Abstract
GenBank® (https://www.ncbi.nlm.nih.gov/genbank/) is a comprehensive, public database that contains 9.9 trillion base pairs from over 2.1 billion nucleotide sequences for 478 000 formally described species. Daily data exchange with the European Nucleotide Archive and the DNA Data Bank of Japan ensures worldwide coverage. Recent updates include new resources for data from the SARS-CoV-2 virus, updates to the NCBI Submission Portal and associated submission wizards for dengue and SARS-CoV-2 viruses, new taxonomy queries for viruses and prokaryotes, and simplified submission processes for EST and GSS sequences.
INTRODUCTION
GenBank (1) is a comprehensive public database of nucleotide sequences and supporting bibliographic and biological annotations built and distributed by the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine (NLM), located on the campus of the US National Institutes of Health (NIH) in Bethesda, MD, USA. After summarizing the growth of GenBank in the past year, this paper will briefly review recent updates and developments.
GROWTH OF THE DATABASE
Handling large sequence records
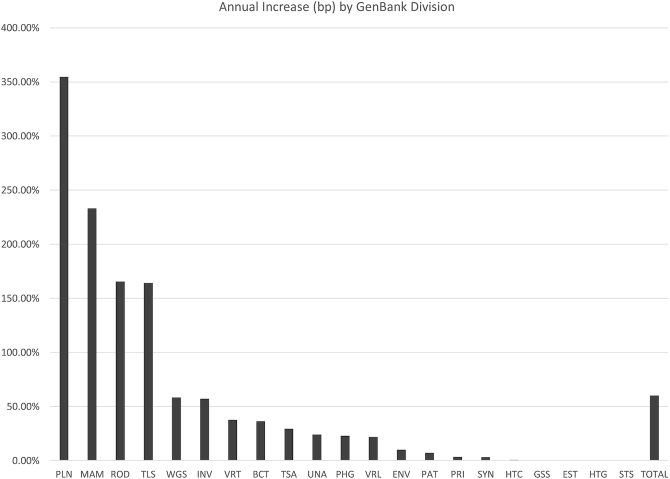
The size and growth of the various divisions of GenBank are shown in Table 1 and Figure 1. Notable increases in the past year occurred in the PLN, MAM and ROD divisions, primarily the result of large, gapped sequences representing single chromosomes. For example, in the PLN division slightly more than 300 sequences (<0.1% of new PLN sequences) accounted for 93% of the annual growth. All of these sequences are longer than 200 Mbp; some are much longer, such as CM022218, a sequence for chromosome 3B from Triticum aestivum with a length of 886 Mbp. As sequencing technologies continue to improve, we expect to see more of these longer records. One looming consequence of this growth will be the need to transition to using 64-bit integers to represent such sequences within databases and analysis software packages, as the current signed 32-bit integers can only represent sequences of about 2.1 Gbp. We have already encountered submitted sequences longer than this limit and have been forced to request that submitters split such records in order to submit them to GenBank. We are in the process of updating our software to handle 64-bit representations, and will continue to update the community on our progress over the coming year.
Table 1.
GenBank divisions
| Division | Description | Base pairsa |
|---|---|---|
| WGS | Whole genome shotgun data | 8 841 649 410 652 |
| TSA | Transcriptome shotgun data | 381 148 464 834 |
| PLN | Plants | 269 438 877 546 |
| BCT | Bacteria | 98 827 135 660 |
| VRT | Other vertebrates | 63 565 835 430 |
| EST | Expressed sequence tags | 43 301 109 577 |
| TLS | Targeted Loci Studies | 27 825 059 498 |
| HTG | High-throughput genomic | 27 781 778 663 |
| PAT | Patent sequences | 26 452 787 091 |
| GSS | Genome survey sequences | 26 378 695 300 |
| MAM | Other mammals | 20 844 388 122 |
| INV | Invertebrates | 19 759 935 222 |
| ROD | Rodents | 12 090 011 771 |
| PRI | Primates | 8 767 435 622 |
| SYN | Synthetic | 7 932 542 985 |
| ENV | Environmental samples | 6 755 612 180 |
| VRL | Viruses | 5 824 026 918 |
| PHG | Phages | 782 571 323 |
| HTC | High-throughput cDNA | 733 210 026 |
| STS | Sequence tagged sites | 640 923 137 |
| UNA | Unannotated | 679 302 |
| TOTAL | All GenBank sequences | 9 890 500 490 859 |
aRelease 239 (8/2020).
Figure 1.
Annual increase in base pairs (bp) for each division of GenBank in release 239 (August 2020) measured relative to GenBank release 233 (August 2019). The ‘TOTAL’ bar indicates the growth for GenBank as a whole. See the text for descriptions of the largest increases.
Acquiring the database
NCBI provides GenBank sequence records in both the traditional flat file format and in a structured ASN.1 format by anonymous FTP at ftp.ncbi.nlm.nih.gov/genbank. For release 239 (15 August 2020) there are 3131 files requiring 1461 GB of uncompressed disk storage. In addition, daily GenBank incremental update files containing new and updated records since the most recent release are available in flat file format at ftp.ncbi.nlm.nih.gov/genbank/daily-nc/.
RECENT DEVELOPMENTS
SARS coronavirus resources
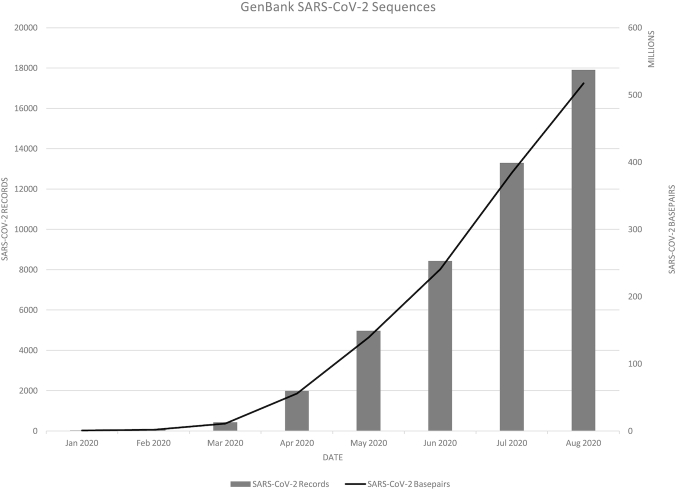
New coronavirus resources
In response to the COVID-19 pandemic that emerged in early 2020 and the accompanying increase in viral sequence data (Figure 2), NCBI made several resources available to assist the community in submitting, analyzing and downloading sequence data for SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). NCBI now offers a customized submission portal for both assembled and unassembled SARS-CoV-2 sequences (https://submit.ncbi.nlm.nih.gov/sarscov2/). On average this portal provides accessions back to submitters in 1–2 h, and assembled sequences will be annotated with VADR (2). Using these portals not only ensures that sequence data are made available through the INSDC databases, but also through the NCBI Virus resource (3), RefSeq (4) and BLAST (5). NCBI collects these and other resources related to SARS-CoV-2 on a new landing page (https://www.ncbi.nlm.nih.gov/sars-cov-2/) that includes links to the above resources in addition to several links to download SARS-CoV-2 data, view relevant literature and much more.
Figure 2.
Growth of SARS-CoV-2 sequence data in GenBank. Each data point represents the cumulative number of records or base pairs at the end of each month.
NCBI virus
Of particular interest is a new section of the NCBI Virus resource devoted to SARS-CoV-2 (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Nucleotide&VirusLineage_ss=SARS-CoV-2,%20taxid:2697049). A link to this page also appears on the SARS-CoV-2 landing page discussed above. This page serves as an information hub for this virus and collects the available genomes and proteins for SARS-CoV-2 in a table that users can browse and filter by 16 attributes including sequence length, the source geographic region, and collection date. Users can then select, download, and align these data, and also build phylogenetic trees.
NCBI Datasets
NCBI Datasets is a new and experimental product that allows users to download complex genomic datasets easily using either a web interface, an API, or a UNIX/LINUX command-line tool (https://www.ncbi.nlm.nih.gov/datasets/). In response to the increasing demand for SARS-CoV-2 data, NCBI Datasets now includes a specialized coronavirus page that provides genome downloads for over 18 000 coronavirus genomes, including over 15 000 complete genomes from SARS-CoV-2 (https://www.ncbi.nlm.nih.gov/datasets/coronavirus/genomes). In addition to the genomic data itself, this interface allows downloads of any combination of annotated SARS-CoV-2 proteins.
Submission process enhancements
Submission portal updates
The NCBI Submission Portal website (https://submit.ncbi.nlm.nih.gov) received several updates in 2020 to improve overall navigation and ease of use. The main page has a new, streamlined design that presents submitters with clear starting points for common data types as well as a suggestion tool that lets submitters enter a data type and quickly find the appropriate process. Part of this new interface is a series of help pages (e.g. https://submit.ncbi.nlm.nih.gov/about/genbank/) that display lists of items submitters should have ready before beginning their submission along with guides for data formatting. Once submitters begin a process, a submission ‘wizard’ will the guide them through the various steps and will provide additional help specific for that process (e.g. https://submit.ncbi.nlm.nih.gov/genbank/help/).
New submission wizards
The Submission Portal for GenBank (https://submit.ncbi.nlm.nih.gov/subs/genbank/) provides three improved wizards to streamline submissions: new wizards for sequences from Dengue virus, mitochondrial cytochrome oxidase (COX1) from metazoans, and an updated wizard for handling diploid genome assemblies. These and similar wizards accelerate the submission process, and the Dengue and COX1 wizards provide automatic feature annotation using VADR (2) and validation functions, relieving submitters of the need to provide their own annotations. The Dengue wizard accepts FASTA formatted sequences and requires the following source information: isolate, serotype/genotype, collection date, host, and country of collection. The COX1 wizard only accepts COX1 gene sequences from metazoans (multicellular animals) without any flanking sequences. Submitters should include an isolate or specimen-voucher for the source organism along with the mitochondrial genetic code if the organism is not in the NCBI taxonomy database. More information about this wizard is available at https://submit.ncbi.nlm.nih.gov/genbank/help/. The WGS wizard (https://submit.ncbi.nlm.nih.gov/subs/genome/) now includes better handling of primary and alternate haplotypes from diploid genome assemblies. These improvements reduce the amount of manual curation previously required for these submissions as well as minimizing the steps required for submitters.
Simplifying EST, GSS and HTG submissions
As previously described (1), EST and GSS sequences are now consolidated in the Nucleotide database with all other GenBank (and INSDC) sequences. Similarly, submitters of EST and GSS sequences can now use the standard BankIt tool that will process EST and GSS submissions as standard GenBank submissions (https://submit.ncbi.nlm.nih.gov/about/bankit/). We expect that submitters of HTG sequences will also be able to use the standard GenBank submission portal in early 2021.
Improved taxonomic searching
Viruses
NCBI Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy/) now supports new Entrez search queries for virus names based on the Baltimore classification that groups viruses based on their nucleic acid (DNA or RNA) strandedness (single-stranded or double-stranded), direction of translation (sense) and method of replication (Table 2). NCBI Taxonomy has replaced the Baltimore classification with a hierarchical classification based on evolutionary relationships provided by the International Committee on Taxonomy of Viruses (ICTV) (6). Although evolutionary relationships are not necessarily reflected, the Baltimore search terms remain in use and can provide functional context. More details on viruses in NCBI Taxonomy are provided elsewhere (7).
Table 2.
New entrez queries for viruses
| Full query | Short version |
|---|---|
| double stranded dna virus[filter] | dsdna virus[filter] |
| single stranded dna virus[filter] | ssdna virus[filter] |
| negative sense single stranded dna virus[filter] | negative sense ssdna virus[filter] |
| positive sense single stranded dna virus[filter] | positive sense ssdna virus[filter] |
| ambisense single stranded dna virus[filter] | ambisense ssdna virus[filter] |
| double stranded dna reverse transcriptase virus[filter] | dsdna rt virus[filter] |
| double stranded rna virus[filter] | dsrna virus[filter] |
| single stranded rna virus[filter] | ssrna virus[filter] |
| negative sense single stranded rna virus[filter] | negative sense ssrna virus[filter] |
| positive sense single stranded rna virus[filter] | positive sense ssrna virus[filter] |
| ambisense single stranded rna virus[filter] | ambisense ssrna virus[filter] |
| single stranded rna reverse transcriptase virus[filter] | ssrna rt virus[filter] |
| ambisense ssdna virus[filter] | N/A |
Prokaryotes
NCBI Taxonomy has also extended Entrez search terms to find bacterial and archaeal names not validly published under the International Code of Nomenclature of Prokaryotes (ICNP) (8). Names of prokaryotes not included in the 1980 List of Approved Names nor published directly in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) can become validated by being included in a Validation List subsequently published in the IJSEM. Until then, they are considered ‘effectively published’ and have no standing in nomenclature under the ICNP (9). These names are displayed in NCBI Taxonomy, but it is now possible to filter them using a search term in Entrez Taxonomy:
effective current name[filter]
Similarly, candidatus names, which are declared for some uncultivated prokaryotic taxa and are not validly published under the Code (8) are now searchable in Entrez Taxonomy as well:
candidatus current name[filter]
A comment is available in a separate column for both sets of names in the nodes.dmp file as part of the expanded taxonomy FTP dump files (ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/new_taxdump).
Advice for submitters
As discussed previously (1), we continue to encourage submitters to provide contextual metadata to support further use and analysis of the data (e.g. country, latitude and longitude of the sampling location) along with other data such as the isolate name or number plus museum/collection identifiers as applicable. We also urge submitters to use evidence tags to provide information about supporting evidence for annotations (https://www.ncbi.nlm.nih.gov/genbank/evidence/). In cases where submitters have used existing public sequencing reads to improve the quality of their assemblies prior to submission, we encourage submitters to cite the accession numbers of these reads within their submission. When submitting prokaryotic genomes, we encourage submitters to either annotate their genomes with the NCBI Prokaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) or request that NCBI annotate the genomes before they are released.
NCBI strongly encourages submitters to register sequencing projects in the BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) and to update their BioProject records after relevant publications are available. Doing so provides reliable linkages between sequencing projects and the data they produce, and may also allow links to the BioSample database (10) that provides additional information about the biological materials used in the study. Finally, we would remind submitters to notify GenBank when their data are published so that we can ensure a timely release of their data.
ELECTRONIC ADDRESSES
www.ncbi.nlm.nih.gov - NCBI Home Page.
gb-sub@ncbi.nlm.nih.gov: Submission of sequence data to GenBank.
update@ncbi.nlm.nih.gov: Revisions to, or notification of release of, ‘confidential’ GenBank entries.
info@ncbi.nlm.nih.gov: General information about NCBI resources.
CITING GENBANK
If you use the GenBank database in your published research, we ask that this article be cited.
Notes
Present address: GenBank, National Center for Biotechnology Information, Building 45, Room 6AN12D-37, 45 Center Drive, Bethesda, MD 20892, USA.
Contributor Information
Eric W Sayers, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Mark Cavanaugh, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Karen Clark, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Kim D Pruitt, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Conrad L Schoch, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Stephen T Sherry, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
Ilene Karsch-Mizrachi, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Building 38A, 8600 Rockville Pike, Bethesda, MD 20894, USA.
FUNDING
Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2020; 48:D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schaffer A.A., Hatcher E.L., Yankie L., Shonkwiler L., Brister J.R., Karsch-Mizrachi I., Nawrocki E.P.. VADR: validation and annotation of virus sequence submissions to GenBank. BMC Bioinform. 2020; 21:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brister J.R., Ako-Adjei D., Bao Y., Blinkova O.. NCBI viral genomes resource. Nucleic Acids Res. 2015; 43:D571–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boratyn G.M., Camacho C., Cooper P.S., Coulouris G., Fong A., Ma N., Madden T.L., Matten W.T., McGinnis S.D., Merezhuk Y. et al.. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013; 41:W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. International Committee on Taxonomy of Viruses Executive Committee The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020; 5:668–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B. et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parker C.T., Tindall B.J., Garrity G.M.. International code of nomenclature of prokaryotes prokaryotic code (2008 Revision). Int. J. Syst. Evol. Microbiol. 2019; 69:S7–S111. [DOI] [PubMed] [Google Scholar]
- 9. Oren A., Garrity G.M., Parte A.C.. Why are so many effectively published names of prokaryotic taxa never validated. Int. J. Syst. Evol. Microbiol. 2018; 68:2125–2129. [DOI] [PubMed] [Google Scholar]
- 10. Barrett T., Clark K., Gevorgyan R., Gorelenkov V., Gribov E., Karsch-Mizrachi I., Kimelman M., Pruitt K.D., Resenchuk S., Tatusova T. et al.. BioProject and BioSample databases at NCBI: facilitating capture and organization of metadata. Nucleic Acids Res. 2012; 40:D57–D63. [DOI] [PMC free article] [PubMed] [Google Scholar]