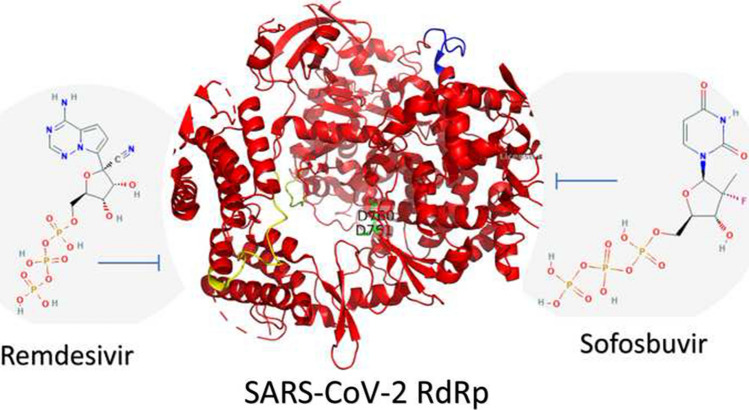
Abstract
Abstract
Coronavirus diseases 2019 (COVID-19) are seriously affecting human health all over the world. Nucleotide inhibitors have promising results in terms of its efficacy against different viral polymerases. In this study, detailed molecular docking and dynamics simulations are used to evaluate the binding affinity of a clinically approved drug, sofosbuvir, with the solved structure of the viral protein RNA-dependent RNA polymerase (RdRp) and compare it to the clinically approved drug, Remdesivir. These drugs are docked onto the three-dimensional structure of the nsp12 protein of SARS-CoV-2, which controls the polymerization process. Hence, it is considered one of the primary therapeutic targets for coronaviruses. Sofosbuvir is a drug that is currently used for HCV treatment; therefore, HCV RdRp is used as a positive control protein target. The protein dynamics are simulated for 100 ns, while the binding is tested during different dynamics states of the SARS-CoV-2 RdRp. Additionally, the drug-protein complexes are further simulated for 20 ns to explore the binding mechanism. The interaction of SARS-CoV-2 RdRp as a target with the active form of sofosbuvir as a ligand demonstrates binding effectiveness. One of the FDA-approved antiviral drugs, such as sofosbuvir, can help us in this mission, aiming to limit the danger of COVID-19. Sofosbuvir was found to bind nsp12 with comparable binding energies to that of Remdesivir, which has been reported for its potential against COVID-19 RdRp and is currently approved by the FDA.
Graphic abstract
Keywords: SARS-CoV-2, COVID-19, Sofosbuvir, Docking, Molecular dynamics simulation, Drug repurposing
Introduction
An outbreak of coronavirus disease (December 2019) caused by a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported for the first time in Wuhan City in China [1]. The novel coronavirus disease (COVID-19) was announced as a global pandemic on the 11th of March 2020 by the World Health Organization (WHO), blankly within seven months of the emergence of its first case. It has kept shifting its epicenter through various continents and has now been exported to 218 countries and territories. It has nearly infected 60 M people worldwide, claimed more than 1.4 M deaths, gravely affected the countries socio-economically, and even the most advanced healthcare systems have crumbled under its weight [2]. It is highly contagious pneumonia caused by severe acute respiratory syndrome coronavirus 2 (SARS CoV-2), and the symptoms involve complications in the gastrointestinal tract, respiratory problems, like fever, dry cough, cold, sore throat, shortness of breath, and may even lead to fatality in 2.4% of the cases. Seven different human coronaviruses (HCoVs) strains have been detected (229E and NL63 (Alphacoronaviruses), and OC43, HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 (Betacoronaviruses)) until now [3, 4].
SARS-CoV and MERS-CoV are firmly bounded strains of such pandemics, each spanning more than 20 countries and killing approximately 1600 people in their wake, with a fatality rate of 10% and 35%, respectively. COVID-19 has a global fatality rate of 2.4%, and as the outbreak reaches anywhere near the previous scales of pandemics, this rate is translating to millions of deaths. These firmly bounded strains are members of the beta coronavirus family and belong to the class of positive-stranded RNA viruses with a huge 30 kb polycistronic genome [5]. The viral replicase polyproteins, namely pp1a and pp1ab, are translated by the proximal two-thirds of the 5′ end of the CoV genome (ORFs 1a and 1b). These two polyproteins are cleaved into 16 nonstructural proteins (nsps), which are a major for viral replication and transcription, thus being regarded as a potential virulence factor and drug targets for the CoV family [6]. ORFs near the 3′ end encode the structural proteins, namely S, M, E, and N, the spike, membrane, envelope, and nucleocapsid proteins. Out of the 16 nsps encoded by the 5′ end, nsp12 includes the RdRp (RNA-dependent RNA polymerase) activity. The central catalytic subunit of the RNA-synthesizing machinery for viral replication and transcription is the RdRp, which has seven catalytic motifs (A-G). Each motif has a particular function from selection and correct positioning to the addition of the newly incorporated NTPs (Nucleoside triphosphates) to the growing chain, which is executed with the help of a set of conserved aspartates amino acid residues. RdRp is a main viral enzyme in the life cycle of RNA viruses and has been targeted in various viral infections, like the hepatitis C virus (HCV), the Zika virus (ZIKV), and human coronaviruses (CoVs) [7–10]. It has continuously served as a principal drug target in different viruses with minimal cytotoxic effects on the host cells as there is no human target that is affected by the drugs. Thus, nsp12 represents an attractive target to develop a possible therapeutic agent against SARS-CoV-2. The previous studies utilized a homology model of the protein for small molecule screening, and with the experimental (cryo-electron microscopy) three-dimensional structure of the protein has been reported in April 2020, the exercise of small molecule screening was necessary to be re-visited.
In the absence of new drugs and vaccines against SARS-CoV-2, the choices for effective COVID-19 treatments are limited. Considering the exigency of the situation, our focus is on identifying the available approved drug as a potential inhibitor against the promising coronavirus drug target, the RdRp, using computer-aided methods. Anti-RdRp drugs against SARS-CoV-2 within the pool of the FDA/WHO-EML approved drugs are expected to be helpful as a possible therapeutic option for the COVID-19; to battle the menace that has threatened the existence of humanity.
In this study, sofosbuvir and remdesivir are docked into the solved structure of SARS-CoV-2 RdRp (PDB ID: 7BTF) and tested using different dynamic states of the protein. Hence, we have used molecular docking and dynamics simulations for 100 ns to identify the mode of interaction capable of inhibiting nsp12 of SARS-CoV-2. The ten different conformations of the SARS-CoV-2 RdRp are used as a target for the small molecules. The results revealed that sofosbuvir might potentially treat COVID-19 patients just like Remdesivir that was recently approved by the FDA against COVID-19.
Materials and methods
Structural retrieval
Protein data bank database is used to retrieve the experimentally solved 3D structures of SARS-CoV-2 RdRp (PDB ID: 7BTF). The selection of the structure is based on the resolution (2.95 Å), the existence of the nsp7 and nsp8 cofactors, and the existence of a divalent cation (Zn+2), which was reported to take part in the interaction with Nucleotide triphosphate (NTP) at the active site of the polymerase [11]. Additionally, the 3D structure of the Hepatitis C Virus (HCV) RdRp (PDB ID: 2XI3) is downloaded for use as a positive control drug target (sofosbuvir approved by the FDA against HCV) [12].
PubChem database is used to retrieve the 3D structure of sofosbuvir triphosphate and remdesivir triphosphate. AutoDockTools-1.5.6 [13] and PyMOL [14] software are used to prepare the drugs to be ready for the docking experiments.
RdRp dynamics
The basic idea of molecular dynamics depends on the solution of Newton’s equations of motion for individual particles (atoms, ions, and molecules) [15]. MDS predicts the properties of assemblies of molecules in terms of their structure and the microscopic interactions between them [15, 16]. The downloaded solved structures for SARS-CoV-2 and HCV RdRps (PDB ID: 7BTF and 2XI3, respectively) are prepared using PyMOL software. Water molecules and ions are removed (except the Zn+2 and Mg+2), while any redundant chains are excluded. Additionally, any missed Hydrogen atoms are added using PyMOL. The High-performance computing unit (Bib Alex) of the Bibliotheca Alexandrina, Alexandria, Egypt, is used to perform the MDS study under the project entitled 'Structural demystifying of some Hepatitis C Virus proteins; An in silico study.' Nanoscale Molecular Dynamics (NAMD 2.13) software is used in the MDS. Its viewer software VMD is used to prepare the input files and analyze the trajectories [15, 17]. CHARMM 36 force field is used in NAMD, and the TIP3P water model is employed [15, 18]. The periodic boundary conditions are used in simulating the system while the temperature is raised from 0 K up to 310 K gradually in the equilibration phase. One hundred nanoseconds MDS production runs are performed for both SARS-CoV-2 and HCV RdRps at the NVT ensemble. After docking, 20 ns MDS runs are performed for the protein-sofosbuvir complexes. Cluster analysis is performed utilizing Chimera software's default parameters (UCSF) [19].
Molecular docking
Molecular docking predicts one molecule's best orientation to a second when bound to each other to form a stable complex that depends on the key and lock theory [20]. The preferred orientation knowledge predicts the strength of the association or binding affinity between two molecules using scoring functions [21, 22]. Sofosbuvir, a potent anti-HCV RdRp inhibitor (approved by FDA in 2013 against HCV), is tested against SARS-CoV-2 RdRp [23–27]. AutoDock Vina 2.4 [21], installed on HP Pavilion g series—precision with Core i3 processor, was employed in this study to assess the binding affinity and mode of interaction between sofosbuvir and remdesivir against SARS-CoV-2 RdRp. A flexible ligand / flexible active site docking scheme is applied with a grid of dimensions of 30 × 30 × 30 Å3. The grid center is maintained at the active site residues, D760 and D761 for SARS-CoV-2 RdRp and D318 and D319 for HCV RdRp, using AutoDock Tools [13]. The centers are at (8.0, 14.7, − 6.4) Å for SARS-CoV-2 RdRp (7BTF) and ( − 17.1, 1.5, 20.6) Å for HCV RdRp (2XI3) with minimal differences in the centers between the different RdRp conformations at spacing 0.375. Moreover, the binding score or affinity of the resulting complexes is detected by the Vina scoring function [21]. Different dynamical states during the Molecular Dynamics Simulation (MDS) run corresponding to the protein's different conformations are used in docking experiments. Additionally, after docking, another MDS run for 20 ns is performed to study the sofosbuvir's binding mode to SARS-CoV-2 and HCV RdRp. Analysis of the docking complexes is done utilizing PyMOL and the Protein–Ligand Interaction Profiler (PLIP) web server (Technical University of Dresden) [28].
Results and discussion
Previously, the effectiveness of some in-market drugs against the COVID-19-causing coronavirus strain, SARS-CoV-2 RdRp homology model, is tested in silico [8, 27, 29, 30]. On the other hand, in this work, we study the binding affinity of sofosbuvir to different dynamic states of the solved structure of SARS-CoV-2 RdRp.
RdRp dynamics study
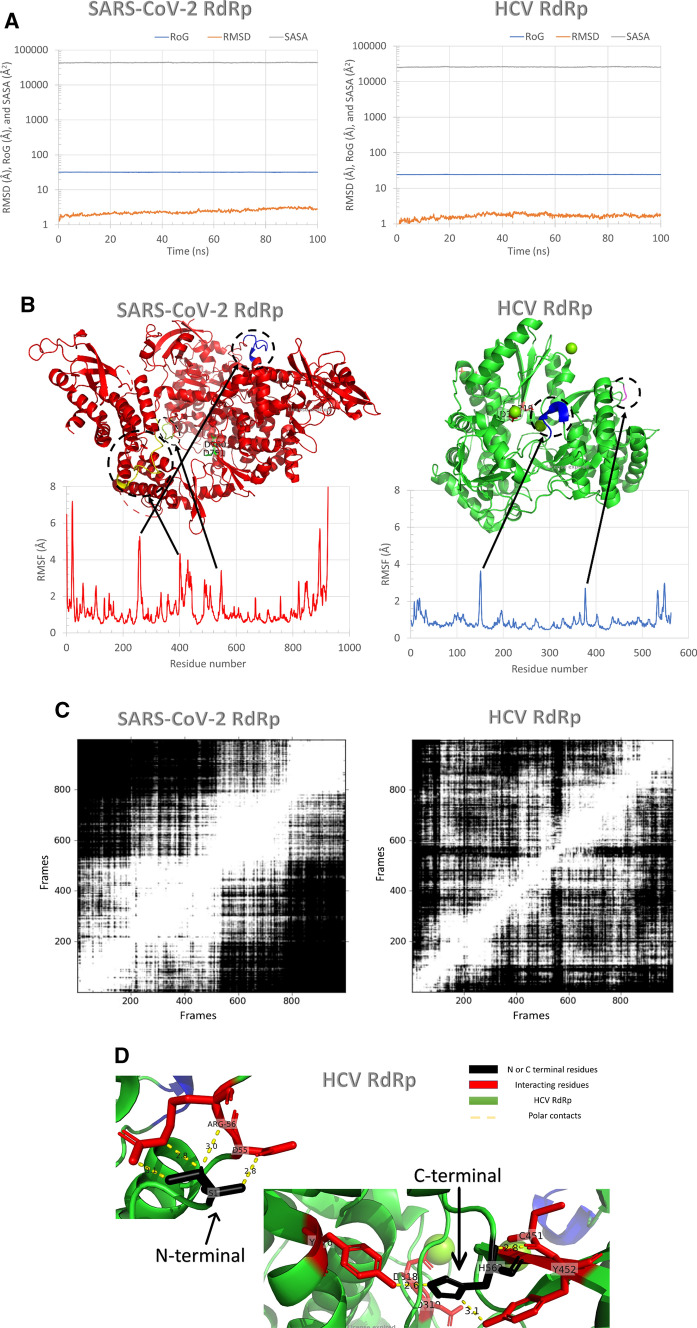
Figure 1 shows the dynamics of the SARS-CoV-2 and HCV RdRp during 100 ns MDS runs. Figure 1a represents the Surface Accessible Surface Area (SASA) in Å2 (gray line), Radius of Gyration (RoG) in Å (blue line), and the Root Mean Square Deviation (RMSD) in Å (orange line) versus time in nanoseconds. As noted from the RMSD graph, the SARS-CoV-2 RdRp and HCV RdRp systems are both equilibrated during the first 20 ns of the simulation. The RMSD values raised from zero up to ~ 1.75 Å for HCV RdRp and ~ 2.5 Å for SARS-CoV-2 RdRp. The SASA and RoG values also indicate that the two systems are equilibrated with the values in the range of ~ 26,000 Å2 and ~ 24.5 Å for HCV RdRp, while the values are ~ 43,000 Å2 and ~ 32 Å for SARS-CoV-2 RdRp. Based on our results, the HCV RdRp system is well stable compared to SARS-CoV-2. This is maybe due to the size of the protein. SARS-CoV-2 RdRp is 924-residue, while HCV RdRp is only 562-residue long.
Fig. 1.
Molecular Dynamics Simulation studies for SARS-CoV-2 and HCV RdRp. a RMSD (orange), RoG (blue), and SASA (gray) for both systems versus time in nanoseconds during the 100 ns simulation. b per residue RMSF during the simulation. RdRp structures are shown in the colored cartoon. c RMSD Map of the dynamics of the SARS-CoV-2 RdRp (left) and HCV RdRp during the 100 ns by using UCSF Chimera software. d N and C terminals of HCV RdRp and its interactions with the protein residues inside the protein
Figure 1b shows the per-residue Root Mean Square Fluctuations (RMSF) in Å for SARS-CoV-2 RdRp (left) and HCV RdRp (write). Each protein is represented in the upper part with the colored ribbons. Each protein's active site is presented in colored sticks and labeled with their one-letter code (D318 and D319 for HCV, while D760 and D761 for SARS-CoV-2). These consecutive aspartates are protruding from the β-turn structure of the polymerase. The most fluctuating regions in each protein are labeled and colored in the ribbon representations. As reflected from the RMSF of HCV RdRp, the most fluctuating regions are the P149-G153 (blue ribbon) and G376-A377 (magenta coil). For SARS-CoV-2 RdRp, the most fluctuating regions are E253-P264 (blue region), S425-F442 (yellow region), and K545-A550 (lemon coil). Noticeably, the N and C terminals of HCV RdRp have very low fluctuations (~1 Å), while for SARS-CoV-2 RdRp, the N and C terminals have high fluctuations (7 Å and 8 Å, respectively). This is maybe due to the embedment of HCV RdRp terminals inside the protein fold (Fig. 1d). The N-terminal residue of HCV RdRp S1 is forming four polar contacts with D55 and R56, while the C-terminal residue H562 form three polar contacts with Y176, C451, and Y452.
Figure 1c shows the RMSD Map of the dynamics of the SARS-CoV-2 RdRp and HCV RdRp during the 100 ns. RMSD Map analysis is a way to analyze the ensemble and calculate root-mean-square deviations between pairs of frames (calculate all pairwise RMSDs between frames and show the result as a map grayscale) [31]. RMSD map shows more similar structures in SARS-CoV-2 RdRp (lighter regions reflecting the frame pairs with lower RMSDs), while less similar frames are detected in HCV RdRp (darker regions reflecting the frame pairs with higher RMSDs).
Cluster analysis of RdRp conformations during the MDS runs was performed using the UCSF Chimera software with default parameters. One representative conformation from each cluster of the first ten most populous clusters is selected for the binding affinity calculations. The SARS-CoV-2 RdRp conformations at 55.7, 93.3, 34.9, 13.3, 28.1, 40.1, 81.3, 67.7, 19.7, and 4.9 ns are the representative conformations from the first 10 clusters, while 64.9, 94.1, 23.3, 14.5, 47.3, 32.9, 28.9, 84.1, 50.9, and 42.5 ns are the representative conformations from the first 10 clusters of HCV RdRp.
RdRp docking study
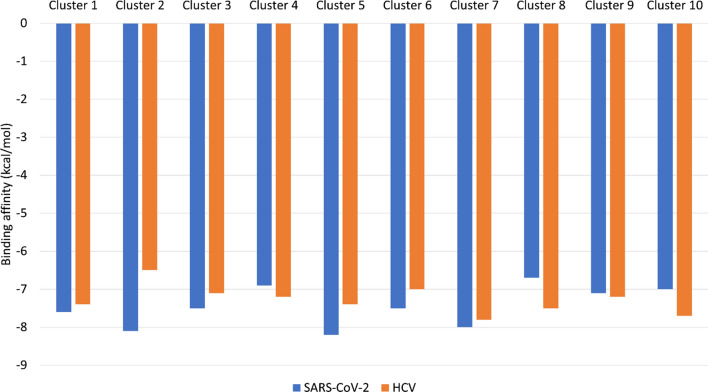
Figure 2 shows the binding energies (in kcal/mol) calculated for the binding of Sofosbuvir against SARS-CoV-2 RdRp (blue) and HCV RdRp (orange) using AutoDock Vina software. The calculated binding affinities are made for ten different conformations for each protein after cluster analysis of the MDS trajectories. The average binding affinity of Sofosbuvir against SARS-CoV-2 RdRp is − 7.46 ± 0.5 kcal/mol, while for the sofosbuvir against HCV RdRp, it is − 7.28 ± 0.35 kcal/mol. The average binding affinities are almost the same for the two RdRp. Hence, sofosbuvir is suggested to effectively bind SARS-CoV-2 RdRp and a possible potential drug against COVID-19.
Fig. 2.
The binding affinity (in kcal/mol) of sofosbuvir against SARS-CoV-2 RdRp (blue) and HCV RdRp (orange) from 10 different conformations for each protein after the cluster analysis of the 100 ns MDS runs. The calculations are done using AutoDock Vina software
The following tables show the details of the binding modes between sofosbuvir and each conformation of the SARS-CoV-2 RdRp (Table 1) and HCV RdRp (Table 2). From the PLIP webserver output, the most stable interactions established between the drug and the proteins are the H-bonding. In contrast, few salt bridges and hydrophobic interactions are reported for some conformations, while only two halogen contacts are reported (red) for SARS-CoV-2 RdRp (Table 1).
Table 1.
Protein–ligand interaction profiler (PLIP) analysis for docking results of the ten clusters of SARS-CoV-2 RdRp
| SARS-CoV-2 RdRp conformation at | AutoDock Vina score (Kcal/mol) | H-bonding | Salt bridges | Hydrophobic interactions | |||
|---|---|---|---|---|---|---|---|
| Number | Residues involved in the interaction | Number | Residues involved in the interaction | Number | Residues involved in the interaction | ||
| 55.7 ns | − 7.6 | 7 | R555(2), Y619, D623(2), N691, and D760 | 3 | R624(3) | 1 | D623 |
| 93.3 ns | − 8.1 | 10 | Y619, K621, R624(2), D760, D761, W800, E811(2), and S814 | 2/1 | K551(2) and D760 | 2 | Y619 |
| 34.9 ns | − 7.5 | 7 | W617, D618, K621, D623, W800, E811, and S814 | 5 | K551(2), R555, K621, and R624 | 1 | D618 |
| 13.3 ns | − 6.9 | 5 | D760, D761(2), E811, and S814 | 2 | R836(2) | ||
| 28.1 ns | − 8.2 | 6 | S549, R555(2), R624(2), and N691 | 2 | R555 and R624 | 4 | Y455, K551, R553, and K621 |
| 40.1 ns | − 7.5 | 12 | K551, W617, D618, Y619, K621, C622, D623(2), D761(2), W800, and E811 | 1 | Y619 | ||
| 81.3 ns | − 8.0 | 8 | R555, Y619, D623(2), D760, D761(2), and W800 | 3 | K551, Y619, and D760 | ||
| 67.7 ns | − 6.7 | 5 | K621, D760, D761, E811, and S814 | 3 | R555(2), and R624 | 1 | Y619 |
| 19.7 ns | − 7.1 | 7 | D761(2), A797, E811, and S814(3) | 1 | E811 | 3 | K798, and E811 |
| 4.9 ns | − 7.0 | 7 | D618, D760, D761, W800, and S814(3) | 1 | R836 | ||
Italics residues are residues interacting through halogen bonds with sofosbuvir
Table 2.
Shows protein–ligand interaction profiler (PLIP) analysis for docking results of ten clusters of HCV(2XI3)
| HCV RdRp conformation at | AutoDock Vina score (Kcal/mol) | H-bonding | Salt bridges | Hydrophobic interactions | |||
|---|---|---|---|---|---|---|---|
| Number | Residues involved in the interaction | Number | Residues involved in the interaction | Number | Residues involved in the interaction | ||
| 64.9 ns | − 7.4 | 11 | R158, F224, D225(2), T287, S288, N291, D318(2), and S556(2) | ||||
| 94.1 ns | − 6.5 | 13 | R158, S218, D225(3), D319, L320(2), S365, C366, and S367(3) | ||||
| 23.3 ns | − 7.1 | 8 | R200(2), D220, D318(2), D319(2), and S367 | 1 | F193 | ||
| 14.5 ns | − 7.2 | 10 | R158, F224, S288, N291, T292, C316, D318(2), and S556(2) | 1 | T287 | ||
| 47.3 ns | − 7.4 | 8 | R158, L159, D225(2), S282(3), and N291 | 1 | R158 | 1 | T287 |
| 32.9 ns | − 7 | 7 | R158, D220, F224, D225, N291, and D318(2) | ||||
| 28.9 ns | − 7.8 | 13 | L159, F224, D225, S288, N291, D318(3), D319(3), and S556(2) | ||||
| 84.1 ns | − 7.5 | 9 | R200, C366, R386, G410, N411, Y448(2), G449, and S556 | 2 | R200 and R386 | 1 | Y448 |
| 50.9 ns | − 7.2 | 9 | K141, R158, L159(2), T221, F224, D318, D319, and C366 | ||||
| 42.5 ns | − 7.7 | 12 | R158, R222, F224, D225, S282(2), S288(2), N291, C316, and D318(2) | 1 | VAL52 | ||
As shown in tables, the active site residues (D760 and D761 for SARS-CoV-2 RdRp and D318 and D319 for HCV RdRp), shown in bold, are interacting with sofosbuvir in most docking trials (9/10 for SARS-CoV-2 and 8/10 for HCV RdRp). An average of 11 interactions are established for each protein when coming in contact with sofosbuvir, most of which are H-bonding. More hydrophobic contacts and salt bridges are found in the case of SARS-CoV-2 RdRp, while only two halogen contacts are formed in SARS-CoV-2 RdRp. The details of the interactions established for SARS-CoV-2 and HCV RdRps are shown in the supplementary figures S1 and S2.
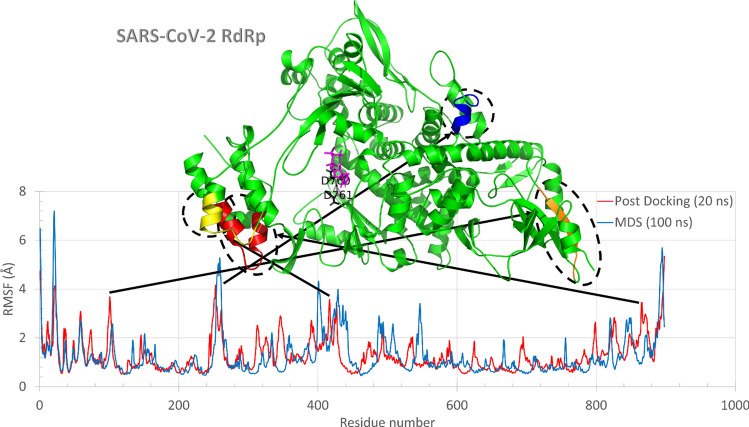
Sofosbuvir, in its active triphosphate form, can enter the nucleotide channel of RdRp and interact with its surface exposed consecutive aspartates (D760 and D761 in SARS-CoV-2 RdRp). To further study the binding mode, a 20 ns MDS is performed for the best complexes of sofosbuvir docked into SARS-CoV-2 RdRp and HCV RdRp using the same conditions. Figure 3 shows the superposition of the per residue RMSF for SARS-CoV-2 RdRp alone (blue line) during 100 ns MDS and the complex of sofosbuvir docked into SARS-CoV-2 RdRp (red line) during 20 ns MDS run. The complex structure is represented (top view) in the green cartoon, where sofosbuvir is depicted in magenta sticks. In addition to the terminals (N and C terminals), the protein–ligand complexes (red line) show some movable regions (RMSF ≤ 3 Å). The movable regions are shown in orange (K98-I106), blue (L251-L261), yellow (D421-V435), and red (D801-L809) cartoons. Figure 3 shows that the movable regions are all apart from the active site aspartates (black sticks) and the drug sofosbuvir (magenta sticks). This may result in the stabilization of the drug in the active site pocket. Hence the binding affinity of sofosbuvir against SARS-CoV-2 is suggested to be stable and not affected by the protein dynamics, just like the case in a previous study on ZIKV [9].
Fig. 3.
The per-residue RMSF for SARS-CoV-2 RdRp (blue line) and sofosbuvir- SARS-CoV-2 RdRp complex (red line). The structure of the complex is represented in the colored cartoon (green). The most fluctuating regions are colored in orange, blue, yellow, and red
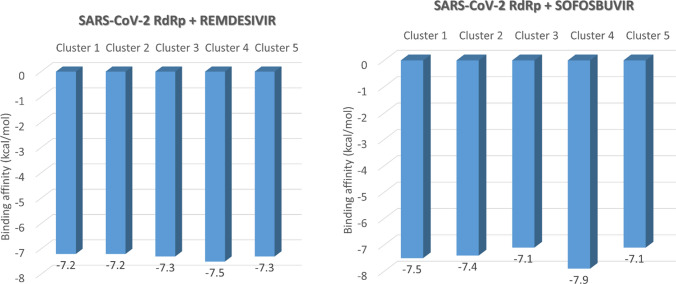
To further check the effect of protein dynamics on the drug binding, cluster analysis is performed for the protein–ligand complex trajectories using UCSF Chimera software. The best five clusters are redocked with Sofosbuvir and Remdesivir using the same protocol used in the previous docking study. Figure 4 shows the two drugs’ binding affinities against SARS-CoV-2 RdRp different conformations, while Tables 3 and 4 list the formed interactions in each docking experiment.
Fig. 4.
The binding affinities of Sofosbuvir (right) and Remdesivir (left) against the different conformations of SARS-CoV-2 RdRp after 20 ns MDS
Table 3.
Protein–ligand interaction profiler (PLIP) analysis for docking results of sofosbuvir against the different clusters of SARS-CoV-2 RdRp
| Cluster number | RdRp conformation at | H-bonding | Salt bridges | Hydrophobic interactions | |||
|---|---|---|---|---|---|---|---|
| Number | Amino acids involved | Number | Amino acids involved | Number | Amino acids involved | ||
| 1 | 5.9 ns | 8 | R555(2), K621, R624(2), T680, T687, and D760 | 3 | R555(3) | ||
| 2 | 13.7 ns | 8 | S549, R555(2), Y619, K621, C622, D623, and N691 | 1 | R555 | ||
| 3 | 18.6 ns | 9 | S549(2), R555(2), K621, D623, D760, and R836(2) | 2 | R555(2) | ||
| 4 | 9.2 ns | 12 | Y619(2), K621, C622, D623(2), N691, D760(4), and D761 | 1 | Y619 | ||
| 5 | 12.7 ns | 12 | K551, N552, R553(2), R555(2), D618, Y619, K621, D623, R624, and D761 | 5 | K551(2), R555, and K621(2) | 1 | D760 |
Table 4.
Protein–ligand interaction profiler (PLIP) analysis for docking results of Remdesivir against the different clusters of SARS-CoV-2 RdRp
| Cluster number | RdRp conformation at | H-bonding | Salt bridges | Hydrophobic interactions | |||
|---|---|---|---|---|---|---|---|
| Number | Amino acids involved | Number | Amino acids involved | Number | Amino acids involved | ||
| 1 | 5.9 ns | 9 | K621, D760, D761(4), W800, and S814(2) | ||||
| 2 | 13.7 ns | 5 | R555, D618, K621, D760, and D761 | 6 | K551(2), R555, and K621(3) | ||
| 3 | 18.6 ns | 5 | R555(2), K621(2), and D760 | 4 | R555(3), and R624 | 1 | R553 |
| 4 | 9.2 ns | 10 | D618, Y619, K621, D760(4), D761(2), and S814 | 4 | K551, R555(2), and K621 | 1 | K551 |
| 5 | 12.7 ns | 7 | K545, Y619, K621, D623, R624, and D760(2) | 3 | R555(2), and K621 | ||
As reported earlier, the active site residues D760 and D761 are involved in the interactions with both Sofosbuvir and Remdesivir. H-bonding is the primary type of interaction that stabilizes the drugs in the protein active site pockets. Additionally, few salt bridges are established upon binding the two drugs into the protein active site, while very few hydrophobic contacts are found in some clusters. The binding affinity of Sofosbuvir is very close to that for Remdesivir against SARS-CoV-2, as shown in Fig. 4. The average binding affinity for sofosbuvir against SARS-CoV-2 RdRp is − 7.4 ± 0.30 kcal/mol, while it is − 7.3 ± 0.11 kcal/mol for Remdesivir. Sofosbuvir proved its safety and anti-HCV activity during the last seven years and maybe a potential SARS-CoV-2 inhibitor. Clinical trials are needed to test its possible COVID-19 activity.
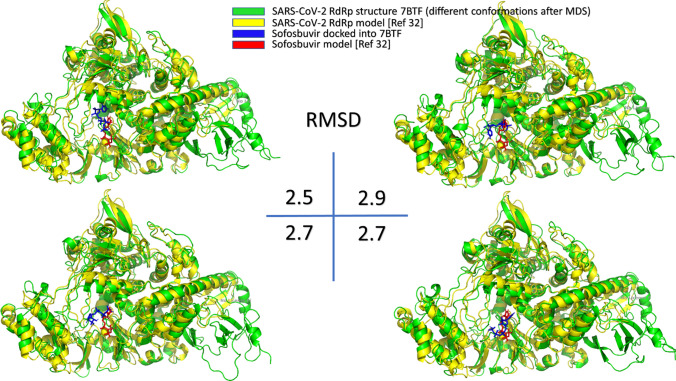
A superposition of the docking complexes for Sofosbuvir against RdRp model for SARS-CoV-2 (reported from a previous study [10]) and the same ligand docked against four different conformations of the solved structure of the same viral protein after MDS and trajectories clustering did in the current study is shown in Fig. 5. Figure illustrates the differences in the binding behavior and energies (− 7.6 kcal/mol for the previous docking study (RdRp model), while − 7.5, − 7.4, − 7.5, and − 7.9 kcal/mol for the four different conformations after clustering analysis (RdRp solved structure)). The root mean square deviations (RMSD) between each pair of the docking complexes lies between 2.5 and 2.9 Å, as shown in Fig. 5. In the two docking experiments, the binding is maintained with the same active site pocket, including the two active site aspartates, while little difference is reported in the binding site to Sofosbuvir [10, 29, 32]. This solidifies the effectiveness of this drug as a possible inhibitor against SARS-CoV-2 RdRp.
Fig. 5.
The superposition of the docking of sofosbuvir (blue and red sticks) into the RdRp model (yellow cartoon) and four different conformations of the RdRp solved structure (PDB ID: 7BTF) subjected to 100 ns MDS (green cartoon). The RMSD values (in Å) for each pair is shown at the center of the figure
Conclusions
Coronavirus diseases 2019 (COVID-19) pandemic seriously threatens human health around the world. The present study attempts to explore the binding affinity of sofosbuvir against the SARS-CoV-2 RdRp during dynamics simulation. Like Remdesivir, Sofosbuvir shows an excellent binding affinity to the RdRp active site and tightly interacts with the binding site; hence, it is supposed to be the right candidate as COVID-19 treatment. It may be used to effectively target SARS-CoV-2 RdRp after confirming the binding assays, in vitro and in vivo.
Acknowledgments
The MDS is done on the Bib Alex HPC, Alexandria, Egypt. This work is supported by the COVID-19 project team of Cairo University in 2020.
Author contributions
AE designed the research study and made the figures. EA performed the calculations and wrote the draft. MS supervises EA All authors have read and approved the final manuscript.
Compliance with ethical standards
Conflict of interest
All the authors declare that there is no conflict of interest for this work.
References
- 1.Organization WH (2020) Coronavirus disease 2019 (COVID-19): situation report, 72.
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malik A, El Masry KM, Ravi M, Sayed F. (2016) middle east respiratory syndrome coronavirus during pregnancy, Abu Dhabi, United Arab Emirates. Emerg Infect Dis. 2013;22(3):515–517. doi: 10.3201/eid2203.151049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for C Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Der Hoek L. Human coronaviruses: what do they cause? Antiviral therapy. 2007;12(4B):651–658. [PubMed] [Google Scholar]
- 7.Elfiky AA. Novel guanosine derivatives against Zika virus polymerase in silico. J Med Virol. 2020;92(1):11–16. doi: 10.1002/jmv.25573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elfiky AA. Reply to a letter to the editor. Life Sci. 2020;252:117715. doi: 10.1016/j.lfs.2020.117715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elfiky AA, Elshemey WM. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanosec. J Med Virol. 2018;90(1):13–18. doi: 10.1002/jmv.24934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253:117592. doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublie S, Ellenberger T. The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol. 1998;8(6):704–712. doi: 10.1016/s0959-440x(98)80089-4. [DOI] [PubMed] [Google Scholar]
- 12.Lawitz E, Mangia A, Wyles D, Rodriguez-Torres M, Hassanein T, Gordon SC, Schultz M, Davis MN, Kayali Z, Reddy KR, Jacobson IM, Kowdley KV, Nyberg L, Subramanian GM, Hyland RH, Arterburn S, Jiang D, McNally J, Brainard D, Symonds WT, McHutchison JG, Sheikh AM, Younossi Z, Gane EJ. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med. 2013;368(20):1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- 13.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.2.4.1 V The PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger, LLC.
- 15.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noorbatcha IA, Khan AM, Salleh HM. Molecular dynamics studies of human ? Glucuronidase. Am J Appl Sci. 2010;7(6):823–828. doi: 10.3844/ajassp.2010.823.828. [DOI] [Google Scholar]
- 17.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 18.Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A. 2001;105(43):9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Leach A (2001) Molecular Modelling: Principles and Applications (2nd Edition). Prentice Hall. doi:citeulike-article-id:571146
- 21.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elfiky AA, Azzam EB. Novel guanosine derivatives against MERS CoV polymerase: an in silico perspective. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N Engl J Med. 2013;368(1):34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- 24.Asselah T. Sofosbuvir for the treatment of hepatitis C virus. Expert Opin Pharmacother. 2014;15(1):121–130. doi: 10.1517/14656566.2014.857656. [DOI] [PubMed] [Google Scholar]
- 25.McQuaid T, Savini C, Seyedkazemi S. Sofosbuvir, a Significant Paradigm Change in HCV Treatment. J Clin Transl Hepatol. 2015;3(1):27–35. doi: 10.14218/JCTH.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfiky AA. The antiviral Sofosbuvir against mucormycosis: an in silico perspective. Future Virol. 2019;14(11):739–744. doi: 10.2217/fvl-2019-0076. [DOI] [Google Scholar]
- 27.Elfiky AA (2020) Natural products may interfere with SARS-CoV-2 attachment to the host cell. J Biomol Struct Dynam (just-accepted):1–16 [DOI] [PMC free article] [PubMed]
- 28.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015;43(W1):W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elfiky AA. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248:117477. doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Z, Lasker K, Schneidman-Duhovny D, Webb B, Huang CC, Pettersen EF, Goddard TD, Meng EC, Sali A, Ferrin TE. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elfiky AA. Corrigendum to "Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARSCoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study". Life Sci. 2020;258:118350. doi: 10.1016/j.lfs.2020.118350. [DOI] [PMC free article] [PubMed] [Google Scholar]