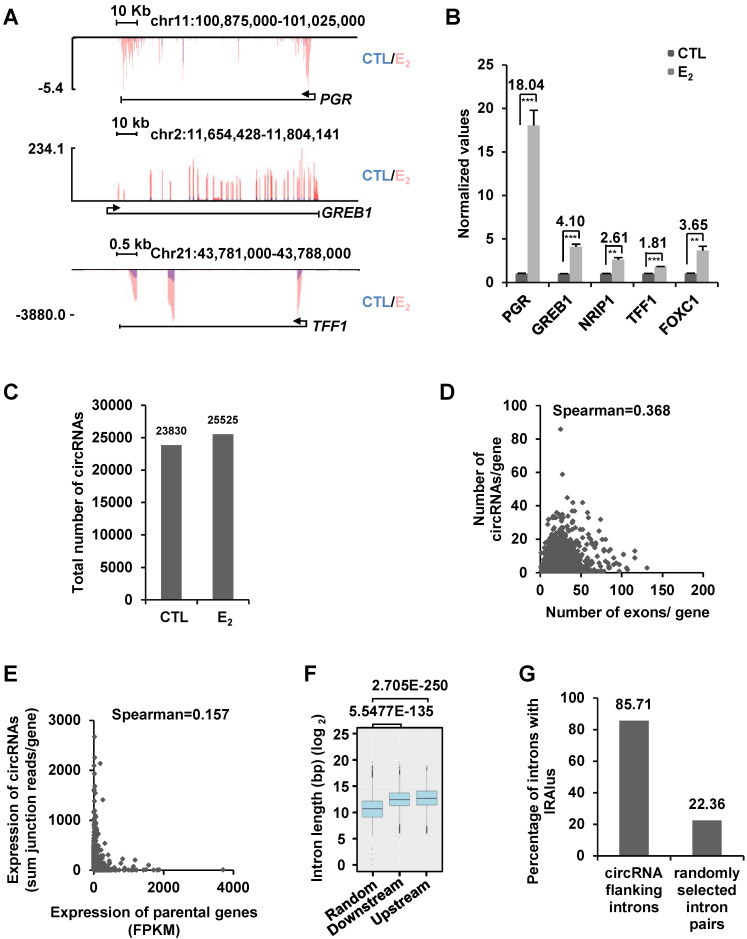
Figure 1.
A large number of circRNAs are detected by circRNA-seq in MCF7 cells. (A) MCF7 cells were cultured in stripping medium for three days and treated with or without estrogen (E2, 10-7 M, 6 h) followed by RNA-seq analysis, and genome browser views of selected genes as indicated were shown. Blue: control (CTL); Red: estrogen (E2). (B) RNA samples as described in (A) were subjected to RT-qPCR analysis to examine the expression of PGR, GREB1, NRIP1, TFF1 and FOXC1. Fold change by estrogen treatment was shown as indicated. Data shown was the relative fold change compared to control samples (CTL) after normalization to actin (± s.e.m, **P < 0.01, ***P < 0.001). (C) RNA samples as described in (A) were subjected to circRNA-seq. Total number of circRNAs predicted from circRNA-seq were shown (unique junction reads ≥ 2). (D) Correlation between the number of exons a gene has and circRNAs originated from that gene was shown. (E) Correlation between the expression of parental gene (FPKM) and circRNA originated from the same gene region (average junction reads/gene) was shown. (F) Box plot showing the length of randomly selected introns or introns flanking, upstream and downstream, circRNA-producing regions. Median: 1,639 bp (random); 5,526 bp (downstream); 6,578 bp (upstream). bp: base pair. (G) Percentage of randomly selected introns or introns flanking circRNA-producing regions containing inverted repeated Alu pairs was shown.