Abstract
Imaging techniques are used to capture anomalies of the human body. The captured images must be understood for diagnosis, prognosis and treatment planning of the anomalies. Medical image understanding is generally performed by skilled medical professionals. However, the scarce availability of human experts and the fatigue and rough estimate procedures involved with them limit the effectiveness of image understanding performed by skilled medical professionals. Convolutional neural networks (CNNs) are effective tools for image understanding. They have outperformed human experts in many image understanding tasks. This article aims to provide a comprehensive survey of applications of CNNs in medical image understanding. The underlying objective is to motivate medical image understanding researchers to extensively apply CNNs in their research and diagnosis. A brief introduction to CNNs has been presented. A discussion on CNN and its various award-winning frameworks have been presented. The major medical image understanding tasks, namely image classification, segmentation, localization and detection have been introduced. Applications of CNN in medical image understanding of the ailments of brain, breast, lung and other organs have been surveyed critically and comprehensively. A critical discussion on some of the challenges is also presented.
Keywords: Classification, Convolutional neural networks, Detection, Image understanding, Localization, Segmentation
Introduction
Loss of human lives can be prevented or the medical trauma experienced in an injury or a disease can be reduced through the timely diagnosis of medical anomalies. Medical anomalies include glaucoma, diabetic retinopathy, tumors [34], interstitial lung diseases [44], heart diseases and tuberculosis. Diagnosis and prognosis involve the understanding of the images of the affected area obtained using X-ray, magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography (PET), single photon emission computed tomography or ultrasound scanning. Image understanding involves the detection of anomalies, ascertaining their locations and borders, and estimating their sizes and severity. The scarce availability of human experts and their fatigue, high consultation charges and rough estimate procedures limit the effectiveness of image understanding. Further, shapes, locations and structures of the medical anomalies are highly variable [55]. This makes diagnosis difficult even for specialized physicians [4]. Therefore, human experts often feel a need for support tools to aid in precise understanding of medical images. This is the motivation for intelligent image understanding systems.
Image understanding systems that exploit machine learning (ML) techniques are fast evolving in recent years. ML techniques include decision tree learning [35], clustering, support vector machines (SVMs) [47], k-means nearest neighbor (K-NN), restricted Boltzmann machines (RBMs) [42] and random forests (RFs) [28]. The pre-requisite for ML techniques to work efficiently is the extraction of discriminant features. And these features are generally unknown and is also a very challenging task especially for applications involving image understanding and is still a topic of research. A logical step to overcome was to create intelligent machines which could learn features needed for image understanding and extract it on its own. One such intelligent and successful model is the convolutional neural network (CNN) model, which automatically learns the needed features and extracts it for medical image understanding. The CNN model is made of convolutional filters whose primary function is to learn and extract necessary features for efficient medical image understanding. CNN started gaining popularity in the year 2012, due to AlexNet [41], a CNN model, which defeated all the others models with a record accuracy and low error rate in imageNet challenge 2012. CNN has been used by corporate giants for providing internet services, automatic tagging in images, product recommendations, home feed personalization and autonomous cars [59]. The major applications of the CNN are in image and signal processing, natural language processing and data analytics. The CNN had a major breakthrough when GoogleNet used it to detect cancer at an accuracy of 89% while human pathologists could achieve the accuracy of only 70% [3].
Motivation and purpose
CNNs have contributed significantly in the areas of image understanding. CNN-based approaches are placed in the leader board of the many image understanding challenges, such as Medical Image Computing and Computer Assisted Intervention (MICCAI) biomedical challenge, Brain Tumor segmentation (BRATS) Multimodal Brain Tumor Segmentation challenge [48], Imagenet classification challenge, challenges of International Conference on Pattern Recognition (ICPR) [31] and Ischemic Stroke Lesion Segmentation (ISLES) challenge [32]. CNN has become a powerful choice as a technique for medical image understanding. Researchers have successfully applied CNNs for many medical image understanding applications like detection of tumors and their classification into benign and malignant [52], detection of skin lesions [50], detection of optical coherence tomography images [39], detection of colon cancer [71], blood cancer, anomalies of the heart [40], breast [36], chest, eye etc. Also CNN-based models like CheXNet [56, 58], used for classifying 14 different ailments of the chest achieved better results compared to the average performance of human experts.
CNNs have also dominated the area of COVID-19 detection using chest X-rays/CT scans. Research involving CNNs is now a dominant topic at major conferences. In addition, there are special issues reserved in reputed journals for solving challenges using deep learning models. The vast amount of literature available on CNNs is the testimonial of their efficiency and the widespread use. However, various research communities are developing these applications concurrently and the dissemination results are scattered in a wide and diverse range of conference proceedings and journals.
A large number of surveys on deep learning have been published recently. A review of deep learning techniques applied in medical imaging, bioinformatics and pervasive sensing has been presented in [60]. A thorough review of deep learning techniques for segmentation of MRI images of brain has been presented in [2]. Survey of deep learning techniques for medical image segmentation, their achievements and challenges involved in medical image segmentation has been presented in [27] Though literature is replete with many survey papers, most of them concentrate on deep learning models which include CNN, recurrent neural network, generative adversial network or on a particular application. There is also no coverage of the application of CNN in early detection of COVID-19 as well as many other areas.
The survey includes research papers on various applications of CNNs in medical image understanding. The papers for the survey are queried from various journal websites. Additionally, arxiv, conference proceedings of various medical image challenges are also included in the survey. Also the references of these papers are checked. The query used are: “CNN” or “deep learning” or “convolutional neural network” or terms related to medical image understanding. These terms had to be present either in title or abstract to be considered.
The objective of this survey is to offer a comprehensive overview of applications and methodology of CNNs and its variants, in the fields of medical image understanding including the detection of latest global pandemic COVID-19. The survey includes overview tables which can be used for quick reference. The authors leverage experiences of their own and that of the research fraternity on the applications of CNNs to provide an insight into various state of the art CNN models, challenges involved in designing CNN model, overview of research trends in the field, and to motivate medical image understanding researchers and medical professionals to extensively apply CNNs in their research and diagnosis, respectively.
Contributions and the structure
Primary contributions of this article are as follows:
To briefly introduce medical image understanding and CNN.
To convey that CNN has percolated in the field of medical image understanding.
To identify the various challenges in medical image understanding.
To highlight contributions of CNN to overcome those challenges
The remainder of this article has been organized as follows: Medical image understanding has been briefly introduced in Sect. 2. A brief introduction of CNN and its architecture has been presented in Sect. 3. The applications of CNN in medical image understanding have been surveyed comprehensively through Sects. 4–7. Finally, concluding remarks and a projection of the trends in CNN applications in image understanding have been presented in Sect. 8.
Medical image understanding
Medical imaging is necessary for the visualization of internal organs for the detection of abnormalities in their anatomy or functioning. Medical image capturing devices, such as X-ray, CT, MRI, PET and ultrasound scanners capture the anatomy or functioning of the internal organs and present them as images or videos. The images and videos must be understood for the accurate detection of anomalies or the diagnosis of functional abnormalities. If an abnormality is detected, then its exact location, size and shape must be determined. These tasks are traditionally performed by the trained physicians based on their judgment and experience. Intelligent healthcare systems aim to perform these tasks using intelligent medical image understanding. Medical image classification, segmentation, detection and localization are the important tasks in medical image understanding.
Medical image classification
Medical image classification involves determining and assigning labels to medical images from a fixed set. The task involves the extraction of features from the image, and assigning labels using the extracted features. Let I denote an image made of pixels and denote the labels. For each pixel x, a feature vector , consisting of values is extracted from the neighborhood N(x) using (1), where for .
| 1 |
A label from the list of labels is assigned to the image based on .
Medical image segmentation
Medical image segmentation helps in image understanding, feature extraction and recognition, and quantitative assessment of lesions or other abnormalities. It provides valuable information for the analysis of pathologies, and subsequently helps in diagnosis and treatment planning. The objective of segmentation is to divide an image into regions that have strong correlations. Segmentation involves dividing the image I into a finite set of regions as expressed in (2).
| 2 |
Medical image localization
Automatic localization of pathology in images is quite an important step towards automatic acquisition planning and post imaging analysis tasks, such as segmentation and functional analysis. Localization involves predicting the object in an image, drawing a bounding box around the object and labeling the object.
The localization function f(I) on an image I computes , which represent respectively, class label, centroid x and y coordinates, and the proportion of the bounding box with respect to width and height of the image as expressed in (3).
| 3 |
Medical image detection
Image detection aims at the classification and the localization of regions of interest by drawing bounding boxes around multiple regions of interest and labeling them. This helps in determining the exact locations of different organs and their orientation. Let I be an image with n objects or regions of interest. Then detection function D(I) computes and these are respectively the class label, centroid x and y coordinates, proportion of the bounding box with respect to width and height of the image I as given in the (4)
| 4 |
Evaluation metrics for image understanding
There are many metrics used for the evaluation of performance of medical image understanding algorithms. The confusion matrix, also known as the error matrix, is the table used for visualizing the performance of an algorithm and for calculation of various evaluation metrics. It provides an insight about the types of errors that are made by the classifier. It is a square matrix in which rows represent the instances of actual results and the columns represent the instances of predicted results of the algorithm. The confusion matrix of a binary classifier is shown in Table 1.
Table 1.
Confusion matrix
| Total test samples | Predicted positive | Predicted negative |
|---|---|---|
| Actual positive | True positives () | False negatives () |
| Actual negative | False positives () | True negatives () |
Here, indicates correctly identified positives, indicates correctly identified negatives, indicates incorrectly identified positives and indicates incorrectly identified negatives. is also known as false error and is known as miss. The sum of correct and incorrect predictions is represented as T and expressed as in (5).
| 5 |
Performance metrics can be determined with the help of confusion matrix and are given in Table 2.
Table 2.
Performance evaluation metrics for image processing
| Evaluation metric | To determine | Formula | Preferred value |
|---|---|---|---|
| Accuracy | Overall how often the result is true | High | |
| Misclassification result (error rate) | Overall how often the result is false | Low | |
| True positive rate () (recall or sensitivity) | When it is actually true, how often does it predict true | High | |
| False positive rate () | When its actually false, how often does it predict true | Low | |
| False negative rate () | Low | ||
| Specificity | When its actually false, how often does it predict false | High | |
| Precision (positive predictive value ()) | When it predicts true, how often is it correct? | High | |
| F-Score (Dice similarity coefficient) | Harmonic mean of recall and precision. | High | |
| Receiver operating characteristic (ROC) | Commonly used graph to summarizes the performance of a classifier over all possible thresholds. | Plot of recall verses FPR | High |
| Area under ROC (AUC) | The area under ROC | Plots versus | High |
A brief introduction to CNNs
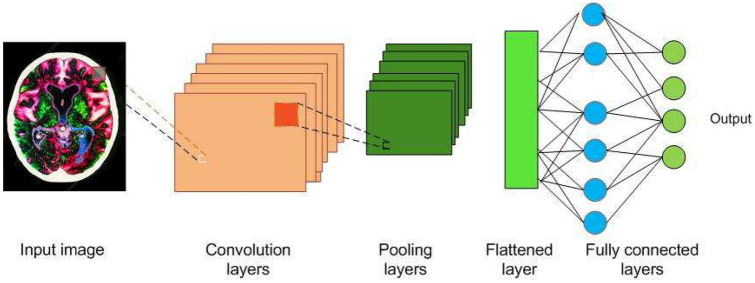
Image understanding by animals is a very fascinating process, and a very simple task for them. But for a machine, to understand an image, there are lot of hidden complexities during the process. What animals feel is the eyes capturing the image, which is processed by the neurons and sent to the brain for interpretation. CNN is a deep learning algorithm inspired by the visual cortex of animal brain [30] and aims to imitate the visual machinery of animals. CNNs represents a quantum leap in the field of image understanding, involving image classification, segmentation, localization, detection etc. The efficacy of CNNs in image understanding is the main reason of its abundant use. CNNs are made of convolutions having learnable weights and biases similar to neurons (nerve cells) of the animal. Convolutional layers, activation functions, pooling and fully-connected layers are the core building blocks of CNNs, as depicted in Fig. 1. Very brief introduction to CNNs has been presented in this paper. Detailed discussions on CNNs are presented in [9, 41].
Fig. 1.
Building blocks of a CNN
Convolution layers (Conv layers)
The visual cortex of the animal brain is made of neuronal cells which extract features of the images. Each neuronal cell extracts different features, which help in image understanding. The conv layer is modeled over the neuronal cells and its objective is to extract features, such as edges, colors, texture and gradient orientation. Conv layers are made of learnable filters called convolutional filters, or kernels, of size , where d is the depth of the image. During the forward pass, the kernels are convolved across the width and height of input volume and dot product is computed between the entries of the filter and the input. Intuitively, the CNN learns filters that gets activated when they come across edge, colors, texture etc. The output of the conv layer is fed into an activation function layer.
Activation functions or nonlinear functions
Since data in real world is mostly nonlinear, activation functions are used for nonlinear transformation of the data. It is used to ensure that the representation in the input space is mapped to a different output space as per the requirements. The different activation functions are discussed in Sects. 3.2.1–3.2.3.
Sigmoid
It takes a real-valued number x and squashes it into range between 0 and 1. In particular, large negative and positive inputs are placed very close to 0 and unity, respectively. It is expressed as in (6).
| 6 |
Tan hyperbolic
It takes a real valued number x and squashes it between to 1 as expressed in (7).
| 7 |
Rectified linear unit (ReLU)
This nonlinear function takes a real valued number x and converts x to 0 if x is negative. ReLU is the most often used nonlinear function for CNN, takes less computation time and hence faster compared to the other two and is expressed in (8).
| 8 |
Pooling
Pooling layer performs a nonlinear down sampling of convolved feature. It decreases the computational power required to process the data through dimensionality reduction. It reduces the spatial size by aggregating data over space or feature type, controls overfitting and overcomes translation and rotational variance of images. Pooling operation results in partitioning of its input into a set of rectangle patches. Each patch gets replaced by a single value depending on the type of pooling selected. The different types are maximum pooling and average pooling.
Fully connected (FC) layer
FC layer is similar to artificial neural network, where each node has incoming connections from all the inputs and all the connections have weights associated with them. The output is sum of all the inputs multiplied by the corresponding weights. FC layer is followed by sigmoid activation function and performs the classifier job.
Data preprocessing and augmentation
The raw images obtained from imaging modalities need to be preprocessed and augmented before sending to CNN. The raw image data might be skewed, altered by bias distortion [55], having intensity inhomogeneity during capture, and hence needs to be preprocessed. Multiple data preprocessing methods exist and the preferred methods are mean subtraction and normalization. CNN needs to be trained on a larger dataset to achieve the best performance. Data augmentation increases the existing set of images by horizontal and vertical flips, transformations, scaling, random cropping, color jittering and intensity variations. The preprocessed, augmented image data is then fed into CNN.
CNN architectures and frameworks
Many CNN architectures have been proposed by researchers depending on kind of task to be performed. A few award-winning architectures are listed in Table 3. CNN frameworks (toolkits) enable the efficient development and implementation of deep learning methods. Various frameworks used by researchers and developers is listed in Table 4.
Table 3.
Various award winning CNN architectures
Table 4.
Various existing CNN frameworks
| Deep learning frameworks | Developed at | Implemented using | Pros | Cons |
|---|---|---|---|---|
| Caffe [33] | Berkeley Vision and Learning Centre | C++ | Cross platform, easy to deploy, fast, has good Matlab and Python interface, models can be trained without writing code | Usability drops outside ConvNets, not very good with new architectures |
| Keras [11] | Francois Chollet, Google | Python | Easiest deep learning framework, clean API, huge active community, easily extensible, user-friendly, works with Theano and TensorFlow | Poor support for multi node training, cannot be used efficiently as an independent framework |
| TensorFlow [1] | Google Brainteam | Python, C++ | Has modular architecture, supports multiple front ends and execution platforms, availability of tensorboard for visualization | Slow, slower than Theano and Torch in terms of execution time, lacks many pre-trained models, not completely open source |
| Torch [15] | New York University | Lua | Modular architecture, easy to setup, displays helpful error messages and has a large amount of tutorials | Difficult to integrate since it is implemented in Lua and its data science stack in R or Python |
| PyTorch [53] | Facebook artificial intelligence group | Python, C++, CUDA | Fast, minimal framework overhead, understanding error messages and stack traces is easy and hence easy to debug, good for research and development | Depends on Matplotlib and Seaborn for visualization, lacks distributed training |
| Theano [3] | Python | Expressive Python syntax, large help on net, supports high level wrappers | Cross platform, needs secondary libraries for deploying, supports single GPU, error messages are not helpful | |
| Neon [51] | Nervana Systems | Python | Very fast due to fast matrix operations, the same code runs on GPU and CPU | Too many errors with scattered dependencies, less material on web, architecture not user friendly, not general-purpose |
| Deeplearning4j [59] | Java | Performance equal to Caffe and better than TensorFlow or Torch, fast | Generally not preferred because Java is unpopular in ML circles | |
| Cognitive Toolkit (CNTK) [64] | Microsoft | C++ | Fast, accurate scalable over GPUs, flexible, allows distributed training, supports C++, Java, Python and C# | Lacks visualizations, crashes with error, no Matlab or Python bindings |
CNN applications in medical image classification
Lung diseases
Interstitial lung disease (ILD) is the disorder of lung parenchyma in which lung tissues get scarred leading to respiratory difficulty. High resolution computed tomography (HRCT) imaging is used to differentiate between different types of ILDs. HRCT images have a high visual variation between different classes and high visual similarity within the same class. Therefore, accurate classification is quite challenging.
Ensemble CNN
Ensemble of rf and overfeat for classification of pulmonary peri fissural nodules of lungs was proposed in [14]. The complexity of the input was reduced by extracting two-dimensional views from three-dimensional volume. The performance was enhanced by using a combination of overfeat followed by rf. The bagging technique of rf boosted the performance of the model. The proposed model obtained an AUC of .
Small-kernel CNN
Low level textual information and more non linear activations enhances performance of classification was emphasized by [4]. The authors shrinked the kernel size to to involve more non linear activations. The receptive fields were kept smaller to capture low level textual information. Also, to handle increasing complexity of the structures, the number of kernels were made proportional to the number of receptive field of its neurons. The model classified the lung tissue image into seven classes (a healthy tissue and six different ILD patterns). The results were compared against AlexNet and VGGNet and the ROC curves. The structure took only 20 s to classify the whole lung area in 30 slices of an average size HRCT scan image. AlexNet and VGG-Net took 136 s and 160 s for classification. The model delivered a classification accuracy of , while the traditional methods delivered an accuracy of .
Whole image CNN
Smaller image patches to prevent loss of spatial information and different attenuation ranges to enhance better visibility was proposed in [18]. Since the images were RGB, the proposed CNN model used three lung attenuation ranges namely, lower attenuation, normal attenuation and higher attenuation. To avoid overfitting, the images were augmented by jitter and cropping. A simple Alexnet model with the above variations was implemented and compared against other CNN models implemented to work on image patches. The performance metrics were accuracy and F-score. The model obtained an F-score of and the average accuracy of .
Multicrop pooling CNN
Limitation of reduced training samples can be overcome by extraction of salient multi scale features. The features were extracted using multicrop pooling for automatic lung nodule malignancy suspicious classification in [68]. The model was a simple 3 layered CNN architecture but with multicrop pooling and randomized leaky ReLu as activation. The proposed method obtained accuracy and AUC of and . Fivefold cross validation was used for evaluation.
The CNN applications in lung classification is summarized in Table 5.
Table 5.
A summary of CNN applications in ILD image classification surveyed in Sect. 4
| References | Preprocessing | Architecture | Dataset | Comparison against | Performance metric |
|---|---|---|---|---|---|
| [44] | Normalized with unit variance and zero-mean | Conv, ReLU, maxpool, FC , backpropagation, batch processing, dropout of 0.6 | ILD | SIFT, LBP, unsupervised feature learning using RBM | Recall, precision |
| [4] | Augmentation by rotating | (Conv, Leaky ReLU, avgpool) , three FC, softmax classifier | ILD from University of Geneva and Bern University hospital | AlexNet, VGGNet | Time, ROC, AUC |
| [18] | Augmentation by adding jitter and cropping | AlexNet architecture | Public ILD database | CNN patch based methods | Accuracy compared to using patch based methods |
| [68] | Augmentation | (Conv, ReLU, maxpool with multicrop) + FC, softmax | LIDC-IDRI dataset | HOG, LBP | Accuracy and AUC |
Coronavirus disease 2019 (COVID-19)
COVID-19 is a global pandemic disease spreading rapidly around the world. Reverse Transcription Polymerase Chain Reaction (RT-PCR) is a commonly employed test for detection of COVID-19 infection. RT-PCR testing is the gold standard for COVID-19 testing, RT-PCR is very complicated, time-consuming and labor-intensive process, sparse availability and not very accurate. Chest X-ray could be used for the initial screening of the COVID-19 in places having shortage of RT-PCR kits and is more accurate at diagnosis. Many researchers have used deep learning to classify if the chest infection is due to COVID-19 or other ailments.
Customized CNN
One of the initial model proposed for detection of COVID-19 was a simple pretrained AlexNet model proposed in [45] and fine tuned on chest X-ray images. The results were very promising with accuracy of classifying positive and negative patients of around . Use of pretrained with transfer learning ResNet and InceptionNet CNN models were also proposed. These models demonstrated that the transfer learning models were also efficient and achieved a test accuracy of .
Bayesian CNN
Uncertainty was explored to enhance the diagnostic performance of classification of COVID-19 datasets in [22]. The primary aim of proposed method was to avoid COVID-19 misdiagnoses. The method explored Monte-Carlo Dropweights Bayesian CNN to estimate uncertainty in deep learning, to better the diagnostic performance of human-machine decisions. The method showed that there is a strong correlation between classification accuracy and estimated uncertainty in predictions. The proposed method used ResNet50v2 model. The softmax layer was preceded by dropweights. Dropweights were applied as an approximation to the Gaussian Process, which was used to estimate meaningful model uncertainty. The softmax layer finally outputs each possible class label’s probability distribution.
PDCOVIDNET
Use of dilation to detect dominant features in the image was explored in [13]. The authors proposed parallel dilated CNN model. The dilated module involved skipping of pixels during convolution process. Parallel CNN branches are proposed with different dilation rates. The features obtained from parallel branches were concatenated and input to the next convolution layer. Concatenation-convolution operation was used to explore feature relationship of dilated convolutions so as to detect dominant features for classification. The model also used Grad-CAM and Grad-CAM++ to highlight the regions of class-discriminative saliency maps. The performance metrics used were accuracy, precision, recall, F1-score with ROC/AUC and are and 0.991 respectively.
CVR-Net
To prevent degrading of final prediction and to compensate for lesser number of datasets, multi scale multi encoder ensemble CNN model for classification of COVID-19 was proposed in [24]. The proposed model ensembled feature maps at different scales obtained from different encoders. To avoid overfitting, geometry based image augmentations and transfer learning was proposed. To overcome vanishing gradients, each encoder consisted of residual and convolutional blocks to allow gradients to pass, like in resNet architecture. Moreover Depth-wise separable convolution was used to create a light weight network. The depth information of feature map was enhanced by concatenating different 2D feature maps of different encoders in channel-wise. The performance metrics for classifying images into positive and negative were recall, precision, f1-score and accuracy. The model showed a very efficient performance with score of nearly for all the metrics.
Twice transfer learning CNN
A denseNet model trained twice using transfer learning approach was proposed in [6]. The denseNet201 model was trained initially on imageNet dataset , followed by chest X-ray 14 dataset and then fine tuned on COVID-19 dataset. Various combinations of training the model first with single transfer learning, twice transfer learning, twice transfer learning with output neuron keeping were experimented. The model with twice transfer learning with output neuron keeping achieved the best performance accuracy of over the other models. Transfer learning on chest X-ray 14 dataset enhanced the result, as the model had learnt most of the features related to chest abnormalities.
Immune response abnormalities
Autoimmune diseases result from an abnormal immune response to a normal body part. The immune system of the body attacks the healthy cells in such diseases. Indirect immunofluorescence (IIF) on human epithelial-2 (HEp-2) cells is used to diagnose an autoimmune disease. Manual identification of these patterns is a time-consuming process.
CUDA ConvNet CNN
Preprocessing using histogram equalization and zero-mean with unit variance increases classification accuracy by an additional with augmentation was proposed in [7]. The experiments also demonstrate that pretraining, followed by fine tuning boosts performance. It achieved an average classification accuracy of which was greater than the previous best of . The authors used Caffe library [33] and CUDA ConvNet model architecture to extract CNN-based features for classification of HEp-2 cells.
Six-layer CNN
Preprocessing and augmentation enhanced the mean classification accuracy of HEp-2 cell images and was shown in [21]. The framework consisted of three stages of image preprocessing, network training and feature extraction with classification. Mean classification accuracy of on ICPR-2012 dataset was obtained. The CNN approaches for HEp-2 cell classification are summarized in Table 6.
Table 6.
A summary of CNN applications in HEp-2 cell classification surveyed in Sect. 4
| References | Preprocessing | Architecture | Dataset | Comparison against | Perfromance metric |
|---|---|---|---|---|---|
| [7] | Augmented by affine transform and intensity variations, green channel images used | Conv, maxpool, ReLU, conv, avgpool, ReLU, conv, avgpool, FC with dropout, weight decay, softmax with multinomial logistic regression | ICPR 2014 | ICPR 2012 benchmark and winners model | Average classification accuracy |
| [21] | Normalized using zero mean and unit variance, and rotated | (Conv, tanh, maxpool) , FC , softmax regression, outputs six classes | ICPR 2014 | ICPR 2012 classification contestants | Accuracy |
Breast tumors
Breast cancer is the most common cancer that affects women across the world. It can be detected by the analysis of mammographs. Two radiologists independently reading the same mammogram has been advocated to overcome any misjudgement.
Stopping monitoring CNN
Stopping monitoring to reduce the computation time is proposed in [52]. Stopping monitoring was performed using AUC on validation set. The model extracted region of interest (ROI) by cropping. The images were augmented to increase the number of samples and also to prevent overfitting. The CNN model proposed was for classification of breast tumors. The result was compared against the state-of-the-art image descriptors HOG and HOG divergence. The proposed method resulted in AUC of compared to for other methods.
Ensemble CNN
Fine tuning enhanced the performance in the case of limited dataset was proposed in [34]. The proposed model was similar to AlexNet and was pretrained on imagenet, followed by fine tuning on breast images due to shortage of breast images. The middle and high level features were extracted from different layers of the network and fed into SVM classifiers for training. The model classified the breast masses into malignant and benign. Due to extraction of efficient features by deep network, simple classifier also resulted in accuracy of . The proposed method was compared against bag-of-words, HOG and SIFT and it outperformed all of them.
Semi-supervised CNN
CNN can also be used in scenarios involving sparse labeled data and abundant unlabeled data. To overcome the sparse labeled data problem, a new graph based semi supervised learning techniques for breast cancer diagnosis was proposed in [73]. For removal of redundancies and feature correlations, dimensionality reduction was employed. The method used four modules, feature extraction to extract 21 features from breast masses, data weighing to minimize the influence of noisy data and division of co-training data labeling followed by the CNN. It involved sub patches extraction of ROIs which were input to three pairs of conv layers, maxpooling and FC layer. Three models CNN, SVM and ANN are compared. The AUC for CNN for mixture of labeled and unlabeled data was compared to of SVM and of ANN and accuracy for CNN with mixed data was compared to for SVM and for ANN. The CNN approaches for breast tumor classification are summarized in Table 7.
Table 7.
A summary of CNN applications in breast medical image classification surveyed in Sect. 4
| References | Preprocessing | Architecture | Comparison against | Performance metric |
|---|---|---|---|---|
| [34] | Normalized by subtracting mean, dividing by standard deviation, augmented by flipping | (Conv, ReLU, maxpool) , (FC + ReLU) + FC, stochastic gradient descent | BOW, HOG, SIFT, VGGNet | ROC and accuracy |
| [52] | Crop, augmentation by flipping and rotating, global contrast normalization, local contrast normalization | (Conv, ReLU, maxpool) + FC | HOG, HGD | AUC |
| [73] | Feature extraction, data weighing, data labeling | (Conv, maxpool) , FC | SVM, ANN, labeled data, unlabeled data and mixed data | Accuracy , AUC |
| [78] | Image oversegmented into atomic regions using superpixel based scheme | (Conv, maxpool) , FC , softmax classifier | Linder, Bianconi, DCNN-Ncut + SVM, DCNN-Ncut + SMC, DCNN-Ncut + SVM, DCNN-SLIC-SVM, DCNN-SLIC-SMC | ROC, AUC, accuracy |
Heart diseases
Electrocardiogram (ECG) is used for the assessment of the electrical activity of the heart to detect anomalies in the heart.
One-dimensional CNN
ECG classification using CNN model demonstrates superior performance with classification accuracy of was proposed in [40]. The model comprised of one-dimensional CNN with three conv layers and two FC layers that fused feature extraction and classification into a single learning body. Once the dedicated CNN was trained for a particular patient, it could be solely used to classify ECG records in a fast and accurate manner.
Fused CNN
Classification of echocardiography videos require both spatial and temporal data. Fused CNN architecture using both spatial and temporal data was proposed in [20]. It used a two-path CNN, one along the spatial direction and the other along temporal direction. Each individual CNN path executed individually and was fused only after obtaining the final classification scores. The spatial CNN learnt the spatial information automatically from the original normalised echo video images. Temporal CNN learnt from acceleration images along the time direction of the echo videos. The outputs of both CNNs were fused and applied to softmax classifier for the final classification. The proposed model achieved an average accuracy of compared to for single path CNN, for three-dimensional KAZE, for three-dimensional SIFT. The long time required for initial training was the disadvantage of this approach. The CNN approaches to heart classification are summarized in Table 8.
Table 8.
A summary of CNN applications in heart classification surveyed in Sect. 4
| References | Preprocessing | Architecture | Dataset | Comparison against | Performance metric |
|---|---|---|---|---|---|
| [40] | ECG signals filtered using bandpass filter at 0.1-100 Hz and digitized at 360 Hz. | One-dimensional CNN with conv , FC | MIT/BIH arrhythmia database | Other existing methods | Accuracy, sensitivity, specificity |
| [20] | Image resized to | Two path CNN of seven layers (conv, ReLU, maxpool) , (conv, ReLU) , (conv, ReLU, maxpool), (conv, dropout) | Tsinghua University Hospital, Beijing and Fuzhou University Hospital, China | SIFT, KAZE | Accuracy |
Eye diseases
Gaussian initialized CNN
Initial training time can be reduced by Gaussian initialization, and overfitting can be avoided by weighted class weights. This was proposed for classifying diabetic retinopathy (DR) in fundus imagery in [57]. The performance was compared with SVM and other methods that required feature extraction prior to classification. The method achieved specificity but less sensitivity of . The trained CNN did a quick diagnosis and gave an immediate response to the patient during screening.
Hyper parameter tuning inception-v4
Automated hyper parameter tuning inception-v4 (HPTI-v4) model for DR in color fundus images classification and detection is proposed in [67]. The images are preprocessed using CLAHE to enhance contrast level, segmented using histogram based segmentation model. Hyper parameter tuning is done using Bayesian optimization method, as Bayesian model has the ability to analyze the previous validation outcome, to create a probabilistic model. Classification is done using HPTI-v4 model followed by multi layer perceptron. The classification is applied on MESSIDOR DR dataset. The CNN model performance was extraordinary with the accuracy, sensitivity, and specificity of , , and respectively.
Colon cancer
Ensemble CNN
Usage of small patches increased the amount of training data and localized the analysis to small nuclei in images. This enhanced the performance of detecting and classifying nuclei in H&E stained histopathology images of colorectal adenocarcinoma. This was proposed in [71]. The model also demonstrated locality sensitive deep learning approach with neighboring ensemble predictor (NEP) in conjunction with a standard softmax CNN and eliminated need of segmentation. The model used dropout to avoid overfitting. The model obtained an AUC of and F-score of
The CNN approaches for colon cancer classification are summarized in Table 9.
Table 9.
A summary of CNN applications in colon medical image classification surveyed in Sect. 4
| References | Preprocessing | Architecture | Comparison against | Performance metric |
|---|---|---|---|---|
| [71] | Augmented using small patches | NEP with standard softmax CNN, ReLU, dropout | AUC , F-score . | |
| [78] | Image subdivided into using sliding window, and border padding to prevent artifacts | (Conv, maxpool) , FC , softmax | Linder, Bianconi, DCNN-SW-SVM, DCNN-SW-SMC | ROC, AUC, accuracy |
| [61] | Normalized by subtracting mean, dividing by standard deviation, augmented by flipping | (Conv, pool) , conv, FC with dropout, softmax | BFD, SSF, DT-CTW, MB-LBP, SIFT | Accuracy |
Brain disorders
Alzheimer’s disease causes the destruction of brain cells leading to memory loss. Classification of Alzheimer’s disease (AD) has been challenging since it involves selection of discriminative features.
Fused CNN
Fusion of two-dimensional CNN and three-dimensional CNN achieves better accuracy was demonstrated in [19]. Information along Z direction acts very crucial for analysis of brain images and three-dimensional CNN was used to retain this information. Since the thickness of brain CT images is thicker than MRI images, geometric normalization of CT images were performed. Output of the last conv layer of two-dimensional CNN was fused with three-dimensional convoluted data to get three classes (Alzheimer’s, lesions, and healthy data). It was compared with two hand-crafted approaches SIFT and KAZE for accuracy and achieved better accuracy of and for AD, lesion and normal class, respectively.
Input cascaded CNN
Lack of training data can be overcome by extensive augmentation and fine tuning was proposed in [62]. Multi-grade brain tumor classification was performed by segmenting the tumor regions from an MR image using input cascaded CNN, extensive augmentation and then fine-tuned using data augmented. The performance was compared against state-of-art methods. It resulted in an accuracy of , sensitivity of and specificity of .
The CNN approaches for medical image classification discussed above are summarized in Table 10.
Table 10.
A summary of CNN applications in medical image classification surveyed in Sect. 4
| References | Organ | Preprocessing | Architecture | Dataset | Performance metric |
|---|---|---|---|---|---|
| [19] | Brain | Geometric normalization of CT images | Conv , (conv + dropout) , fusion with 3-D convoluted data | 3-D datasets from 282 subjects (51 with AD, 118 with lesions, 117 normal) | Accuracy and for AD, lesion and normal class |
| [63] | Brain | Motion correction, skull stripping, spatial smoothing | LeNet | ADNI dataset | Accuracy |
| [62] | Brain | Data augmentation | Input cascaded CNN for segmentation, VGG-19 with transfer learning | Radiopedia and brain tumor | Accuracy , sensitivity , specificity |
| [57] | Eye, retinopathy | Color normalization, image size reduced to | Conv , FC with softmax classifier | Kaggle | Specificity but less sensitivity |
| [82] | Digestive organs | (Conv, maxpool) , FC , mini batch stochastic gradient descent | Real clinical data of wireless capsule endoscopy images | Accuracy |
CNN applications in medical image segmentation
CNNs have been applied to implement efficient segmentation of images of brain tumors, hearts, breasts, retina, fetal abdomen, stromal and epithelial tissues.
Brain tumors
MRI is used to obtain detailed images of the brain to diagnose tumors. Automatic segmentation of a brain tumor is very challenging because it involves the extraction of high level features.
Small kernel CNN
Patch-wise training and use of small filter sizes () was proposed for segmentation of gliomas in [54]. This provided an advantage of deep architecture, while retaining the same receptive fields. Two separate models were trained for high and low gliomas. High glioma model consisted of eight conv layers and three dense layers. Low glioma model contained four conv layers and three dense layers. Maxpooling was used along with dropout for dense layers. It ranked fourth in the BRATS-2015 challenge. Data augmentation was achieved by rotation which enhanced the performance of segmentation of gliomas.
Fully blown CNN
Fully blown MRI two-dimensional images enhances performance of segmentation of sub-cortical human brain structure. This was shown in [66]. The proposed model applied Markov random field on CNN output to impose volumetric homogeneity to the final results. It outperformed several state-of-the-art methods.
Multipath CNN
Two pathways, one for convolution and the other for deconvolution, enhances segmentation output was shown in [8]. The model was used for automatic MS lesions segmentation. The model had convolutional pathway consisting of alternating conv, pool layers, and a deconvolutional pathway consisting of alternate deconv layer and unpooling layer. The pretraining was performed by convolutional RBMs (convRBM). Both pretraining and fine training were performed on a highly optimized GPU-accelerated implementation of three-dimensional convRBMs and convolutional encoder networks (CEN). It was compared with five publicly available methods and established as comparison reference points. The model performance was evaluated using evaluation metrics DSC, TPR and FPR. TPR and FPR achieved were comparatively better than the previous models developed. However, it achieved lesser DSC in comparison to other methods.
Cascaded CNN
In case of imbalanced label distributions, two phase training could be used. Global contextual features and local detailed features can be learned simultaneously by two-pathway architecture for brain segmentation and was proposed in [25]. The advantage of two-pathway was, it could recognize fine details of the tumor at a local scale and correct labels at a global scale to yield a better segmentation. Slice-by-slice segmentation from the axial view due to less resolution in the third dimension was performed. The cascaded CNN achieved better rank than two-pathway CNN and was ranked second at the MICCAI BRATS-2013 challenge. The evaluation metrics used were DSC, specificity and sensitivity and the obtained values were and . The time taken for segmentation was between 25 s and 3 min.
Multiscale CNN
In case of brain tumor segmentation, a multiscale CNN architecture for extracting both local and global features at different scales was proposed in [80]. The model performed better due to different features extracted at various resolution. The computation time was reduced by exploiting a two-dimensional CNN instead of a three-dimensional CNN. Three patch sizes , and were input to three CNNs for feature extraction. All the features extracted were input to the FC layer. Evaluation of the model was by DSC and accuracy. The model performance was almost as stable as the best method with an accuracy of nearly .
Multipath and multiscale CNN
Twopath and multiscale architecture were also explored for brain lesion segmentation by [37]. The model exploited smaller kernels to get local neighbour information and employed parallel convolutional pathways for multiscale processing. It achieved highest accuracy when applied on patients with severe traumatic brain injuries. It could also segment small and diffused pathologies. Three-dimensional CNN produced accurate segmentation borders. FC three-dimensional CRF imposed regularization constraints on CNN output and produced final hard segmentation labels. Also, due to its generic nature, it cold be applied to different lesion segmentation tasks with slight modifications. It was ranked first in the stroke lesions ISLES-SISS-2015 challenge.
Advantages of multipath and multiscale CNN was exploited for automatic segmentation of analytical brain images in [49]. The bigger kernel was used for spatial information. A separate network branch was used for each patch size, and only the output layer was shared. Mini batch learning and RMSprop were used to train the network with ReLU and cross entropy as the cost function. Automatic segmentation was evaluated using the DSC and mean surface distance between manual and automatic segmentation. It achieved accurate segmentation in terms of DSC for all tissue classes. The CNN approaches for brain segmentation discussed above are summarized in Table 11.
Table 11.
A summary of CNN applications in brain medical image segmentation surveyed in Sect. 5
| References | Preprocessing | Architecture | Performance metric |
|---|---|---|---|
| [55] | N4ITK bias field correction, Nyul’s intensity normalization | Seven layers with conv and ReLU at 1, 3, 5, 6 and maxpool at 2 and 4, layer seven was FC with softmax | Precision and accuracy |
| [66] | Augmented by flipping and translation | Shallow network, (conv, max pool layers) . First two layer outputs are subsampled, five output classes | DSC, Haudsroff distance, contour mean distance |
| [25] | Removed higher and lower intensity, normalization | Two path CNN with cascaded architecture with maxout, followed by maxpool, softmax. optimization using stochastic gradient descent, regularization using L1, L2 and dropout. | DSC , sensitivity and specificity |
| [37] | 2 pathway, 11 layers architecture called deep medic, with three-dimensional CNN, FC three-dimensional CRF and smaller kernels | DSC, precision and sensitivity | |
| [49] | Classified brain tissue pixels into classes, using different kernel sizes | Separate network branch was used for each patch size, and only output layer was shared. Mini batch learning and RMSprop to train the network with ReLU and cross entropy as the cost function. | DSC and mean surface distance between manual and automatic segmentation |
Breast cancer
Breast cancers can be predicted by automatically segmenting breast density and by characterizing mammographic textural patterns.
FCNN
Redundant computations in conv and max pool layers can be avoided by using ROI segmentation in a fast scanning deep CNN (FCNN). The above technique was applied for segmentation of histopathological breast cancer images and was proposed in [72]. The proposed work was compared against three texture classification methods namely raw pixel patch with large scale SVM, local binary pattern feature with large scale SVM and texton histogram with logistic booting. The evaluation metrics used were accuracy, efficiency and scalability. Ihe proposed method was robust to intra-class variance. It achieved an F-score of , whereas the other methods delivered the maximum F-score of . It took only 2.3 s to segment image of resolution of .
Probability map CNN
Probability maps were explored for iterative region merging for shape initialization along with compact nucleus shape repository with selection-based dictionary learning algorithm in [77]. The model resulted in better automatic nucleus segmentation using CNN. The framework was tested on three types of histopathology images namely, brain tumor, pancreatic neuro endocrine tumor and breast cancer. The parameters for comparison were precision, recall and F-score. It achieved better performance when compared to SVM, RF and DBN, especially for breast cancer images. Pixel-wise segmentation accuracy measured using DSC, HD and MAD resulted in superior performance when compared to other methods.
Patch CNN
Advantages of patch-based CNN was exploited in [78]. The method also exploited super pixel method to over segment breast cancer H&E images into atomic images. The result was natural boundaries with errors being subtle and less egregious, whereas sliding window methods resulted in zigzag boundaries. Both patch-based CNN and superpixel techniques were combined for segmenting and classifying the stromal and epithelial regions in histopathological images for detection of breast and colorectal cancer. The proposed model outperformed CNN with SVM. The comparison was done against methods using handcrafted features. It achieved accuracy and Deep CNN-Ncut-SVM had better AUC than other CNN.
Eye diseases
Greedy CNN
The architecture of conventional CNNs was tweaked by making the filters of the CNN learn sequentially using a greedy approach of boosting instead of backpropagation. Boosting was applied to learn diverse filters to minimize weighted classification error. The ensembling learning was proposed for the automatic segmentation of optic cup and optic disc from retinal fundus images to detect glaucoma in [81]. The model performed entropy sampling to identify informative points on landmarks such as edges, blood vessels. etc. The weight updates were done considering final classification error instead of back propagation error. The method operated on patches of image taken around a point. A F-score of was obtained which was comparatively better to normal CNN whose best F-score was .
Multi label inference CNN
Retinal blood vessel segmentation was dealt as multi-label inference problem and solved using CNN in [16]. The model extracted green channel from RGB fundus image, as blood vessels manifest high contrast in green channel. The model was upsampled at the sixth layer to increase spatial dimension for structured output. The output of CNN model was modeled as vector instead of a scalar, due to multiple labels. It achieved precision of , sensitivity of , specificity of , accuracy of and AUC of .
Lung
U net
Lung segmentation and bone shadow exclusion techniques for analysis of lung cancer using U-net architecture is proposed in [23]. The images were preprocessed to eliminate bone shadow and a simple U-net architecture was used to segment the lung ROI. The results obtained were very promising and showed a good speed and precise segmentation. The CNN approaches for medical image segmentation discussed above are summarized in Table 12.
Table 12.
A summary of CNN applications in medical image segmentation surveyed in Sect. 5
| References | Organ | Dataset | Comparison against | Performance metric |
|---|---|---|---|---|
| [8] | MS lesion | ISBI 2015 and MICCAI 2008 datasets | Five publicly available methods | DSC, TPR and FPR. Achieved better TPR and FPR, but less DSC values |
| [36] | Breast | Dutch breast cancer screening dataset | State of the art methods | AUC |
| [72] | Breast | The Cancer Genome Atlas breast cancer dataset | Three texture classification methods namely RPLSVM, LBPLSVM and THLB | Accuracy, efficiency and scalability and F-score of |
| [77] | Nucleus | Brain tumor, pancreatic NET and breast cancer | SVM, RF and DBN | Precision, recall and F-score |
| [83] | Heart | CCTA scans of 60 patients | Manual segmentation | Sensitivity and specificity DSC and mean absolute surface distance |
| [81] | Eye glaucoma | DRISHTI-GS, Messidor | Manual | Precision, recall, F-score |
CNN applications in medical image detection
Breast tumors
A Camelyon grand challenge for automatic detection of metastatic breast cancer in digital whole slide images of sentinel lymph node biopsies is organised by the International Symposium of Biomedical Imaging.
GoogLeNet CNN
The award-winning system with performance very closer to human accuracy was proposed in [76]. The computation time was reduced by first excluding the white background of digital images using Otsu’s algorithm. The method exploited advantages of patch-based classification to obtain better results. Also the model trained extensively on misclassified image patches, to decrease classification error. The results of the patches were embedded on heatmap image and heatmaps were used to compute evaluation scores. AUC of was obtained, and was the top performer in the challenge. In case of lesion based detection, the system achieved the sensitivity of , whereas the second ranking score was .
Eye diseases
Dynamic CNN
Random assignment of weights, speeds up the training and improves the performance. This was proposed for hemorrhage detection in fundus eye images in [75]. Also the samples were dynamically selected at every training epoch from a large pool of medical images. Pre-processing was performed using image contrast using gaussian filters. To prevent overfitting, the images were augmented. For correct classification of hemorrhage, the result was convolved with gaussian filter to smoothen the values. It achieved sensitivity, specificity and ROC of , and , whereas non selective sampling obtains sensitivity, specificity and ROC of and for Messidor dataset. AUC was used to monitor overfitting during training and when AUC value reached a stable maximum, the CNN training was stopped.
Ensemble CNN
An ensemble performs better than a single CNN and can be used to achieve higher performance. The ensemble model for detection of retinal vessels in fundus images is proposed in [46]. The model was an ensemble of twelve CNNs. Each CNN’s output probability was averaged to get the final vessel’s probability of each pixel. The probability was used to discriminate between vessel pixels from non-vessel ones for detection. The performance measures, accuracy and Kappa score were compared with existing state of the art methods. It stood second in terms of accuracy as well as in kappa score. The model obtained a FROC score of 0.928.
Cell division
LeNet CNN
Augmentation and shifting the centroid of the object enhanced the performance, and was proposed in [69]. The model was for automatic detection of mitosis (cell divisions) and quantification of mitosis occurring during scratch assay. The positive example training samples was augmented by mirroring and rotating by and centering by shifting the centroid of the object to the patch center. A random additional sampling of negative samples was added in the same amount as positive examples. The performance parameters used were sensitivity, specificity, AUC and F-score and compared with SVM. The results indicated significant increase in F-score (for SVM , for CNN ). The model concluded that both positive and negative samples are needed for better performance. The CNN applications in medical image detection reviewed in this paper are summarized in Table 13.
Table 13.
A summary of CNN applications in medical image detection surveyed in Sect. 6
| References | Organ | Preprocessing | Architecture | Performance metric |
|---|---|---|---|---|
| [76] | Breast | Threshold tissue from white background and patch-based classification | GoogLeNet | AUC for slide based classification , lesion based detection sensitivity |
| [50] | Skin (melanoma) | Illumination correction step, removal of noise using gaussian filters | (Conv, ReLU, maxpool) , FC, softmax | Sensitivity, specificity, true positive, false positive and accuracy |
| [75] | Hemorrhage (eye) | Image contrast using gaussian filters, augment using vertical, horizontal flips | (Conv, ReLU, maxpool) , FC, softmax | Sensitivity, specificity and ROC |
| [46] | Eye retina | Ensemble of twelve CNN having (conv, ReLU) , (FC, dropout) | Accuracy, FROC of 0.928 | |
| [69] | Cell | Extraction of grayscale patches, segmentation and binarization | LeNet | Sensitivity, specificity, AUC and F-score. F-score (SVM , CNN ) |
CNN applications in medical image localization
Breast tumors
Semi-supervised deep CNN
To overcome challenges of sparsely labeled data, a model was proposed in [73]. The unlabeled data was first automatically labeled using labeled data. Newly labeled data and initial labeled data were used to train the deep CNN. The method used semi-supervised deep CNN for breast cancer diagnosis. The performance of CNN was compared with SVM and ANN using different numbers of labeled data like 40, 70 and 100. The model produced comparable results even with sparse labeled data with accuracy of and AUC of .
Heart diseases
Pyramid of scales localization
Pyramid of scales (PoS) localization leads to better performance, specially in cases, where the size of the organ varies between the patients. The size of the heart is not consistent among human, and hence PoS was proposed for localization of left ventricle (LV) in cardiac MRI images in [17]. The model also exploited patch based training. Evaluation metrics used were accuracy, sensitivity and specificity and were respectively and . The limitation of the approach was the computing time of 10 s/image.
Fetal abnormalities
Transfer learning CNN
Transfer learning uses the knowledge of low layers of a base CNN trained on a large cross domain of dataset of images. Transfer learning advantages include saving of training time, and need of less data for training. This reduces overfitting and enhances the classification performance. Domain transferred deep CNN for fetal abdominal standard plane (FASP) localization in fetal ultrasound scanning was proposed in [10]. The base CNN was trained on 2014 ImageNet detection dataset. The metrics accuracy, precision, recall and F-score were the highest when compared to R-CNN and RVD. The drawback of the system was that it took more time to locate FASP from one ultra sound video. The CNN methods for image localization previewed in this paper are summarized in Table 14.
Table 14.
A summary of CNN applications in medical image localization surveyed in Sect. 7
| References | Organ | Architecture | Dataset | Comparison against | Performance Metric |
|---|---|---|---|---|---|
| [17] | Heart | (Conv, ReLU, maxpool) , FC | Public dataset of York University | Accuracy , sensitivity , specificity 99.07% | |
| [10] | Fetal abdominal | (Conv, ReLU, maxpool) conv , maxpool, FC | Imagenet 2014, Shenzen Maternal and Child Healthcare Hospital, China | R-CNN and RVD | Accuracy, precision, recall, F-score |
| [73] | Breast | Semi-supervised deep CNN | SVM, ANN | Accuracy , AUC |
The papers reviewed for medical image understanding are summarized in Table 15.
Table 15.
Summary of papers reviewed for CNN applications in medical image understanding
| Image understanding tasks | Organs targeted | References |
|---|---|---|
| Medical image classification | Lung | [4, 18, 44, 68] |
| COVID-19 | [6, 13, 22, 24, 45] | |
| HEp-2 | [7, 21] | |
| Breast | [34, 52, 73, 78] | |
| Heart | [20, 40] | |
| Eye | [57] | |
| Digestive organs | [82] | |
| Brain | [19, 63] | |
| Colon | [61, 71, 78] | |
| Medical image segmentation | Brain | [25, 37, 49, 55, 66] |
| Breast | [36, 72] | |
| MS lesion | [8] | |
| Nucleus | [77] | |
| Heart | [83] | |
| Eye | [81] | |
| Medical image detection | Breast | [5, 76] |
| Melanoma | [50] | |
| Eye | [46, 75] | |
| Cell | [69] | |
| Medical Image localization | Heart | [17] |
| Fetal Abdomen | [10] |
Critical review and conclusion
CNNs have been successfully applied in the areas of medical image understanding and this section provides a critical review of applications of CNNs in medical image understanding. Firstly, the literature contains a vast number of CNN architectures. It is difficult to select the best architecture for a specific task due to high diversity in architectures. Moreover, the same architecture might result in different performance due to inefficient data preprocessing techniques. A prior knowledge of data for applying the correct preprocessing technique is needed. Futhermore, hyper parameters optimization (dropout rate, learning rate, optimizer etc) help in enhancing or declining the performance of a network.
For training, CNNs require exhaustive amounts of data containing the most comprehensive information. Insufficient information or features leads to underfitting of the model. However, augmentation could be applied in such scenarios as it results in translation variance and increases the training dataset, thereby enhancing the CNNs efficiency. Furthermore, transfer learning and fine-tuning could also be used to enhance the efficiency in case of sparse availability of data. These enhance the performance since the low level features are nearly the same for most of the images.
Small-sized kernels could be used to enhance the performance by capturing low-level textual information. However, it is at the cost of increased computational complexity during training. Moreover, multiple pathway architecture could be used to enhance performance of CNN. The performance is enhanced due to simultaneous learning of global contextual features and local detailed features, but this in turn, increases the computational burden on the processor and memory.
One of the challenge involved in medical data is the class imbalance problem, where the positive class is generally under-represented and most of the images belong to the normal class. Designing CNNs to work on imbalanced data is a challenging task. However, researchers have tried to overcome this challenge by applying augmentation of the under-represented data. Denser CNNs could also lead to the vanishing gradient problem which could be overcome by using skip connections as in the inceptionNet architecture.
Furthermore, CNNs’ significant depth and enormous size require huge memory and higher computational resources for training. The deeper CNNs involves millions of training parameters which could lead the model to overfit and also inefficient at generalization, especially in the case of limited dataset. This calls for models which are lightweight and which could also extract critical features like the dense models. Lightweight CNNs could be explored further.
Medical image Understanding would be more efficient in the presence of background context or knowledge about the image to be understood. In this context, CNNs would be more efficient if the data consists of not only images, but also patient history. Hence, the next challenging task would be to build models, which take as input both images and patient history to make a decision and this could be the next research trend.
Interpreting CNNs is challenging due to many layers, millions of parameters, and complex, nonlinear data structures. CNN researchers have been concentrating on building accurate models without quantifying uncertainty in the obtained results. The need for successful utilization of the CNN model in medical diagnosis lies in providing confidence and this confidence needs the ability of the model to ascertain its uncertainty or certainty or explain the results obtained. This field needs further exploration. Although, researchers have proposed heat maps, class activation maps (CAM), grad CAM, grad CAM++ for visualization of CNN outputs, the area of visualization is still a challenge.
The various challenges and methods of overcoming some of the challenges of medical image understanding are summarized in Table 16. Further, efficient architectures to overcome some of the challenges as per the survey are summarized in Table 17.
Table 16.
Ways of addressing challenges of medical image understanding
| Challenges in medical image understanding | Solutions for overcoming challenges |
|---|---|
| Less training samples | Augmentation |
| Sparsity of labeled data | Semi supervised deep CNN |
| Noise in images | Gaussian filtering |
| Thicker CT images of brain | Geometric normalization |
| Extract local and global features | Multipathway CNN |
| Avoid redundant computations | ROI segmentation using fCNN |
| Reduce classification errors | Train on misclassified image patches |
| Heart classification | Two path CNN, for spatial and temporal features |
| Imbalanced label distributions | Two phase training |
| Less resolution in brain images | Slice by slice segmentation |
| Increase accuracy and AUC in eye images (retinal eye vessel) | Ensemble learning (boosting instead of back propagation, RF instead of FC) |
Table 17.
Efficient CNN architectures For medical image understanding
| Organ (modality) | Diseases | Tasks | Best architecture |
|---|---|---|---|
| Brain MRI | Alzheimer | Segementation | Lenet-5, GoogleNet |
| Detection | Multipath CNN | ||
| Epilepsy | Classification | CNN with seven conv and | |
| Schizophrenic, bipolar disorder | Segementation | Patch based CNN with 4 conv, 2 maxpool and softmax regression | |
| Breast mammogram | Tumour | Segementation | (Conv + maxpool) FC, Softmax |
| Detection | Scaled VGGnet or GoogleNet | ||
| Classification | Alexnet | ||
| Localization | Semi Supervised Deep CNN | ||
| Heart (CT) | Coronary artery calcium scoring | Segmentation | Two path CNN |
| Heart (ECG) | ECG | Classification | Two path CNN, one along spatial and the other along temporal and fused together finally |
| Heart (MRI) left ventricle | Localization | (Conv, maxpool) , FC | |
| Lung (CT) | Cancer | Classification | Ensemble of overfeat across axial, saggital and coronal plane |
| Nodule | Detection | Lenet, Overfeat | |
| COVID-19 | Classification | multi scale multi encoder ensemble CNN model | |
| ILD | Classification | Any CNN architecture like Alexnet | |
| Hep-2 cell | Classification | (Conv, maxpool) , 2 FC, softmax regressor | |
| Eye | Haemorrhage | Detection | (Conv, ReLu, maxpool) , FC, softmax classifier |
| Glaucoma | Detection | (Conv, ReLu, maxpool) , FC with adaboost | |
| Retinopathy | Segmentation | (Conv, ReLu, maxpool) , 3 FC, softmax classifier | |
| Colon | Polyp | Classification | Any simple CNN architecture |
| Polyp | Detection | Ensemble of CNN | |
| Skin | Melanoma | Detection | (Conv, ReLu, maxpool) , FC |
| Liver (CT) | Classification | (Conv, maxpool) , 3 conv, max pool, 3 FC, softmax | |
| Abdomen (US scan) | Fetus | Localization | (Conv, ReLU, maxpool) conv , maxpool, FC |
Deep learning includes methods like CNN, recurrent neural network and generative adversial networks. The review of these methods and applications have not been included, as these methods by themselves become topics of research and there is lot of research happening in those areas. Moreover, all the aspects involved in medical image understanding are also not included since it is an ocean and the focus in the paper is only on a few important techniques involved.
Conclusion
The heterogeneous nature of medical anomalies in terms of shape, size, appearance, location and symptoms poses challenges for medical anomaly diagnosis and prognosis. Traditional methods of using human specialists involve fatigue, oversight and high cost and also sparse availability. ML-based healthcare systems need efficient feature extraction methods. But efficient features are still unknown and also, the methods available for feature extraction are not very efficient. This calls for intelligent healthcare systems that automatically extract efficient features for medical image understanding that aids diagnosis and prognosis. CNN is a popular technique for solving medical image understanding challenges due to its highly efficient methods of feature extraction and learning low level, mid level and high level discriminant features of an input medical image.
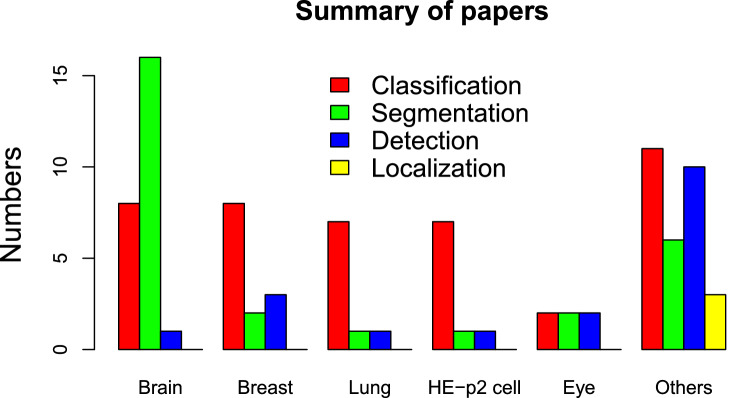
The literature reviewed in this paper underscores that researchers have focused their attention on the use of CNN to overcome many challenges in medical image understanding. Many have accomplished the task successfully. The CNN methods discussed in this paper have been found to either outperform or compliment the existing traditional and ML approaches in terms of accuracy, sensitivity, AUC, DSC, time taken etc. However, their performance is often not the best due to a few factors. A snapshot summary of the quantum of research articles surveyed in this article is presented in the Fig. 2.
Fig. 2.
Bar chart summarizing the number of papers surveyed
The challenges in image understanding with respect to medical imaging have been discussed in this paper. Various image understanding tasks have been introduced. In addition, CNN and its various components have been outlined briefly. The approaches used by the researchers to address the various challenges in medical image understanding have been surveyed.
CNN models have been described as black boxes and there is a lot of research happening in terms of analyzing and understanding output at every layer. Since medical images are involved, we need an accountable and efficient prediction system which should also be able to articulate about a decision taken. Researchers are also working on image captioning (textual representations of the image) [29]. This will enable physicians to understand the perception of the network at both output layer and intermediate levels. Researchers have tried Bayesian deep learning models which calculates the uncertainty estimates [38]. This would help physicians assess the model. All these could further accelerate medical image understanding using CNNs among physicians.
Acknowledgements
The authors acknowledge with gratitude the support received from REVA University, Bengaluru, and M. S. Ramaiah University of Applied Sciences, Bengaluru, India.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
D. R. Sarvamangala, Email: sarvamangala.dr@reva.edu.in
Raghavendra V. Kulkarni, Email: arvie@ieee.org
References
- 1.Abadi M, Agarwal A, Barham P (2016) Tensorflow: large-scale machine learning on heterogeneous distributed systems. CoRR Arxiv: 1603.04467
- 2.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging. 2017;30(4):449–459. doi: 10.1007/s10278-017-9983-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Rfou R, Alain G, Almahairi A, Angermueller C, Bahdanau D, Ballas N, Bengio Y (2016) Theano: a python framework for fast computation of mathematical expressions. arXiv:1605.02688
- 4.Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou SG. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1207–1216. doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 5.Apou G, Schaadt NS, Naegel B. Structures in normal breast tissue. Comput Biol Med. 2016;74:91–102. doi: 10.1016/j.compbiomed.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Bassi PAS, Attux R (2020) A deep convolutional neural network for COVID-19 detection using chest X-rays
- 7.Bayramoglu N, Kannala J, Heikkilä J (2015) Human epithelial type 2 cell classification with convolutional neural networks. In: Proceedings of the 15th IEEE international conference on bioinformatics and bioengineering, BIBE, pp 1–6
- 8.Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam RC. Deep 3-D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2016;35(5):1229–1239. doi: 10.1109/TMI.2016.2528821. [DOI] [PubMed] [Google Scholar]
- 9.CS231N convolutional neural network for visual recogntion (2019). http://cs231n.stanford.edu/. Accessed 24 June 2019
- 10.Chen H, Ni D, Qin J. Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE J Biomed Health Inform. 2015;19(5):1627–1636. doi: 10.1109/JBHI.2015.2425041. [DOI] [PubMed] [Google Scholar]
- 11.Chollet F (2018) Keras: the Python deep learning library. https://keras.io/. Accessed 24 June 2019
- 12.Chollet F (2017) Xception: deep learning with depthwise separable convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, (CVPR), pp 1800–1807
- 13.Chowdhury NK, Rahman MM, Kabir MA (2020) Pdcovidnet: a parallel-dilated convolutional neural network architecture for detecting COVID-19 from chest X-ray images [DOI] [PMC free article] [PubMed]
- 14.Ciompi F, de Hoop B, van Riel SJ. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2-D views and a convolutional neural network out-of-the-box. Med Image Anal. 2015;26(1):195–202. doi: 10.1016/j.media.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Collobert R, Kavukcuoglu K, Farabet C (2011) Torch7: a Matlab-like environment for machine learning. In: BigLearn, NIPS workshop, EPFL-CONF-192376
- 16.Dasgupta A, Singh S (2017) A fully convolutional neural network based structured prediction approach towards the retinal vessel segmentation. In: Proceedings of the 14th IEEE international symposium on biomedical imaging (ISBI), pp 248–251
- 17.Emad O, Yassine IA, Fahmy AS (2015) Automatic localization of the left ventricle in cardiac MRI images using deep learning. In: Proceedings of the 37th IEEE annual international conference on engineering in medicine and biology society (EMBC), pp 683–686 [DOI] [PubMed]
- 18.Gao M, Bagci U, Lu L, Wu A, Buty M. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. CMBBE Imaging Vis. 2018;6(1):1–6. doi: 10.1080/21681163.2015.1124249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao XW, Hui R (2016)A deep learning based approach to classification of CT brain images. In: Proceedings of the SAI computing conference, pp 28–31
- 20.Gao XW, Li W, Loomes M, Wang L. A fused deep learning architecture for viewpoint classification of echocardiography. Inf Fusion. 2017;36:103–113. doi: 10.1016/j.inffus.2016.11.007. [DOI] [Google Scholar]
- 21.Gao Z, Zhang J, Zhou L, Wang L. HEp-2i cell image classification with deep convolutional neural networks. IEEE J Biomed Health Inform. 2014;21:416–428. doi: 10.1109/JBHI.2016.2526603. [DOI] [PubMed] [Google Scholar]
- 22.Ghoshal B, Tucker A (2020) Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection arXiv:2003.10769
- 23.Gordienko Y, Gang P, Hui J, Zeng W, Kochura Y, Alienin O, Rokovyi O, Stirenko S (2017) Deep learning with lung segmentation and bone shadow exclusion techniques for chest X-ray analysis of lung cancer. CoRR arXiv:1712.07632
- 24.Hasan MK, Alam MA, Elahi MTE, Roy S, Wahid SR (2020) CVR-NET: a deep convolutional neural network for coronavirus recognition from chest radiography images
- 25.Havaei M, Davy A, Warde-Farley D. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. doi: 10.1016/j.media.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 26.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, (CVPR), pp 770–778
- 27.Hesamian M, Jia W, He X. Deep learning techniques for medical image segmentation: achievements and challenges. J Digit Imaging. 2019;32:582–596. doi: 10.1007/s10278-019-00227-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho TK (1995) Random decision forests. In: Proceedings of the 3rd IEEE international conference on document analysis and recognition, vol 1, pp 278–282
- 29.Hossain MZ, Sohel F, Shiratuddin MF, Laga H (2018) A comprehensive survey of deep learning for image captioning. CoRR arXiv:1810.04020
- 30.Hubel D, Wiesel T. Receptive fields of single neurones in the cat’s striate cortex. J Physiol. 1959;148(3):574–591. doi: 10.1113/jphysiol.1959.sp006308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ICPR 2018 international conference on pattern recognition (2019). http://www.icpr2018.org. Accessed 24 June 2019
- 32.ISLES challenge 2018 ischemic stroke lesion segmentation (2018). http://www.isles-challenge.org. Accessed 24 June 2019
- 33.Jia, Y., Shelhamer E, Donahue J (2014) Caffe: convolutional architecture for fast feature embedding. In: Proceedings of the 22th ACM international conference on multimedia, MM ’14, pp 675–678. ACM, New York
- 34.Jiao Z, Gao X, Wang Y, Li J. A deep feature based framework for breast masses classification. Neurocomputing. 2016;197:221–231. doi: 10.1016/j.neucom.2016.02.060. [DOI] [Google Scholar]
- 35.Jusman Y, Ng SC, Abu Osman NA. Intelligent screening systems for cervical cancer. Sci World J. 2014;2014:810368. doi: 10.1155/2014/810368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallenberg M, Petersen K, Nielsen M. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016;35(5):1322–1331. doi: 10.1109/TMI.2016.2532122. [DOI] [PubMed] [Google Scholar]
- 37.Kamnitsas K, Ferrante E, Parisot S (2016) Deepmedic for brain tumor segmentation. In: Proceedings of the international workshop on brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Springer, pp 138–149
- 38.Kendall A, Gal Y (2017) What uncertainties do we need in bayesian deep learning for computer vision? CoRR arXiv:1703.04977
- 39.Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Kiranyaz S, Ince T, Gabbouj M. Real-time patient-specific ECG classification by 1-D convolutional neural networks. IEEE Trans Biomed Eng. 2016;63(3):664–675. doi: 10.1109/TBME.2015.2468589. [DOI] [PubMed] [Google Scholar]
- 41.Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems 25: proceedings of the 26th annual conference on neural information processing systems, pp 1106–1114
- 42.Larochelle H, Bengio Y (2008) Classification using discriminative restricted Boltzmann machines. In: Proceedings of the 25th international conference on machine learning, pp 536–543
- 43.LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 44.Li Q, Cai W, Wang X, Zhou Y, Feng DD, Chen M (2014) Medical image classification with convolutional neural network. In: Proceedings of the 13th international conference on control, automation, robotics and vision (ICARCV), pp 844–848
- 45.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK (2020)Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. CoRR arXiv:2004.00038
- 46.Maji D, Santara A, Mitra P (2016) Ensemble of deep convolutional neural networks for learning to detect retinal vessels in fundus images. arXiv preprint arXiv:1603.04833 [DOI] [PubMed]
- 47.Mavroforakis ME, Georgiou HV, Dimitropoulos N. Mammographic masses characterization based on localized texture and dataset fractal analysis using linear, neural and support vector machine classifiers. Artif Intell Med. 2006;37(2):145–162. doi: 10.1016/j.artmed.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Menze BH, Jakab A, Bauer S. The multimodal brain tumor image segmentation benchmark (BRATS) IEEE Trans Med Imaging. 2015;34(10):1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJNL, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016;35(5):1252–1261. doi: 10.1109/TMI.2016.2548501. [DOI] [PubMed] [Google Scholar]
- 50.Nasr-Esfahani E, Samavi S, Karimi N (2016) Melanoma detection by analysis of clinical images using convolutional neural network. In: Proceedings of the 38th IEEE annual international conference on engineering in medicine and biology society, EMBC, pp 1373–1376 [DOI] [PubMed]
- 51.Nervana Systems Inc (2018) neon. http://neon.nervanasys.com/docs/latest/. Accessed 24 June 2019
- 52.Ovalle JEA, González FA, Ramos-Pollán R, Oliveira JL, Guevara-López MÁ. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Prog Biomed. 2016;127:248–257. doi: 10.1016/j.cmpb.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 53.Paszke A, Gross S, Chintala S, Chanan G (2018) Tensors and dynamic neural networks in Python with strong GPU acceleration. https://pytorch.org/. Accessed 24 June 2019
- 54.Pereira S, Pinto A, Alves V, Silva CA (2015) Deep convolutional neural networks for the segmentation of gliomas in multi-sequence MRI. In: Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries—first international workshop, Brainles 2015, held in conjunction with MICCAI 2015, Munich, Germany, October 5, 2015, revised selected papers, pp 131–143
- 55.Pereira S, Pinto A, Alves V, Silva CA. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging. 2016;35(5):1240–1251. doi: 10.1109/TMI.2016.2538465. [DOI] [PubMed] [Google Scholar]
- 56.Phillips NA, Rajpurkar P, Sabini, M, Krishnan R, Zhou, S, Pareek, A, Phu NM, Wang C, Ng AY, Lungren MP (2020) Chexphoto: 10,000+ smartphone photos and synthetic photographic transformations of chest X-rays for benchmarking deep learning robustness. CoRR arXiv:2007.06199
- 57.Pratt H, Coenen F, Broadbent DM, Harding SP, Zheng Y (2016) Convolutional neural networks for diabetic retinopathy. In: Proceedings of the 20th conference on medical image understanding and analysis, MIUA, pp 200–205
- 58.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding DY, Bagul A, Langlotz C, Shpanskaya KS, Lungren MP, Ng AY (2017) Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. CoRR arXiv:1711.05225
- 59.Ranzato M, Hinton GE, LeCun Y. Guest editorial: deep learning. Int J Comput Vis. 2015;113(1):1–2. doi: 10.1007/s11263-015-0813-1. [DOI] [Google Scholar]
- 60.Ravi D, Wong C, Deligianni F, Berthelot M, Pérez JA, Lo B, Yang G. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21(1):4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 61.Ribeiro E, Uhl A, Häfner M (2016) Colonic polyp classification with convolutional neural networks. In: Proceedings of the 29th IEEE international symposium on computer-based medical systems, (CBMS), pp 253–258
- 62.Sajjad M, Khan S, Muhammad K, Wu W, Ullah A, Baik SW. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J Comput Sci. 2019;30:174–182. doi: 10.1016/j.jocs.2018.12.003. [DOI] [Google Scholar]
- 63.Sarraf S, Tofighi G (2016) Classification of Alzheimer’s disease using fMRI data and deep learning convolutional neural networks. Computer Research Repository. arXiv:1603.08631
- 64.Seide F, Agarwal A (2016) CNTK: Microsoft’s open-source deep-learning toolkit. In: Proceedings of the 22nd ACM international conference on knowledge discovery and data mining, p 2135
- 65.Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y (2013) Overfeat: integrated recognition, localization and detection using convolutional networks. Computing Research Repository. arXiv:1312.6229
- 66.Shakeri M, Tsogkas S, Ferrante E, Lippe S, Kadoury S, Paragios N, Kokkinos I (2016) Sub-cortical brain structure segmentation using F-CNNs. In: Proceedigns of the IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, pp 269–272
- 67.Shankar K, Zhang Y, Liu Y, Wu L, Chen CH. Hyperparameter tuning deep learning for diabetic retinopathy fundus image classification. IEEE Access. 2020;8:118164–118173. doi: 10.1109/ACCESS.2020.3005152. [DOI] [Google Scholar]
- 68.Shen W, Zhou M. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit. 2017;61:663–673. doi: 10.1016/j.patcog.2016.05.029. [DOI] [Google Scholar]
- 69.Shkolyar A, Gefen A, Benayahu D, Greenspan H (2015) Automatic detection of cell divisions (mitosis) in live-imaging microscopy images using convolutional neural networks. In: Proceedings of the 37th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 743–746 [DOI] [PubMed]
- 70.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. Computer Research Repository. arXiv:1409.1556
- 71.Sirinukunwattana K, Raza SEA, Tsang Y, Snead DRJ, Cree IA, Rajpoot NM. Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging. 2016;35(5):1196–1206. doi: 10.1109/TMI.2016.2525803. [DOI] [PubMed] [Google Scholar]
- 72.Su H, Liu F, Xie Y (2015) Region segmentation in histopathological breast cancer images using deep convolutional neural network. In: Proceedings of the 12th IEEE international symposium on biomedical imaging (ISBI), pp 55–58
- 73.Sun W, Tseng TB, Zhang J, Qian W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graph. 2017;57:4–9. doi: 10.1016/j.compmedimag.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 74.Szegedy C, Liu W, Jia Y, Sermanet P, Reed SE (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), pp 1–9
- 75.van Grinsven MJ, van Ginneken B, Hoyng CB. Fast convolutional neural network training using selective data sampling: application to hemorrhage detection in color fundus images. IEEE Trans Med Imaging. 2016;35(5):1273–1284. doi: 10.1109/TMI.2016.2526689. [DOI] [PubMed] [Google Scholar]
- 76.Wang D, Khosla A, Gargeya R, Irshad H, Beck AH (2016) Deep learning for identifying metastatic breast cancer. Computer Research Repository. arXiv:1606.05718
- 77.Xing F, Xie Y, Yang L. An automatic learning-based framework for robust nucleus segmentation. IEEE Trans Med Imaging. 2016;35(2):550–566. doi: 10.1109/TMI.2015.2481436. [DOI] [PubMed] [Google Scholar]
- 78.Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing. 2016;191:214–223. doi: 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeiler MD, Fergus R (2013) Visualizing and understanding convolutional networks. Computer Research Repository. arXiv:1311.2901
- 80.Zhao L, Jia K. Multiscale CNNs for brain tumor segmentation and diagnosis. Comput Math Methods Med. 2016;2016:8356294:1–8356294:7. doi: 10.1155/2016/8356294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zilly JG, Buhmann JM, Mahapatra D. Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation. Comput Med Imaging Graph. 2017;55:28–41. doi: 10.1016/j.compmedimag.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 82.Zou Y, Li L, Wang Y (2015) Classifying digestive organs in wireless capsule endoscopy images based on deep convolutional neural network. In: Proceedings of the IEEE international conference on digital signal processing, DSP, pp 1274–1278
- 83.Zreik M, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Isgum I (2016) Automatic segmentation of the left ventricle in cardiac CT angiography using convolutional neural networks. In: Proceedings of the 13th IEEE international symposium on biomedical imaging (ISBI), pp 40–43