0103 vs 0.0246±0.0039, P<0.05).
Figure 3.
Expression of CENP-A mRNA in GC tissues (0.0445±0.0173) and non-cancerous tissues (0.0138±0.0124). CENP-A mRNA was significantly overexpressed in GC tissues compared with non-cancerous tissues (P<0.05).
Expression of CENP-A Protein in GC by IHC
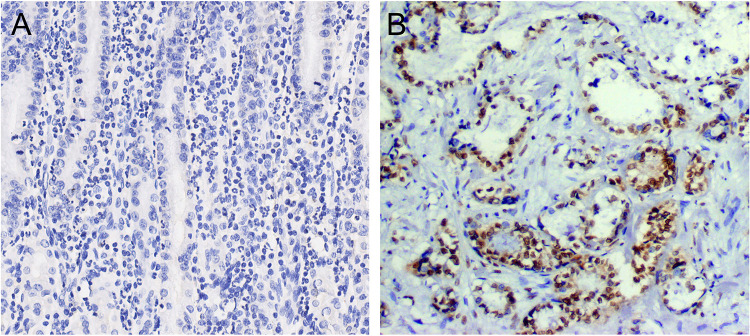
To initially detect CENP-A expression in GC, we collected 120 GC samples and 30 noncancerous samples. IHC was applied to investigate the expression levels of CENP-A protein in GC specimens. Among 120 cancer samples, 85 GC samples (70.8%) had a high expression of CENP-A, with a score defined as positive. Absent (8/30; 26.67%) or weak (22/30; 73.33%) CENP-A protein expression was found in the mucosa of non-cancerous tissues (Figure 4). Additionally, the protein expression level of CENP-A in GC patients with stage III/IV (64/81; 79.01%) was significantly higher than GC patients with stage I/II (21/39; 53.85%) (Table 2).
Figure 4.
Immunohistochemical staining for CENP-A in noncancerous tissues and gastric cancer lesions. (A) CENP-A negative in noncancerous tissues. (B) CENP-A was overexpressed in gastric cancer (magnifications were×400).
Table 2.
Relationship Between CENP-A Expression and Clinicopathological Characteristics of GC Patients
| Clinicopathologic Features | Cases (n) | CENP-A Level | P value | |
|---|---|---|---|---|
| Positive | Negative | |||
| Gender | 0.187 | |||
| Male | 76 | 57 | 19 | |
| Female | 44 | 28 | 16 | |
| Age | 0.629 | |||
| ≥50Y | 86 | 62 | 24 | |
| <50Y | 34 | 23 | 11 | |
| Tumor | 0.946 | |||
| ≥5cm | 68 | 48 | 20 | |
| <5cm | 52 | 37 | 15 | |
| Tumor classification | 0.001* | |||
| Well+ Moderate | 54 | 30 | 24 | |
| Poor | 66 | 55 | 11 | |
| TNM Stage | 0.005* | |||
| I, II | 39 | 21 | 18 | |
| II1, IV | 81 | 64 | 17 | |
| Local invasion | 0.062 | |||
| T1–2 | 43 | 26 | 17 | |
| T3–4 | 77 | 59 | 18 | |
| Lymph node metastasis | 0.049* | |||
| Yes | 87 | 66 | 21 | |
| No | 33 | 19 | 14 | |
| Distant metastasis | 0.008* | |||
| Yes | 34 | 30 | 4 | |
| No | 86 | 55 | 31 | |
| Lauren type | 0.000* | |||
| Intestinal | 48 | 25 | 23 | |
| Diffuse | 72 | 60 | 12 | |
Notes: P values are based on χ2 or Fisher’s exact test. *Significant difference.
Correlations Between CENP-A Expression Level and Clinical Features of GC Patients
In order to investigate the impact of CENP-A on the prognosis of GC patients, we looked into the correlation between CENP-A expression and clinicopathological characteristics in the 120 GC patients. We found upregulation of CENP-A was significantly correlated with TNM stage, histological differentiation, lymph node metastasis, Lauren type and distant metastasis (P<0.05). There were no correlations, however, between CENP-A protein expression and age, gender, or tumor size (P>0.05) (Table 2). Multivariate regression analysis revealed that TNM stage, high CENP-A protein expression, lymph node metastasis and distant metastasis were independent prognostic factors for gastric cancer patients (P<0.05) (Table 3).
Table 3.
Univariate and Multivariate Analysis of Overall Survival (OS) of Gastric Carcinoma (GC)
| Variables | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P value | OR | 95% CI | P value | |
| Gender (M/F) | 0.838 | 0.386–1.822 | 0.656 | NR | ||
| Age (≤55Y/>55Y) | 0.890 | 0.421–1.881 | 0.761 | NR | ||
| Tumor classification (W+M/P) | 1.621 | 0.765–3.433 | 0.207 | NR | ||
| TNM Stage (I–II/III–IV) | 9.533 | 3.890–23.364 | 0.000* | 2.405 | 1.063–4.648 | 0.025* |
| Size of tumor (<5cm/≥5cm) | 1.370 | 0.647–2.903 | 0.411 | NR | ||
| Lauren type (Intestinal/diffuse) | 1.054 | 0.502–2.215 | 0.890 | NR | ||
| Local invasion (T1–2/T3–4) | 4.425 | 1.909–10.254 | 0.001* | 1.401 | 0.564–2.968 | 0.435 |
| Lymph node metastasis (+/–) | 5.308 | 2.290–12.301 | 0.000* | 3.654 | 1.075–6.965 | 0.009* |
| Distant metastasis (+/–) | 3.130 | 0.078–0.218 | 0.000* | 3.134 | 1.454–4.875 | 0.000* |
| CENP-A expression (+/–) | 3.389 | 1.494–7.689 | 0.004* | 1.976 | 1.021–3.865 | 0.039* |
Note: *Significant difference.
Abbreviations: OR, odds ratio; CI, confidence interval; M, male; F, female; W, well; M/P, Moderate/Poor; +, positive; –, negative; NR, variable was not included in the result model.
Prognostic Significance of CENP-A for GC
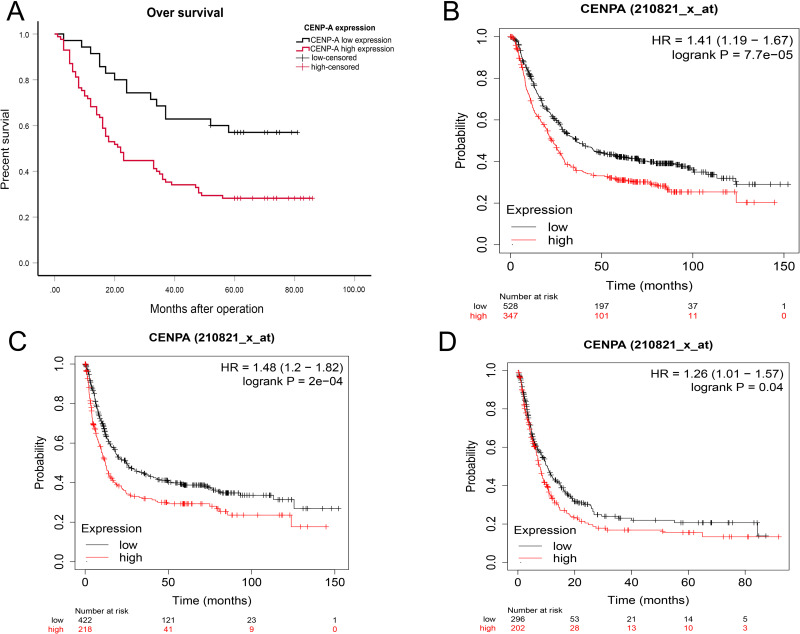
The relationship between CENP-A expression levels and the survival condition of 120 GC patients was analyzed. The Kaplan-Meier survival curves revealed that GC patients with overexpressed CENP-A exhibited significantly worse OS than those who did not overexpress (Figure 5A, P < 0.05). CENP-A was identified as an independent risk factor for the prognosis of GC patients.
Figure 5.
Prognostic roles of CENP-A in gastric cancer. (A) Survival curves are plotted for gastric cancer patients (n =120). (B) The Kaplan-Meier plotter indicated that the GC patients with overexpressed CENP-A had significantly worse OS. (C) The Kaplan-Meier plotter showed the CENP-A high expression group had worse first progress (FP). (D) The Kaplan-Meier plotter revealed CENP-A upregulation correlated with worse post-progression survival (PPS).
Prognostic Analysis in the Kaplan-Meier Plotter Database
Using the KM plotter database, we further investigated the correlation between CENP-A expression and GC patients’ prognosis. Observations show that high CENP-A mRNA expression was significantly associated with worse overall survival (OS), first progress (FP) and post-progression survival (PPS) in GC patients (Figure 5B–D, P < 0.05).
Discussion
Despite great advances in cancer diagnosis and comprehensive therapy, the prognosis of gastric cancer patients is still far from optimistic. Biomarkers that can predict the metastasis or prognosis of GC are also scarce. Thus, further study of GC development and the metastatic molecular mechanisms to explore novel biomarkers, allowing early diagnosis and drug targets for tumor therapy, has provoked significant attention.23
CENP family members have been considered as vital functional genes in cancer development. For instance, members of CENP, including CENP-H, CENP-M, and CENP-F, were all revealed to be correlated with tumors. Specifically, CENP-H is known to contribute to poor prognoses in tongue cancer patients,24 while overexpression of CENP-M promoted hepatocarcinogenesis.25 The development and progression of nasopharyngeal carcinoma, gastrointestinal stromal tumors, and breast cancer are often attributed to CENP-F.26,27
In this study, we focused on one major component of the CENP family, CENP-A; an important histone H3 variant, exhibiting a typical core histone tissue consisting of a structured histone folding domain with flexible N-terminal and C-terminal tails on both sides.10 The C terminal location of CENP-A is an important centromere region. Its histone domain, known as the CENP-A targeting domain (CATD), consists of a loop L1 structure plus an adjacent 2 helix domain.5,28 CATD is of great significance for CENPA recruitment and normal assembly of centromeres. As a centromere-specific variant, CENP-A is deemed a key epigenetic mark for centromere identity and propagation.6 The transcription of CENP-A begins in the G1-S stage, with the peak of CENP-A synthesis in the stage of G2. It possesses two main functions: first, it is a necessary and sufficient condition for the formation and maintenance of centromeres. Second, it forms a platform for kinetochore assembly and mediates chromosome separation.5,9
CENP-A, along with CENP-C and CENP-T contributes to the maintenance of centromere integrity in human cells;29 too much or too little CENP-A may destroy genome integrity. Defects in kinetochore function may lead to chromosome missegregation and tumorigenesis.
During our work we identified the upregulated DEGs via GEO2R and a Venn diagram, and selected CENP-A, previously unreported in GC, for study. Microarray experiments were conducted on the Affymetrix U133 plus 2.0 Gene Chip Array. Through microarray, we discovered the significantly high expression of CENP-A in GC tissues. Moreover, we employed bioinformatic databases (Oncomine, GEPIA and Ualcan) to assess the mRNA expression level of CENP-A; the results indicated that CENP-A expression in GC was far in excess of normal levels. Next, we validated these findings using immunohistochemistry (IHC) and Real-Time quantitative PCR (RT-qPCR) in GC and adjacent normal tissues. We confirmed that CENP-A is overexpressed in GC. Besides, CENP-A expression levels in GC patients with late stage (stage III/IV) were significantly higher than GC patients with early stage (stage I/II), both at the level of mRNA and protein. The results demonstrated that CENPA might play an important role in GC progression.
We also analyzed the relationship between CENP-A expression and clinicopathologic characteristics. We demonstrated that up-regulation of CENP-A was closely correlated with TNM stage, types of histological differentiation, lymph node metastasis, Lauren type and distant metastasis (P<0.05). There was no correlation between CENP-A protein expression and age, gender, tumor size or local invasion (P>0.05). Further multivariate regression analysis revealed that CENP-A upregulation may serve as an independent predictor for the poor outcome of GC patients. The survival analysis demonstrated that patients with upregulation of CENP-A displayed a significantly poorer overall survival (OS) rate than those who had CENP-A down-regulation. Finally, we employed the Kaplan-Meier plotter to verify the levels of CENP-A mRNA expression and prognostic significance of CENP-A for GC. Consistent with our results, the KM plotter demonstrated that high CENP-A mRNA expression had a significant association with a poor prognostic outcome in GC patients.
In summary, our findings and previous results corroborate the overexpression of CENP-A in gastric cancer, with its atypical increased expression forecasting a poor prognosis. CENP-A may play a significant role on tumor progression, metastasis and prognosis, serving as a valuable survival marker to improve outlook and conceivably promote the development of novel treatments for GC.
There are, however, several limitations to this research. The mechanism of CENP-A in GC is far from clear; further investigation is required to fully elucidate, while translation of its functions into clinical practice will need in-depth study and additional experimental validation.
Funding Statement
No specific funding was disclosed.
Disclosure
The authors report no conflicts of interest for this work.
References
- 1.Ho SWT, Tan P. Dissection of gastric cancer heterogeneity for precision oncology. Cancer Sci. 2019;110(11):3405–3414. doi: 10.1111/cas.14191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 3.Thapa S, Chetry M, Huang K, et al. Significance of aquaporins’ expression in the prognosis of gastric cancer. Biosci Rep. 2018;38(3):BSR20171687. doi: 10.1042/BSR20171687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh HL, Tsai MM. Tumor progression-dependent angiogenesis in gastric cancer and its potential application. World J Gastrointest Oncol. 2019;11(9):686–704. doi: 10.4251/wjgo.v11.i9.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding WY, Huang D, He J. Research progress of correlation between CENP-A and malignant tumors. J Gannan Med Univ. 2018;38:1273–1276. [Google Scholar]
- 6.Remnant L, Booth DG, Vargiu G, et al. In vitro BioID: mapping the CENP-A microenvironment with high temporal and spatial resolution. Mol Biol Cell. 2019;30(11):1314–1325. doi: 10.1091/mbc.E18-12-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong P, Pidoux AL, Toda NRT, et al. Interspecies conservation of organisation and function between nonhomologous regional centromeres. Nat Commun. 2019;28(1):2343. doi: 10.1038/s41467-019-09824-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zasadzińska E, Huang J, Bailey AO, et al. Inheritance of CENP-A nucleosomes during DNA replication requires HJURP. Dev Cell. 2018;47(3):348–362. doi: 10.1016/j.devcel.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinshaw SM, Harrison SC. Kinetochore function from the bottom up. Trends Cell Biol. 2018;28(1):22–33. doi: 10.1016/j.tcb.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 10.Sharma AB, Dimitrov S, Ali H, et al. Centromeric and ectopic assembly of CENP-A chromatin in health and cancer: old marks and new tracks. Nucleic Acids Res. 2019;20(47):1051–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdivia MM, Hamdouch K, Ortiz M, Astola A. CENPA a genomic marker for centromere activity and human diseases. Curr Genomics. 2009;10(5):326–335. doi: 10.2174/138920209788920985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahlke MA, Nechemia-Arbely Y. Guarding the genome: CENP-A-chromatin in health and cancer. Genes (Basel). 2020;16(11):810. doi: 10.3390/genes11070810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deyter GM, Hildebrand EM, Barber AD, et al. Histone H4 facilitates the proteolysis of the budding yeast CENP-A Cse4 centromeric histone variant. Genetics. 2017;205(1):113–124. doi: 10.1534/genetics.116.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Karpen GH, Zhang Q. Exploring the role of CENP-A Ser18 phosphorylation in CIN and Tumorigenesis. Cell Cycle. 2017;16(24):2323–2325. doi: 10.1080/15384101.2017.1387698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Clermont PL, Jiao W, et al. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int J Cancer. 2016;139(4):899–907. doi: 10.1002/ijc.30133 [DOI] [PubMed] [Google Scholar]
- 16.Liu WT, Wang Y, Zhang J, et al. A novel strategy of integrated microarray analysis identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic biomarkers in lung adenocarcinoma. Cancer Lett. 2018;425:43–53. doi: 10.1016/j.canlet.2018.03.043 [DOI] [PubMed] [Google Scholar]
- 17.Rahman MR, Islam T, Gov E, et al. Identification of prognostic biomarker signatures and candidate drugs in colorectal cancer: insights from systems biology analysis. Medicina (Kaunas). 2019;55(1):20. doi: 10.3390/medicina55010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes DR, Yu J, Shanker K, et al. A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/S1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;8(19):649–658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45(3):461–469. doi: 10.1016/j.ejca.2008.10.032 [DOI] [PubMed] [Google Scholar]
- 22.Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;1(17):1850–1857. doi: 10.1158/1078-0432.CCR-10-2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun T, Wang D, Ping Y, et al. Integrated profiling identifies SLC5A6 and MFAP2 as novel diagnostic and prognostic biomarkers in gastric cancer patients. Int J Oncol. 2020;56:460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao WT, Yu CP, Wu DH, et al. Upregulation of CENP-H in tongue cancer correlates with poor prognosis and progression. J Exp Clin Cancer Res. 2009;5(28):74. doi: 10.1186/1756-9966-28-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao Y, Najeeb RM, Ma D, et al. Upregulation of CENPM promotes hepatocarcinogenesis through mutiple mechanisms. J Exp Clin Cancer Res. 2019;38(1):458. doi: 10.1186/s13046-019-1444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int. 2019;11(19):264. doi: 10.1186/s12935-019-0986-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An F, Zhang Z, Xia M. Functional analysis of the nasopharyngeal carcinoma primary tumor-associated gene interaction network. Mol Med Rep. 2015;12(4):4975–4980. doi: 10.3892/mmr.2015.4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arimura Y, Tachiwana H, Takagi H, et al. The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism. Nat Commun. 2019;10(1):576. doi: 10.1038/s41467-019-08314-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giunta S, Funabiki H. Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc Natl Acad Sci U S A. 2017;114(8):1928–1933. doi: 10.1073/pnas.1615133114 [DOI] [PMC free article] [PubMed] [Google Scholar]