Abstract
Prostate cancer (PCa) is one of the most common epithelial malignant tumors and the fifth leading cause of cancer death in men. An increasing number of studies have demonstrated that N6-methyladenosine (m6A) plays a crucial role in tumorigenesis and tumor development. However, little is known about the role and levels of common m6A regulators and m6A levels in PCa. In this study, we analyzed the characteristic expression of m6A regulators in PCa and castration-resistant prostate cancer (CRPC). UALCAN and cBioPortal were used to estimate the clinical value and genetic alterations of m6A regulators, respectively. The correlation between m6A regulators and androgen receptor (AR) was assessed using Gene Expression Profiling Interactive Analysis (GEPIA) by Pearson correlation statistics. Total m6A levels were detected in transgenic adenocarcinoma of the mouse prostate (TRAMP) mice and PCa cell lines. Results showed that the expression of methyltransferase-like 3 (METTL3) and YTH domain family members, namely, YTHDC2, YTHDF1, and YTHDF2 were generally upregulated in PCa, whereas those of fat mass and obesity-associated protein (FTO), AlkB homolog 5 (ALKBH5), and methyltransferase-like 14 (METTL14) were downregulated. The expression of METTL3, METTL14, Wilms' tumor 1-associating protein (WTAP), YTHDC2, YTHDF1, and YTHDF2 were remarkably higher in CRPC with lymph node metastasis than that in CRPC with bone metastasis, whereas ALKBH5, FTO, and YTHDF3 significantly decreased in CRPC with lymph node metastasis tissues. YTHDF1, YTHDF2, and YTHDC2 were positively correlated with the Gleason grades of PCa, and METTL14, FTO, and ALKBH5 were negatively associated with the Gleason classification. M6A regulators were positively correlated with AR. Patients with a genomic alteration of m6A were associated with poor disease-free survival (DFS). The total m6A levels in TRAMP mice increased dramatically compared with those in tumor-free mice, and m6A levels in LNCaP cell lines were higher than DU145 and PC3 cell lines. In summary, METTL3, METTL14, ALKBH5, FTO, YTHDC2, YTHDF1, and YTHDF2 were abnormally expressed in PCa and related to Gleason classification. Changes in m6A levels maybe contributed to the development and progression of PCa.
Keywords: prostate cancer, N6-methyladenosine, castration-resistant prostate cancer, tumorigenesis and progression, androgen receptor
Introduction
More than 160 posttranscriptional modifications of cellular RNAs have been discovered 1, and N6-methyladenosine (m6A) is the most prevalent RNA modification of mRNA and non-coding RNA in eukaryotic cells 2. M6A methylation regulators include methyltransferase complex, RNA demethylases, and specific RNA-binding proteins, which are also known as “writers,” “erasers,” and “readers,” respectively 3. Methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms' tumor 1-associating protein (WTAP) act as writers in mammalian cells 4, 5. AlkB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are m6A demethylases that mediate the reversal of m6A methylation 6, 7. The RNA modifications of m6A are recognized by “reader” proteins. These proteins are the YTH domain family members, including YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 8. Growing evidence demonstrates that the aberrant expression of m6A RNA methylation is closely related to various cancers, such as hepatocellular carcinoma (HCC) 9, gastric cancer (GC) 10, 11, endometrial cancer 12, and glioblastoma 13. Targeting FTO/m6A/MYC/CEBPA signaling exerts an anti-tumor effect in most leukemia samples 14. The knockout of METTL14 substantially decreases the growth of Epstein-Barr virus-associated tumorigenesis in a xenograft model 15. These studies indicate that m6A modification plays a critical role in cancer and maybe a potential target for cancer therapy.
Prostate cancer (PCa) is one of the most common epithelial malignant tumors in aging males worldwide and the fifth leading cause of cancer death in men 16, 17. Androgen deprivation therapy (ADT) is the primary treatment for patients with advanced PCa, however, most patients eventually develop resistance to castration-resistant prostate cancer (CRPC) 18. Despite the emergence of new therapeutic approaches, patients with metastatic PCa have a low quality of life and a low 5-year survival rate of less than 30% 19, 20. The high morbidity and mortality of PCa necessitate more work to reveal its underlying mechanisms. Recent evidence suggests that m6A modification is closely connected to PCa. VIRMA and YTHDF3 are increased in stages III/IV than in stage II in PCa 21. Li et al. 22 found that YTHDF2 is highly expressed in PCa and the downregulation of YTHDF2 considerably increases the level of m6A and inhibits the proliferation and migration of PCa cell lines. Cai et al. 23 reported that the silencing of METTL3 by shRNA inhibits cell proliferation, colony formation, and invasion of PCa cells and inhibits the growth of tumors in vivo. However, little is known about the role of different m6A regulators in the development and progression of PCa. In this study, we evaluated the expression of m6A regulators, the correlation between m6A regulators and clinical outcomes, and the genetic mutations of these regulators in PCa. We also analyzed the role of m6A levels in PCa by in vivo and in vitro experiments.
Materials and methods
Expression data sets
The Cancer Genome Atlas (TCGA)-Prostate adenocarcinoma (TCGA-PRAD) cohort data and the gene expression of 495 patients with PCa (499 prostate tissues) and 52 normal samples were obtained from the National Cancer Institute Genomic Data Commons (https://portal.gdc.cancer.gov/). R version 3.6.3 software was utilized to normalize and process the expression data. Oncomine database (https://www.oncomine.org/) is a powerful data-mining platform that contains 715 independent datasets and the data of 86,733 tumor tissues and adjacent normal tissues 24. In this study, we analyzed the mRNA expression levels of m6A regulators in different types of cancer by Oncomine data mining. The search parameters were as follows: type of analysis: cancer versus normal tissues; type of data: mRNA; P value: 0.05. The Human Cancer Metastasis Database (HCMDB, http://hcmdb.i-sanger.com/index) 25 is an online integrated database for storing and analyzing the large-scale expression data of cancer metastasis. We selected the data set of EXP00339 in HCMDB (dataset ID: GSE74685), which contains 89 CRPC samples that comprise 20 samples of bone metastasis and 69 of lymph node metastasis, to explore the expression of m6A regulators in CRPC with different metastasis sites.
M6A regulators and Gleason classification
UALCAN (http://ualcan.path.uab.edu) is an interactive online portal providing the gene expression and clinical analysis of tumor subsets, using TCGA level 3 RNA sequencing and clinical data from 31 cancer types 26. UALCAN was implemented to analyze the relative expression of m6A regulators in different Gleason classifications to understand the clinical importance of m6A regulators in PCa.
M6A regulators and androgen receptor (AR)
The Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn/index.html) 27 is a website for TCGA cancer and the Genotype-Tissue Expression profile analysis and interaction analysis. GEPIA can be used to perform the correlation analysis of gene pairs by Spearman, Pearson, or Kendall correlation statistics. The correlation between m6A regulators and AR was analyzed using GEPIA by Pearson correlation statistics.
Genetic alteration of m6A regulators in PCa
CBioPortal (http://www.cbioportal.org/) is an open-access website that explores, visualizes, and analyzes multidimensional cancer genomics data. We selected eight studies (Prostate Adenocarcinoma [Broad/Cornell, Nat Genet 2012], Prostate Adenocarcinoma [Fred Hutchinson CRC, Nat Med 2016], Prostate Adenocarcinoma [MSKCC, PNAS 2014], Prostate Adenocarcinoma [TCGA, Firehose Legacy], Prostate Adenocarcinoma [CPC-GENE, Nature 2017], Prostate Adenocarcinoma [MSKCC, Cancer Cell 2010], Prostate Adenocarcinoma [MSKCC/DFCI, Nature Genetics 2018], and Prostate Cancer [DKFZ, Cancer Cell 2018]) to analyze the genetic alterations of m6A regulators in PCa. Disease-free survival (DFS) curve was drawn by the Kaplan-Meier method, and the difference in DFS was evaluated by log-rank test.
Cell lines and culture conditions
Three human PCa cell lines were used in this study: PC3, DU145, and LNCaP. PC3 and DU145 cell lines were obtained from the Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences. LNCaP cell line was acquired from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The basal medium was RPMI-1640 medium (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco), 1% penicillin and 1% streptomycin (Invitrogen). LNCaP cells were cultured in basic medium supplemented with 1.0 mM L-glutamine and 1.0 mM sodium pyruvate (Invitrogen). All cells were incubated in a humidified incubator at 37°C and 5% CO2 (Thermo Scientific).
TRAMP mice
Hemizygous male transgenic adenocarcinoma of the mouse prostate (TRAMP) mice (8247 Ng/J) and wild-type C57BL/6 females were obtained from the Model Animal Research Center of Nanjing University (MARCNU, Nanjing, Jiangsu, China). The mice were reared and propagated by the MARCNU protocol. Male offspring were genotyped by polymerase chain reaction (PCR) according to protocol. All animal experiments were performed following institutional guidelines and approved by the Institutional Animal Care and Use Committee of Guangxi Medical University.
Detection of total m6A levels in TRAMP mice and PCa cell lines
Twelve TRAMP mice were randomly divided into four groups with three mice in each group. The prostate tissues of the mice at 8, 16, 26, and 36 weeks were collected for RNA extraction. Total RNA was extracted from the prostate tissues and PCa cell lines by TRIzol Reagent (Thermo Fisher Scientific). EpiQuik m6A RNA Methylation Quantification Kit (EpiGentek) was used to detect the total m6A RNA methylation levels.
Statistical analysis
Statistical data were analyzed using GraphPad Prism version 7.0 (GraphPad Software, USA). An independent t-test was used to evaluate the expression of m6A regulators between PCa tissues and normal tissues, and the results were presented as the mean ± SD. Differences were considered statistically significant if P <0.05.
Results
M6A regulators in PCa were distinct from normal tissues
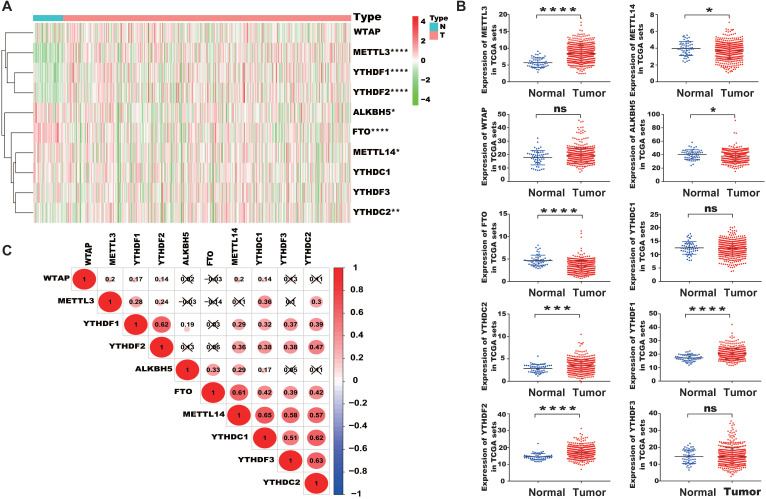
We extracted the mRNA expression matrix of 10 common m6A regulators from TCGA-PRAD to evaluate their expression in PCa. The results showed that METTL3, a chief component of m6A methyltransferase, was remarkably increased in PCa tissues than in normal controls (P < 0.05, Figure 1A, 1B). YTHDC2, YTHDF1, and YTHDF2 were also substantially higher in PCa tissues than in normal controls, whereas the expression of FTO, ALKBH5, and METTL14 decreased in PCa. Then, the correlation among m6A regulators was analyzed. Results indicated that METTL14 was positively correlated with YTHDC1, YTHDF2, and YTHDC2, and YTHDC1, YTHDC2, YTHDF2, and YTHDF1 were positively correlated with each other (Figure 1C).
Figure 1.
Characteristic expression of m6A regulators in PCa. (A) Heatmap of the expression of different m6A regulators between PCa tissues and normal tissue samples. (B) Scatter plots of the expression of m6A regulators in PCa. (C) Spearman correlation analysis of m6A regulators in PCa (Red and green colors in the plots symbolized relatively high and low expression, respectively).
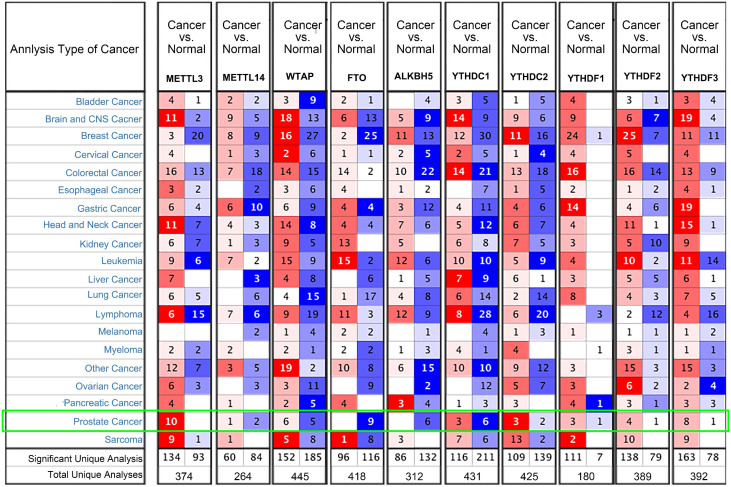
We examined the mRNA levels of m6A regulators in different PCa datasets based on the Oncomine database to confirm the expression patterns of the m6A regulators in PCa. The results showed that the expression trends of the 10 common m6A regulators were consistent with the TCGA-PRAD (Figure 2). METTL3 was highly expressed in 10 PCa datasets, and FTO and ALKBH5 were downregulated in nine and six datasets, respectively.
Figure 2.
Transcriptional levels of m6A regulators in PCa (Oncomine). Arabic numbers in the figure showed the numbers of datasets in which m6A regulator expression was significantly upregulated (red) or downregulated (blue) in PCa tissues compared with adjacent normal tissues.
Differential expression profiles of m6A regulators in CRPC
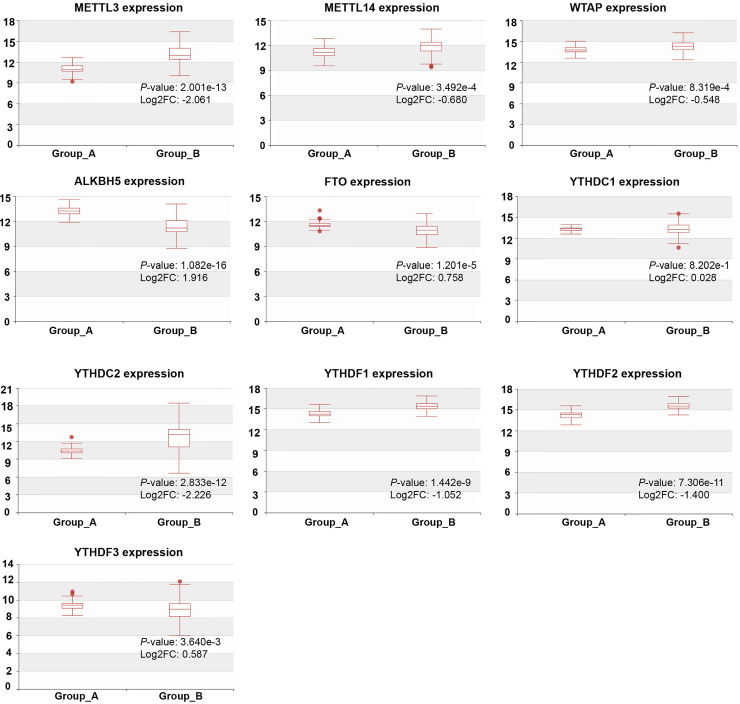
We detected the mRNA levels of m6A regulators in CRPC with different metastasis sites using HCMDB to further examine their expression in PCa. The results showed that, except YTHDC1, the expression of m6A regulators was statistically significant at different CRPC metastasis sites (P < 0.01, Figure 3). The expression of METTL3, METTL14, WTAP, YTHDC2, YTHDF1, and YTHDF2 was remarkably higher in CRPC with lymph node metastasis than in CRPC with bone metastasis, whereas ALKBH5, FTO, and YTHDF3 were substantially decreased in CRPC with lymph node metastasis.
Figure 3.
Expression of m6A regulators in CRPC with different metastasis sites. Group_A, metastasis tumor of CRPC with bone metastasis; Group_B, metastasis tumor of CRPC with lymph node metastasis experiment.
Correlation between the expression of m6A regulators and Gleason classification
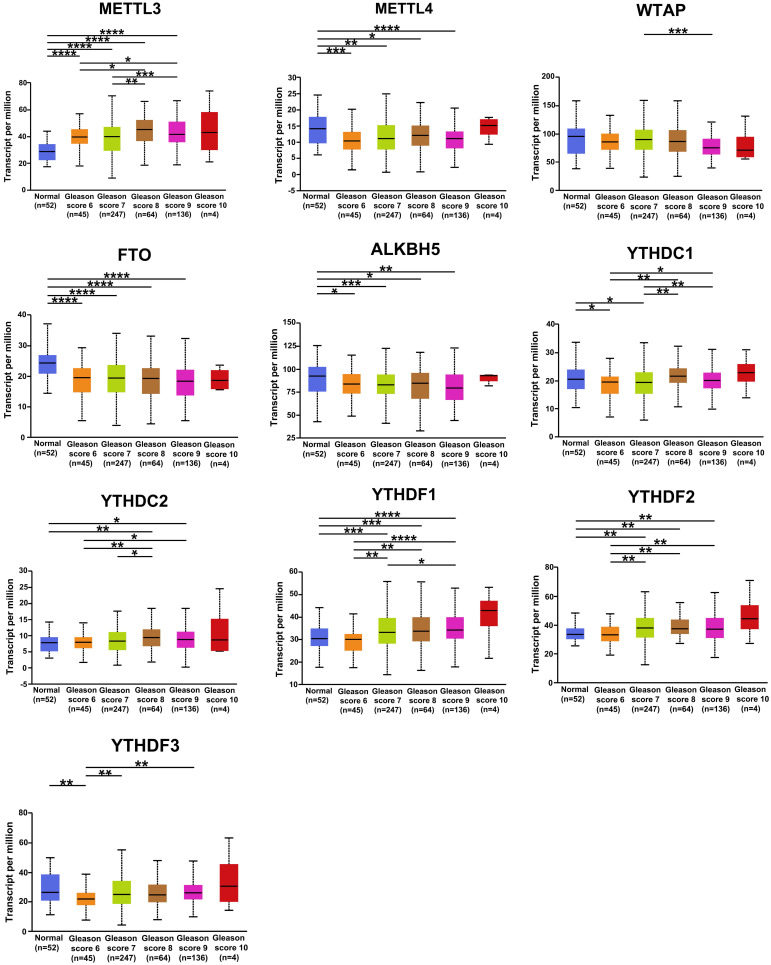
The correlation between the expression of m6A regulators in PCa and Gleason classification was assessed using the UALCAN database. The results were shown in Figure 4. YTHDF1, YTHDF2, and YTHDC2 were positively correlated with Gleason grade, and the mRNA level of METTL3 was higher in patients with different Gleason grades of PCa than in normal samples. METTL14, FTO, and ALKBH5 were negatively associated with the Gleason classification of PCa. These results suggested that the mRNA expression of m6A regulators was related to the clinical features of patients with PCa.
Figure 4.
Relative expression of m6A regulators in normal and PCa tissues with different Gleason classifications. * P < 0.05, ** P < 0.01, ***P < 0.001.
Correlation of m6A regulators and AR
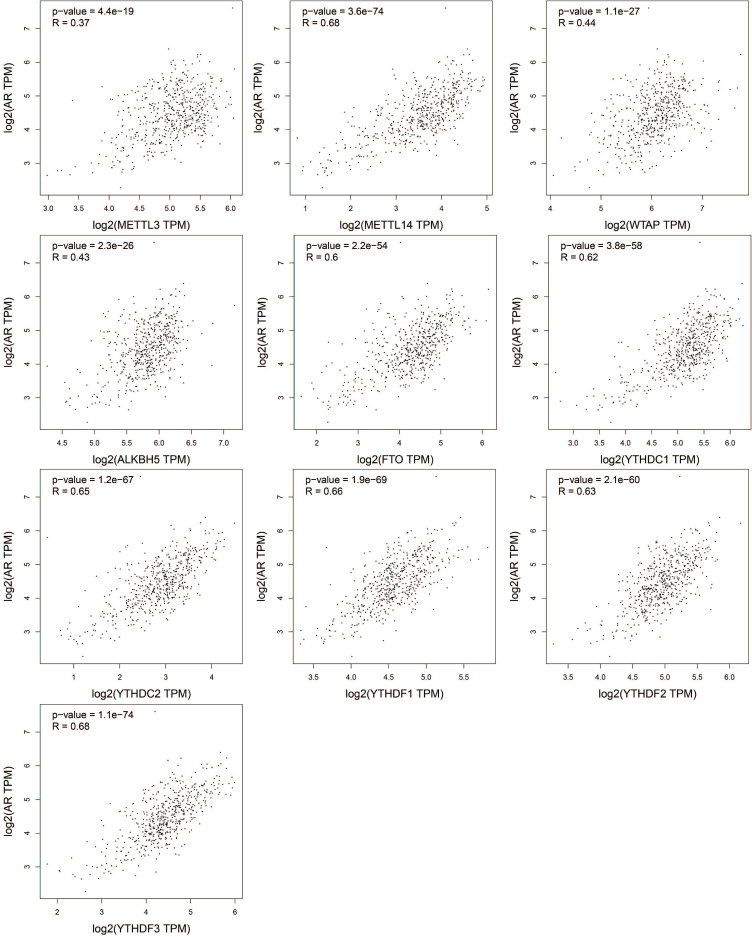
AR is a primary oncogene driver of PCa and plays a key role in the development and progression of PCa 28, 29. The progress of CRPC still depends on continuous AR signals 30. The relationship of the expression of m6A regulators and AR was assessed by GEPIA, and the results were presented in the scatter diagrams in Figure 5. Surprisingly, all the m6A regulators were positively correlated with AR (P<0.05). METTL14, FTO, YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3 were remarkably related to AR (P < 0.0001, R > 0.6).
Figure 5.
Correlation between the mRNA expression of m6A regulators and AR in PCa.
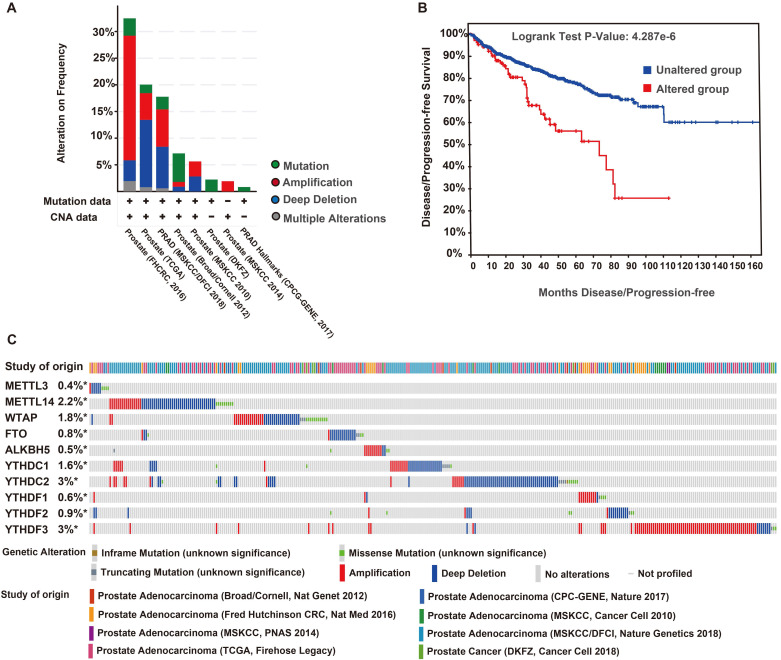
Genetic mutations of m6A regulators and their associations with the DFS of patients with PCa
The genetic variations of the m6A regulators were analyzed using the cBioPortal database. We found 343 (12%) alterations from 2797 queried patients, including mutation, multiple alterations, amplification, and deep deletions. Deep deletion was the most common type of genomic alteration among these variations (Figure 6A). The DFS curves indicated that patients with a genomic alteration of m6A had remarkably worse DFS (Figure 6B). The alterations frequencies of YTHDC2, YTHDF3, METTL14, WTAP, YTHDC1, YTHDF2, FTO, YTHDF1, ALKBH5, and METTL3 were 3%, 3%, 2.2%, 1.8%, 1.6%, 0.9%, 0.8%, 0.6%, 0.5%, and 0.4%, respectively (Figure 6C).
Figure 6.
Genomic alteration and prognosis of m6A regulators in eight PCa studies (cBioportal). (A) OncoPrint of 10 m6A regulators in PCa. (B) DFS/ progression-free survival (PFS) of patients with PCa who had altered (red) and unaltered (blue) mRNA expression of the m6A regulators. (C) Genetic alteration frequencies of the 10 m6A regulators in PCa.
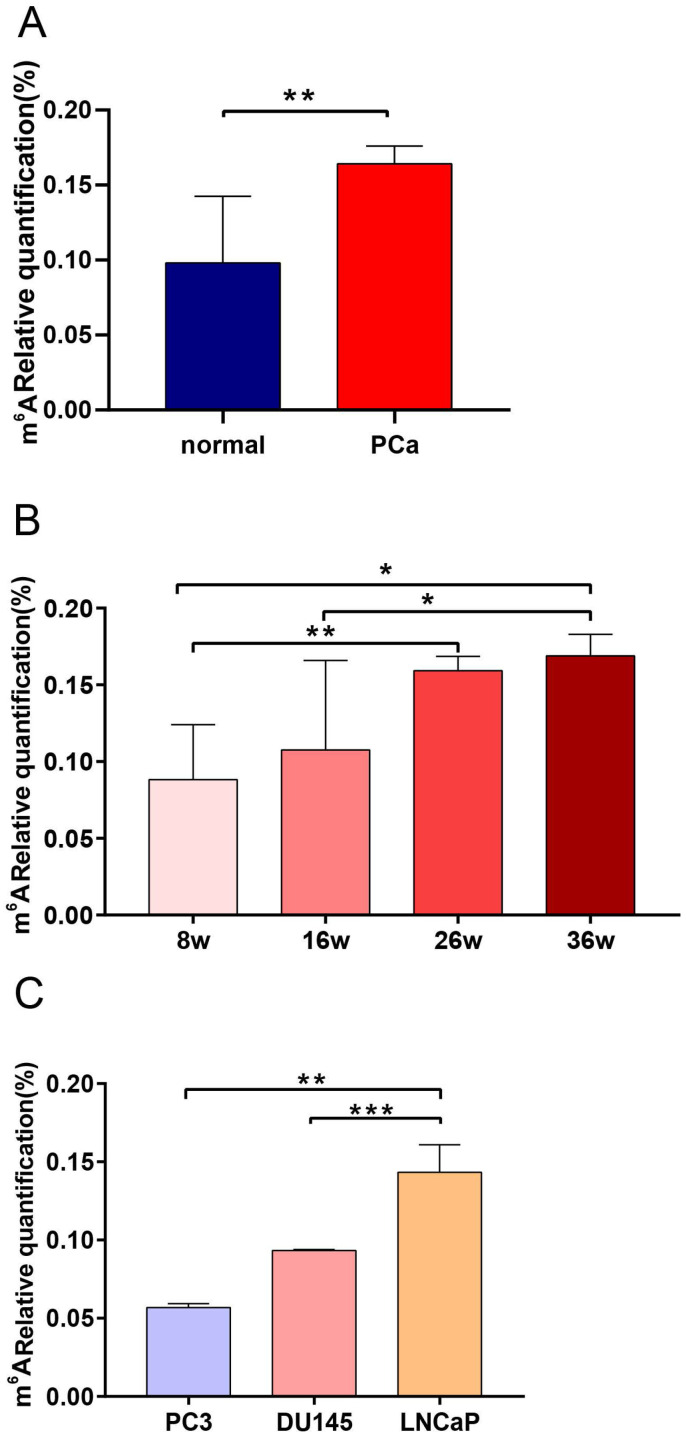
Expression of total m6A levels in TRAMP mice and PCa cell lines
TRAMP mice can spontaneously develop PCa and their pathological progression is similar to that of human PCa 31. We determined the total m6A expression of TRAMP mice at different stages of PCa to characterize the specific m6A level profile. Results showed that the m6A level of TRAMP mice increased significantly compared with that of tumor-free mice (P < 0.01; Figure 7A). We also found that m6A expression level was significantly upregulated with the increase in cancer progression (P < 0.05; Figure 7B). To evaluate the functional relevance of m6A between PCa and CRPC, we observed the m6A expression levels of AR-independent (DU145 and PC3) and AR-dependent PCa cell lines (LNCaP). The results showed that the m6A level of LNCaP cells increased significantly compared with those of DU145 and PC3 cells (Figure 7C).
Figure 7.
Expression levels of the m6A in TRAMP mice and PCa cell lines. (A) Expression of normal prostate tissues and PCa tissues in TRAMP mice. (B) Expression of m6A at different stages of TRMAP mice. (C) Expression of m6A in AR-independent and AR-dependent PCa cell lines.
Discussion
M6A modification is involved in tumorigenesis, metastasis, and drug resistance 32. Wang et al. 10 found that increased expression of METTL3 in GC promotes tumor angiogenesis, supporting that METTL3 is a promising prognostic biomarker and therapeutic target for GC. It was reported that the downregulation of METTL14 is related to poor prognosis in patients with colorectal cancer (CRC), and the knockdown of METTL14 promotes the proliferation and invasion of CRC cells 33. Low expression of ALKBH5 was found to be closely linked to the occurrence and development of non-small cell lung cancer (NSCLC) 34. Elevated expression of some subtypes of FTO has a critical oncogenic role in acute myeloid leukemia (AML) 35. The upregulation of FTO promotes the development of melanoma and its resistance to PD-1 2. However, little is known about the role of multiple m6A regulators in PCa 23.
In this study, the expression, clinical value, and mutation of 10 m6A regulators in PCa, as well as their correlation with AR, were analyzed. Total m6A expression levels were determined in PCa cell lines and TRAMP mice. The results of our study showed that FTO and ALKBH5 had low- expression in PCa and were also significantly decreased in CRPC with bone metastasis tissues than in CRPC with lymph node metastasis. METTL3, YTHDC2, YTHDF1, and YTHDF2 had a remarkably high expression in PCa and CRPC with lymph node metastasis. METTL14 decreased in PCa and CRPC with bone metastasis tissues. The Gleason grading system is an effective predictor of the prognosis of patients with PCa because it can evaluate the differentiation and malignancy of PCa 36. We assessed the relationship of m6A regulators and their Gleason classifications. We found that METTL3, YTHDF1, YTHDF2, and YTHDC2 were generally upregulated with different Gleason grades, whereas METTL14, FTO, and ALKBH5 had low expression in patients with different Gleason classifications of PCa. We also found that patients with a genomic alteration of m6A were associated with poor DFS. These findings suggest that METTL3, METTL14, ALKBH5, FTO, YTHDC2, YTHDF1, and YTHDF2 were abnormally expressed in PCa and related to Gleason classification, and these m6A modulators were the potential molecular targets for the diagnosis and treatment for PCa.
TRAMP mice developed varying degrees of prostate hyperplasia at 6-12 weeks, progressed to severe hyperplasia and adenocarcinoma at 18 weeks, and presented primary tumors at 24-30 weeks 37. M6A methylation level contributes to the progression of human cancer 38, 39. We hypothesized that changes in m6A level promoted the development of PCa, thus, we detected the m6A levels in TRAMP mice and PCa cell lines. We found that the m6A level of TRAMP mice increased with ages. These results indicated that changes in m6A levels were possibly involved in the occurrence and development of PCa.
Our study had some limitations. First, although we found that m6A regulators were abnormally expressed in PCa, the potential diagnostic and therapeutic effects were not evaluated, hence, further research is necessary to explore whether m6A regulators could be used as diagnostic markers or therapeutic targets in PCa. Second, we did not elucidate the molecular mechanisms of m6A regulators in PCa. The detailed mechanism of m6A regulators in PCa warrants further investigation.
In conclusion, we analyzed the expression of m6A regulators, their relationship with Gleason grades and AR, and their genomic variations in PCa, and detected m6A levels in TRAMP mice and PCa cell lines. The results indicated that the abnormal expression of m6A regulators and abnormal m6A levels contribute to the development of PCa. This study provides a further understanding of epigenetic RNA modification in PCa.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81472414), Guangxi Medical University Training Program for Distinguished Young Scholars.
Authors' Contributions
Yanling Hu, Qunying Wu, and Huifeng Wang designed the study and wrote the article. Xing Xie, Yiming Huang, and Huifeng Wang performed the experiments in vitro and in vivo. Qunying Wu, Huifeng Wang, Xing Xie, Yiming Huang interpreted the results and revised the manuscript. Shanshan Meng and Youcheng Li helped to interpret the data. All authors have read and approved the final manuscript.
Abbreviations
- PCa
Prostate cancer
- m6A
N6-methyladenosine
- CRPC
castration-resistant prostate cancer
- AR
androgen receptor
- DFS
disease-free survival
- TRAMP
transgenic adenocarcinoma of the mouse prostate
- METTL3
Methyltransferase-like-3
- METTL14
Methyltransferase-like-14
- WTAP
Wilms tumor 1 associated protein
- ALKBH5
AlkB homolog 5
- FTO
fat mass and obesity-associated protein
- HCC
hepatocellular carcinoma
- GC
gastric cancer
- ADT
Androgen deprivation therapy
- NCI-GDC
National Cancer Institute Genomic Data Commons
- HCMDB
Human Cancer Metastasis Database
- GEPIA
Gene Expression Profiling Interactive Analysis
- NSCLC
non-small cell lung cancer
- AML
acute myeloid leukemia
- OV
ovarian cancer
References
- 1.Chen Z, Zhao P, Li F. et al. Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences. Brief Bioinform. 2019;11:bbz112. doi: 10.1093/bib/bbz112. [DOI] [PubMed] [Google Scholar]
- 2.Yang S, Wei J, Cui YH. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panneerdoss S, Eedunuri VK, Yadav P. et al. Cross-talk among writers, readers, and erasers of m(6)A regulates cancer growth and progression. Sci Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Yue Y, Han D. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Li Y, Toth JI. et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mauer J, Luo X, Blanjoie A. et al. Reversible methylation of m(6)Am in the 5' cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao S, Sun H, Xu C. YTH Domain: A Family of N(6)-methyladenosine (m(6)A) Readers. Genomics Proteomics Bioinformatics. 2018;16:99–107. doi: 10.1016/j.gpb.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Peng C, Chen J. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Chen C, Ding Q. et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 11.Yue B, Song C, Yang L. et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Eckert MA, Harada BT. et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S, Zhao BS, Zhou A. et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su R, Dong L, Li C. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang F, Singh RK, Pei Y. et al. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019;15:e1007796. doi: 10.1371/journal.ppat.1007796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 17.Bray F, Ferlay J, Soerjomataram I. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Breijo S, Chantada-Abal V, Aller-Rodriguez M. et al. [Castration resistance mechanisms in prostate cancer.] Arch Esp Urol. 2018;71:628–638. [PubMed] [Google Scholar]
- 19.Tian C, Deng Y, Jin Y. et al. Long non-coding RNA RNCR3 promotes prostate cancer progression through targeting miR-185-5p. Am J Transl Res. 2018;10:1562–1570. [PMC free article] [PubMed] [Google Scholar]
- 20.Burki TK. Abiraterone and castration-resistant prostate cancer. Lancet Oncol. 2013;14:e48. doi: 10.1016/s1470-2045(12)70585-5. [DOI] [PubMed] [Google Scholar]
- 21.Lobo J, Barros-Silva D, Henrique R. et al. The Emerging Role of Epitranscriptomics in Cancer: Focus on Urological Tumors. Genes (Basel) 2018;9:552. doi: 10.3390/genes9110552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Meng S, Xu M. et al. Downregulation of N(6)-methyladenosine binding YTHDF2 protein mediated by miR-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget. 2018;9:3752–3764. doi: 10.18632/oncotarget.23365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai J, Yang F, Zhan H. et al. RNA m(6)A Methyltransferase METTL3 Promotes The Growth Of Prostate Cancer By Regulating Hedgehog Pathway. Onco Targets Ther. 2019;12:9143–9152. doi: 10.2147/OTT.S226796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V. et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng G, Ma Y, Zou Y. et al. HCMDB: the human cancer metastasis database. Nucleic Acids Res. 2018;46:D950–D955. doi: 10.1093/nar/gkx1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandrashekar DS, Bashel B, Balasubramanya SAH. et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang Z, Li C, Kang B. et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson D, Van Allen EM, Wu YM. et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Coleman I, Morrissey C. et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–378. doi: 10.1038/nm.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welti J, Sharp A, Yuan W. et al. Targeting Bromodomain and Extra-Terminal (BET) Family Proteins in Castration-Resistant Prostate Cancer (CRPC) Clin Cancer Res. 2018;24:3149–3162. doi: 10.1158/1078-0432.CCR-17-3571. [DOI] [PubMed] [Google Scholar]
- 31.Kaplan-Lefko PJ, Chen TM, Ittmann MM. et al. Pathobiology of autochthonous prostate cancer in a pre-clinical transgenic mouse model. Prostate. 2003;55:219–237. doi: 10.1002/pros.10215. [DOI] [PubMed] [Google Scholar]
- 32.Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270–288. doi: 10.1016/j.ccell.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang X, Zhang S, He C. et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin D, Guo J, Wu Y. et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer. 2020;19:40. doi: 10.1186/s12943-020-01161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Weng H, Su R. et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tilki D, Chen MH, Wu J. et al. Surgery vs Radiotherapy in the Management of Biopsy Gleason Score 9-10 Prostate Cancer and the Risk of Mortality. JAMA Oncol. 2019;5:213–220. doi: 10.1001/jamaoncol.2018.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kido LA, de Almeida Lamas C, Marostica MR Jr. et al. Transgenic Adenocarcinoma of the Mouse Prostate (TRAMP) model: A good alternative to study PCa progression and chemoprevention approaches. Life Sci. 2019;217:141–147. doi: 10.1016/j.lfs.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 38.He Y, Xing J, Wang S. et al. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med. 2019;7:797. doi: 10.21037/atm.2019.12.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge L, Zhang N, Chen Z. et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin Chem. 2020;66:342–351. doi: 10.1093/clinchem/hvz004. [DOI] [PubMed] [Google Scholar]