Abstract
The Integrated Microbial Genomes & Microbiomes system (IMG/M: https://img.jgi.doe.gov/m/) contains annotated isolate genome and metagenome datasets sequenced at the DOE’s Joint Genome Institute (JGI), submitted by external users, or imported from public sources such as NCBI. IMG v 6.0 includes advanced search functions and a new tool for statistical analysis of mixed sets of genomes and metagenome bins. The new IMG web user interface also has a new Help page with additional documentation and webinar tutorials to help users better understand how to use various IMG functions and tools for their research. New datasets have been processed with the prokaryotic annotation pipeline v.5, which includes extended protein family assignments.
INTRODUCTION
The Integrated Microbial Genomes & Microbiomes (IMG/M: https://img.jgi.doe.gov/m/) is a user driven data management resource that enables users worldwide to analyze microbial genomes and metagenomes in a comparative context. IMG includes genomes of cultivated and uncultivated archaea, bacteria, eukarya, plasmids, viruses, as well as genome fragments (genomic regions of interest generated by targeted sequencing), metagenomes and metatranscriptomes. First, sample, sequencing and analysis project information is registered in the Genomes OnLine Database (GOLD) (1), including environmental metadata, sampling and sequencing technology, as well as data processing protocols. GOLD metadata follow the standards defined by the Genomics Standards Consortium (2), and provide valuable context for downstream search and analysis of sequence data in IMG. After the required metadata is collected, the sequence data, which can come from one of the three main sources described below are processed by the IMG annotation pipeline v.5 (3). The status of the processing is tracked through the IMG submission system (https://img.jgi.doe.gov/submit/).
The bulk of the sequence data included in IMG is generated by the JGI, including isolate and single-cell genomes, and microbiomes. NCBI is another major source of data for IMG, either from GenBank (4) which remains the main source of reference isolate genomes, prioritized based on phylogenetic diversity, or from the Sequence Read Archive (SRA) (5) for selected microbiomes, which are then assembled and annotated by the JGI processing pipelines. The third source of IMG data is through external user submissions of assembled sequences, including genomes, metagenomes and metatranscriptomes generated by any sequencing technology. On a case-by-case basis, error-corrected unassembled data generated by long-read sequencing technologies, such as PacBio (6) and ONT (7) can be also supported. Currently only assembled but unannotated prokaryotic genome and metagenome submissions are accepted through external submissions to IMG, with sequence data provided in FASTA format.
De novo annotation of sequences submitted in FASTA format starts with identification of encoded structural features such as protein-coding genes (CDSs) and non-coding RNAs, regulatory RNA features and binding motifs, as well as CRISPR elements. Briefly, CRISPR elements are detected using a modified CRT (8), tRNAs are predicted using tRNAscan-SE 2.0.6 (9), ribosomal RNAs, non-coding RNAs and RNA regulatory features are predicted using Rfam covariance models and Infernal tools (10–12), and protein-coding genes are called by Prodigal v2.6.2 (13) and GeneMark (14). CDSs undergo functional annotation, which involves protein assignment to various protein and domain classifications, such as an updated 2014 version of COGs (15), version 30 of Pfam-A (16), version 15.0 of TIGRFAM (17), version 1.75 of SUPERFAMILY (18), version 01_06_2016 of SMART (19) and version 4.2.0 of CATH-FunFam (20). All these assignments are performed using a thread-optimized hmmsearch from the HMMER v3.1b2 package (21,22). Proteomes are also associated with KEGG Orthology (KO) terms (23) using LAST v1066 (24), with KEGG pathways based on KO term assignments and with MetaCyc pathways (25) based on gene annotations with Enzyme Commission (EC) numbers derived from KO terms. Best LAST hits between CDSs and IMG reference proteomes derived from high quality public genomes are computed for placing the sequences in phylogenetic context through Phylogenetic Distribution of Best Hits tool.
In addition, CDSs encoded in isolate genomes undergo prediction of signal peptides and transmembrane regions using SignalP v4.1 (26) and TMHMM 2.0c (27) and Bidirectional Best Hits (BBH) between newly loaded proteomes and IMG reference proteomes are computed using LAST (24). Other computations available for genome sequences include Average Nucleotide Identity (ANI) (28) distance matrix computations, and biosynthetic clusters (29,30), as previously described. The detailed descriptions of IMG processing of genomes and metagenomes or metatranscriptomes is provided elsewhere (31).
Due to the size of IMG data (currently over 65 billion genes), it is impossible to upgrade all genome and metagenome annotations to the latest version of the annotation pipeline. Since pipeline differences can lead to annotation discrepancies, which may confound downstream analysis, users can find the detailed information about the pipeline used to annotate particular datasets on the Genome/Metagenome Details pages of the respective datasets in the ‘IMG Release/Pipeline Version’ field. In addition, a user can request reannotation of a specific set of genomes or metagenomes, in which case they will be processed using the most recent version of the pipeline. The reannotated versions can replace older datasets or alternatively both versions can be kept for comparison.
DATA CONTENT
Genomics data and microbiome samples
As of August 2020, IMG included 364.3 million genes from isolate genomes, which represents about 34% data growth since July 2018 (32). There were also 64.66 billion metagenome genes, which represents a 19.7% growth over the past 2 years. Table 1 shows the current IMG database content compared with the same database 2 years ago.
Table 1.
IMG dataset content comparison
| Total (8/2020) | Public (8/2020) | Total (7/2018) | Public (7/2018) | |
|---|---|---|---|---|
| Archaea | 3011 | 1967 | 2453 | 1762 |
| Bacteria | 99 004 | 83 768 | 75 130 | 63 736 |
| Eukaryota | 746 | 710 | 733 | 697 |
| Virus | 9 804 | 8 392 | 9 674 | 8 388 |
| Plasmid | 1208 | 1188 | 1215 | 1190 |
| Metagenome | 26 488 | 21 813 | 18 907 | 13 232 |
| Metatranscriptome | 6371 | 6174 | 4605 | 2423 |
| Cell enrichment | 2357 | 2110 | 1333 | 801 |
| Single particle sort | 5806 | 5378 | 3954 | 3486 |
| Metagenome bin | 85 565 | 83 287 | 78 253 | 76 337 |
The number of IMG submissions from external users also enjoys a healthy growth. As of August 2020, there are 20 940 external isolate genome submissions and 13 708 external metagenome submissions. Among these, 22% (4203 isolate genomes and 3471 microbiomes) were submitted during the last two years. These numbers exclude JGI-generated data, as well as public genomes and microbiomes imported from NCBI. All datasets imported from NCBI are publicly available to all IMG users as soon as they are loaded into IMG. JGI-generated data follow the JGI Data Release and Usage policy, which is described on the JGI website (https://jgi.doe.gov/user-program-info/pmo-overview/policies/). The visibility settings for externally submitted datasets follow the IMG Data Release policy, which is described on the IMG submission website (https://img.jgi.doe.gov/submit).
IMG has two specialized data marts, which include additional data and analysis tools: IMG/ABC (https://img.jgi.doe.gov/abc/) for biosynthetic gene clusters (BGCs) and secondary metabolites (29,30), and IMG/VR (https://img.jgi.doe.gov/vr/) for viral genomes (33). IMG/ABC v.5.0 was released in 2019 (29) to include new biosynthetic gene clusters predicted by a new version of antiSMASH (antibiotics and Secondary Metabolite Analysis SHell) v.5. This IMG/ABC version also includes new analysis tools for BGCs and a new viewer for users to browse antiSMASH results. IMG/VR has also been updated to include new viral data (33). The two data marts provide access and custom analysis tools for additional features (biosynthetic clusters and viruses, respectively). They share the content of Analysis Carts and Workspace with core IMG enabling navigation between data marts, so that the data of interest (genomes, genes, contigs) are found in the core IMG, the results are saved to the Carts and/or Workspace, and then additional analyses are performed in a specialized data mart. For instance, a user can compare groups of genomes, such as Butyrivibrio and Pseudobutyrivibrio spp. described below, in terms of their biosynthetic gene cluster (BGC) profiles. These Workspace sets created in IMG/MER can be loaded into the IMG/ABC Genome Cart and ‘Browse BCs by BGC Type’ menu option can be used to view a heat map or tabular display of the counts of various BGC types in these genomes.
DATA ANALYSIS
IMG allows users to query and browse the data, and perform many analyses through the IMG User Interface (UI) (https://img.jgi.doe.gov/m/), which continues to be augmented and improved to support the increasing growth and new types of data. IMG’s data and analysis capabilities for microbial genomes were recently contrasted with other analogous publicly available portals pointing to IMG’s unique strengths in terms of genomic tools, metadata-driven search capabilities, large number and breadth of genome data (34). Several improvements of the IMG UI and additional new tools are discussed below.
An updated Find Genes menu
Find Genes menu, which was available since IMG’s inception has been redeveloped to help users find genes and proteins of interest based on their attributes and sequence similarity. Find Genes menu now includes a new Gene Search interface, which is similar to the previously developed Genome Search framework (32). The new Gene Search interface has a tab for Quick Gene Search, which allows users to perform a simple keyword search with a limited set of parameters, and an Advanced Search Builder tab, which enables construction of complicated queries using a variety of gene and protein attributes.
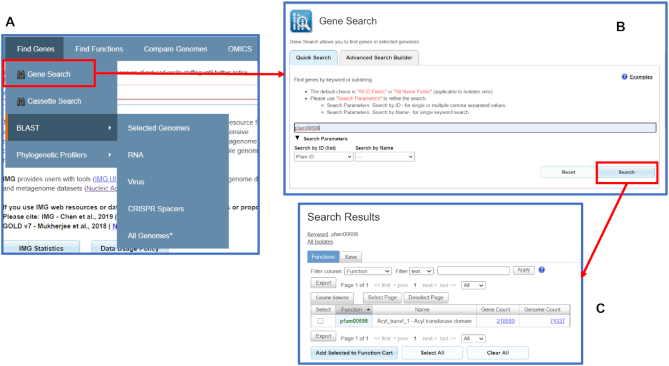
The Quick Search option allows users to find genes and proteins of interest based on numerical identifiers, external accessions or keywords in isolate genomes only (Figure 1A). A user can provide one or more of IMG gene IDs, Genbank accessions, or any of the protein family identifiers supported by IMG such as COG, Pfam ID, or Enzyme Commission number. For example, typing in ‘pfam00698’ with the ‘Search by ID’ option will retrieve all proteins assigned PF00698 (Acl_trans_1) in isolate genomes (Figure 1B, C). ‘Search by Name’ option enables keyword search on a variety of protein family names as well as gene symbols. For instance, to find all proteins with a potential function of ‘enolase’ in isolate genomes, a user can type ‘enolase’ and select the ‘All Name fields.’ The results show all protein families that have ‘enolase’ in their names or definitions. Specifically, there are three Pfams including two domains of enolase enzyme, pfam00113 (Enolase_C - Enolase, C-terminal TIM barrel domain) and pfam03952 (Enolase_N - Enolase, N-terminal domain), as well as an enolase-like protein family described by pfam13378 (MR_MLE_C - Enolase C-terminal domain-like). After clicking on the count of protein families with the keywords a user can view the counts of proteins assigned to these families and navigate further to individual proteins or select protein families of interest to add them to Function Cart.
Figure 1.
Gene Search. (A) The Find Genes menu includes the new Gene Search function and BLAST. (B) Quick search allows users to search genes using function IDs such as pfam00698 or using names. (C) Quick search result of pfam00698 shows the numbers of genes and genomes with this particular function.
As discussed above, the Quick Search runs on all isolate genomes, but not on metagenomes or metatranscriptomes due to the large data size. Even though it is limited to genomes only, for ubiquitous protein families it may still retrieve very large counts. To enable search in a specific set of isolate genomes and/or metagenomes/metatranscriptomes, users can go to the Advanced Search Builder. This option also enables search on a wide range of gene and protein attributes and their combinations. As an example, consider a query to retrieve all 16S rRNA genes longer than 500nt in the freshwater sediment metagenomes in IMG. First a user would identify all freshwater sediment metagenomes in IMG, which can be done using the Advanced Search Builder in Genome Search and the following query conditions:
Taxonomy – Domain: *Microbiome
Study Dataset Names – Genome Name / Sample Name: Freshwater
Environmental Classification – GOLD Ecosystem Type: Sediment
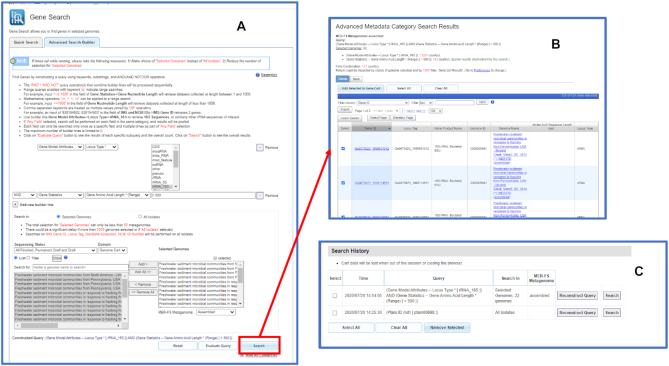
This Genome Search query retrieves a total of 22 metagenomes, which can be selected and added to the Genome Cart. In order to find all 16S rRNA genes longer than 500nt in these metagenomes a user would go to the Advanced Search Builder in the Gene Search interface and add the following query conditions (Figure 2A):
Figure 2.
Advanced Search Builder option for the new Gene Search feature. (A) The advanced search option allows users to search all 16s rRNA genes with length greater than 500nt in the freshwater sediment metagenomes previously saved in the Genome Cart. (B) The search result shows that there are 131 genes satisfying the search criteria. (C) Users can view and reuse previously constructed query conditions.
Gene Model Attributes – Locus Type: rRNA_16S
Gene Statistics – Gene Amino Acid Length: > 500
and add all metagenomes in the Genome Cart to the set of ‘Selected Genomes.’ The query retrieves 131 16S rRNA genes (Figure 2B), which can be selected and added to Gene Cart for further analysis. Gene Cart also allows exporting selected gene sequences in a FASTA format. Similar to the Genome Search interface, Gene Search also records all search history to allow users to view and to reuse their previous queries (Figure 2C).
An updated sequence similarity search interface within the Find Genes menu is available under BLAST section (Figure 1A). BLAST options now include genomes (all isolates or selected datasets, including metagenomes and metatranscriptomes), RNA, viruses and CRISPR spacers. The options for BLAST against RNA collections have been expanded to all types of rRNAs including 5S, 16S, 18S, 23S, 28S, as well as other RNA genes. There are separate databases for isolate and metagenomic/metatranscriptomic RNA sequences. The display options of BLAST results have been expanded to include both raw results and a table with alignment details and selection capabilities. In addition, registered IMG users have an option of submitting a computation job from the Expert Review IMG site (https://img.jgi.doe.gov/mer/), which allows them to search against larger collections of IMG sequences (up to 500 genomes and/or metagenomes in one search).
Metagenome bin browse and search
Starting in 2018, IMG incorporates the results of automated metagenome binning using MetaBAT (35) along with CheckM (36) and other quality assessment metrics for ensuring bin data quality (32). As of August 2020, IMG has a total of 85 565 (83 287 public) high-quality and medium-quality metagenome bins. IMG’s v.5.0 UI had very few options for querying and analyzing metagenome bins, mainly limited to browsing the Metagenome Statistics section of the Microbiome Details page, where Metagenome Bins counts for the bins found in this metagenome were displayed. IMG v.6.0 has additional visualization, searching and analysis tools for the bins.
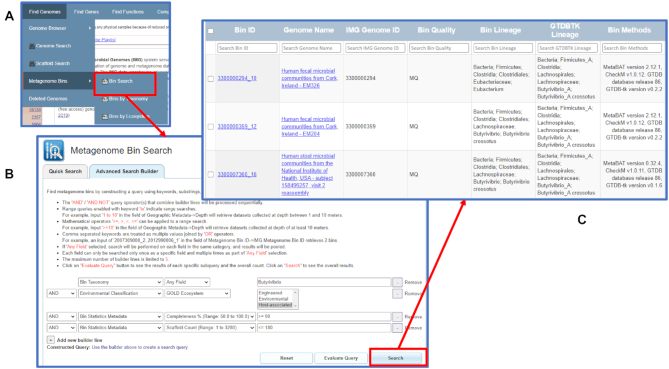
The new menu item Metagenome Bins under Find Genomes provides an option to search metagenome bins together with two browsing options to view the bins by taxonomy or by ecosystem. Users can select the Bins by Ecosystem option to view all metagenome bins organized by the GOLD ecosystem hierarchy (see Figure 3A). Users can expand the graphic display by clicking on any of the cells. For example, clicking on Plants ecosystem category will expand the display to show all types and subtypes under this category (Figure 3B). Clicking on a ‘breadcrumb’ (Figure 3C) opens a list of metagenome bins in this particular ecosystem type or subtype. Figure 3D shows all metagenome bins in the Nodule ecosystem type. Users can select any or all of the six bins to add to the scaffold analysis cart. Registered IMG users can also save copies of the bins as Workspace Scaffold Sets for further analysis and editing. The Bins by Taxonomy option is similar to the Bins by Ecosystem option except that bins are organized based on the predicted bin lineage based on the scaffold NCBI lineage assigned by IMG.
Figure 3.
Browse metagenome bins by Ecosystem. (A) From the Find Genomes menu item, users can select Metagenome Bins to find Bins by Ecosystem browse option. (B) Users can click on any cell to expand. This example shows the expansion of Plants ecosystem category under Host-associated ecosystem. (C) Users can click on the ‘breadcrumb’ to view the detailed list. (D) The list shows all metagenome bins in the Nodule ecosystem type. Users can select any or all of the six bins to add to the scaffold analysis cart or workspace scaffold dataset.
The Bin Search interface in the Metagenome Bins menu allows users to search bins based on a variety of attributes across all or specific sets of metagenomes (Figure 4A). Similar to the Genome Search and Gene Search interfaces, Bin Search also has a Quick Search option and an Advanced Search Builder option. Quick Search allows users to search bins by entering metagenome bin IDs, IMG metagenome IDs, or GOLD IDs. It also allows users to search bins based on their predicted NCBI or GTDB-Tk (37) lineages. For example, one can search all metagenome bins that are classified as Cyanobacteria by entering the term in the keyword search and selecting ‘NCBI Phylum’ in the Search by Name dropdown list.
Figure 4.
Advanced Search Builder option of the new Metagenome Bin Search feature. (A) The Bin Search option in metagenome bin search allows users to search all metagenome bins in the IMG database using quick search or advanced search. (B) Users can build a complex query to find Butyrivibrio and Pseudobutyrivibrio bins with completeness of at least 90% and in no more than 180 contigs or scaffolds. Users can click the Search button to run the query. (C) Result shows that 11 bins satisfying this query condition. (Only 3 rows are displayed here.)
The Advanced Search Builder option allows users to find bins based on a combination of environmental parameters, their quality, predicted lineage, and other characteristics. This capability is illustrated by an example of finding Butyrivibrio and Pseudobutyrivibrio bins with completeness of at least 90% and in no more than 180 contigs or scaffolds, which were found in host-associated metagenomes, such as human fecal samples or animal rumen. Such query can be constructed by a combination of the following four builder lines (Figure 4B):
Bin Taxonomy – All Field: Butyrivibrio
Environmental Classification – GOLD Ecosystem: Host-associated
Bin Statistics Metadata – Completeness: > = 90
Bin Statistics Metadata – Scaffold Count: < = 180
Eleven bins satisfying this search condition (Figure 4C) can be reviewed in the results table, which lists the predicted lineage of the bins, as well as other statistics, such as total size and number of scaffolds. In our example, predicted NCBI and GTDB-tk taxonomy for the majority of bins is in agreement up to the family level (Lachnospiraceae). However, one bin (3300000294_10) has different family assignments according to NCBI and GTDB-tk taxonomy (Eubacteriaceae and Lachnospiraceae, respectively). Users can click on the bin ID to view the Bin Details page and investigate the reason for this discrepancy, which turns out to be because of a large number of relatively short scaffolds with the predicted lineage of Eubacteriaceae/Eubacterium. Their combined length exceeds 10% of the total bin size, suggesting that CheckM may have underestimated the contamination of this bin. Bins retrieved by the query can be selected and saved to the Scaffold Cart as a combined set of scaffolds or to Workspace as copies of individual bins. Similar to other search interfaces Metagenome Bin Search also saves query history to be reused in the future searches.
New analysis tool for comparison of mixed sets of genomes and metagenome bins
Since each metagenome bin can be considered as an equivalent of a population genome (with the caveats of possible contamination and lower levels of completeness), a natural extension of IMG genome analysis tools is to allow users to analyze metagenome bins against isolates (comparing ‘uncultivated’ versus ‘cultivated’) or together with isolate genomes (e.g., to potentially expand the phylogenetic diversity of available genomes to include uncultivated ones). Similarly, users may wish to compare the sets of scaffolds that they have created and saved in the Workspace to metagenome bins or genomes. Some possible use cases include comparison of bins from the same lineage, but from different environments to find possible environment-driven functional adaptations or comparison of bins from the same environment but from different lineages to identify potential functional complementarities.
For this purpose, a new Analysis Data Group (ADG) option under Workspace has been implemented. It allows registered IMG users to create analysis data groups, which may include any or all of the following: (i) metagenome bins (scaffold sets created by selecting IMG metagenome bins and saved to Workspace as Scaffold Sets), (ii) user-created scaffold sets (e.g., generated by searching metagenomes of interest for scaffolds with specific lineage assignment, GC content or read depth), (iii) isolate genomes, (iv) metagenomes. Two viewers for analysis data groups are available: simple list allows dataset selection for group creation, editing and deletion, while a graphical viewer provides a tree view of various sets within each group. After creating two or more of such groups, users can then perform statistical analysis of feature abundance using one of the five statistical methods provided. Features that can be used in this comparison include protein families (COG, Pfam, KEGG Orthology Terms, etc.) as well as functional groupings of protein families (COG Functional Categories, KEGG Modules) and taxonomic categories. Statistical tests include Fisher's Exact (38), Mann–Whitney (39) and Welch's T-test (40) to compare two analysis data groups, while Analysis of Variance or ANOVA (41) and Kruskal-Wallis (42) can be used to compare 3–10 analysis data groups. In general, since the groups are heterogeneous, collecting the data to perform statistical analysis on feature counts, as well as performing statistical testing itself, is too computationally intensive to finish in real time. Therefore, this new analysis tool is only available as an on-demand computation to the registered IMG users of the Expert Review IMG site (https://img.jgi.doe.gov/mer/).
Running a functional comparison of isolate genomes and bins using the ADG tool
We illustrate how users can apply the new analysis feature by building upon the functional analysis of rumen microbiome members described in Seshadri et al. (43). This study, which included only isolate genomes, found that many ruminal isolates from the genera Butyrivibrio and Pseudobutyrivibrio have lost an enolase gene, which encodes a ubiquitous glycolytic enzyme conserved in all domains of life. In the example below, we attempt to identify the functional signatures of enolase-positive and enolase-negative Butyrivibrio and Pseudobutyrivibrio strains by comparing these two sets of isolate genomes to the taxonomically equivalent bins of comparable sequence quality.
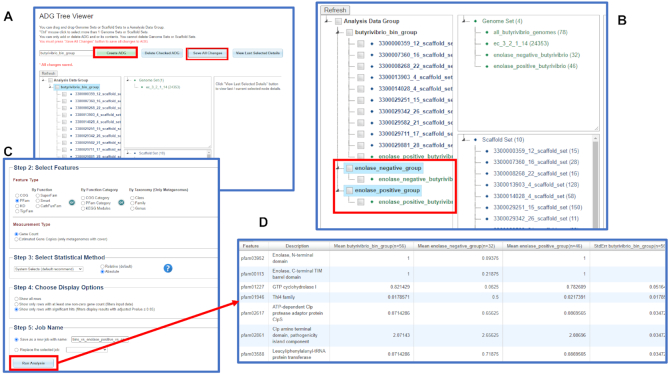
First, an analysis group is created consisting of Butyrivibrio bins from host-associated metagenomes with completeness of at least 90% and no more than 180 contigs. These can be found by a query described in the section about Advanced Search Builder in Metagenome Bin Search (Figure 4). We will exclude the bin 3300000294_10 with discordant assignments by NCBI and GTDB-Tk taxonomy from the analysis. All bins in the resulting set of ten are coming from human stool samples; we can select them and save to the Workspace. The default names of scaffold sets consist of bin ID with an added extension ‘scaffold_set’. Next, we create an Analysis Data Group by going to Workspace → Analysis Data Group menu, clicking on ‘Create Group’ tab, selecting the bins in the list under ‘Step 2: Select Scaffold Sets’ menu and saving the set under the name ‘butyrivibrio_bin_group.’ Alternatively, users can also click the ADG Tree Viewer button to use drag and drop to create a new group. To use the latter option, first enter the name ‘butyrivibrio_bin_group’ in the blank text field, and then click the Create ADG button. Then drag and drop all 10 bins to this newly created group and finally click the Save All Changes button to save the change (Figure 5A).
Figure 5.
New analysis tool for users to analyze genomes together with metagenome bins. (A) In the Workspace, a user can create new Analysis Data Groups (ADGs) by dragging and dropping genome sets and scaffold sets. A new ADG butyrivibrio_bin_group is created to include all the 10 butyrivibrio metagenome bins. (B) Two additional ADGs enolase_positive_group and enolase_negative_group can be created to include genome sets enolase_positive_butyrivibrio and enolase_negative_butyrivibrio, respectively. (C) The user can then select the three ADGs to analyze Pfam distribution using the suggested analysis method, and submit the statistical analysis to be a computation job. (D) The user will receive an email notification when the job has been completed. The result can be viewed following the link included in the email or from a corresponding link in the Workspace My Jobs list.
The isolate genomes of Butyrivibrio and Pseudobutyrivibrio spp. are retrieved using an Advanced Search Builder in Genome Search interface with the following query conditions:
Taxonomy – Genus: butyrivibrio
Sequencing Assembly Annotation – Is Public: Yes
Sequencing Assembly Annotation – GOLD Analysis Project Type: Genome Analysis (Isolate)
Sequencing Assembly Annotation – GOLD Sequencing Strategy: Whole Genome Sequencing
This query returns 78 genomes (as of July 2020), which can be selected and added to Genome Cart. In addition, we save them to Workspace as a Genome set ‘all_butyrivibrio_genomes.’ Users can review their metadata using Table Configuration options in Genome Cart and selecting fields such as ‘Host name’ and ‘Isolation.’ Notably, only two strains with known host or isolation source (Butyrivibrio fibrisolvens 16/4 and Butyrivibrio crossotus DSM 2876) originate from human stool samples, while the remainder are from the forestomach of different ruminants.
In order to identify enolase-positive genomes, we use pfam00113 (Enolase_C - Enolase, C-terminal TIM barrel domain) and pfam03952 (Enolase_N - Enolase, N-terminal domain); these can be found by running a Quick Search query for protein families with ‘enolase’ in the name or definition, as described in the section about Gene Search interface. As reported in Seshadri et al. (43), some enolase-negative genomes have enolase pseudogenes (shortened and/or fragmented genes). Therefore, we can find enolase-positive genomes by searching for proteins that have both Pfam domains and are nearly full length (400 amino acids or longer). We use Advanced Search Builder in Gene Search interface selecting the genomes in our Genome Cart and the following query conditions:
Function IDs – Pfam ID: pfam00113, pfam03952
Gene Statistics – Gene Amino Acid Length: > = 400
This query returns 46 genes in 46 genomes; there are seven more genes with query Pfams that do not satisfy our length criteria and are likely pseudogenes. We can create a set of enolase-positive genomes by selecting the genes in the results table, adding them to Gene Cart, then emptying the Genome Cart and adding the genomes of genes in the Gene Cart to Genome Cart. Of the two strains isolated from human feces, only Butyrivibrio crossotus DSM 2876 falls into the enolase-positive group.
We can select the 46 genomes of enolase-positive strains in the Genome Cart and save them as a Workspace Genome set ‘enolase_positive_butyrivibrio.’ To construct the set of enolase-negative Butyrivibrio and Pseudobutyrivibrio spp. we go to the ‘Set Operations’ tab in the Genome Set Workspace, which allows users to create new sets by finding a union or intersection of sets or subtracting one set from another. Subtracting ‘enolase_positive_butyrivibrio’ from ‘all_butyrivibrio_genomes’ results in a set of 32 genomes, which we will save as ‘enolase_negative_butyrivibrio.’ In order to perform a three-way statistical analysis of differential abundance of protein families, we construct two more analysis data groups by going to the Workspace → Analysis Data Group menu. Following a procedure described above for bins, we create a new group ‘enolase_positive_group’ to include the genome set ‘enolase_positive_butyrivibrio’ and another group ‘enolase_negative_group’ to include the genome set ‘enolase_negative_butyrivibrio’ (Figure 5B).
After creating three analysis data groups, we click on the ‘ADG Statistical Analysis Tool’ button, select the groups butyrivibrio_bin_group, enolase_positive_group, and enolase_negative_group, choose ‘Pfam’ as a feature type in ‘Function’ category, ‘Gene Count’ as measurement type, and ‘Absolute’ for the count type, and ‘Show only rows with significant hits’ as display option. We will keep the default of ‘System select’ as a statistical method. After providing the job name ‘bins_vs_enolase_positive_vs_enolase_negative,’ we submit the job by clicking on the Run Analysis button (Figure 5C). In this analysis, bins are treated the same way as isolate genomes: the per-genome or per-bin counts of genes assigned to each Pfam are retrieved and the mean counts of Pfams for each analysis data group are computed together with their standard deviations. The statistical significance is tested using the Kruskal–Wallis test, which is a non-parametric test for three or more groups. Benjamini–Hochberg method is used to control false discovery rate (FDR) control at 1%.
Interpreting the results of a functional comparison of isolate genomes and bins
The users receive an email notification upon the completion of the analysis. They can also monitor the progress of their jobs by going to the Workspace → My Jobs menu. In the IMG UI users can view the results limited to the top 1000 rows, subject to web browser limitations (Figure 5D). Users can also download the complete results for further analysis including raw counts for all features in each genome, scaffold set or metagenome in the analysis data groups. In our three-way comparison, 1,084 Pfams have statistically significant differences of mean counts between at least two analysis groups, examples of which are provided in Tables 2 and 3.
Table 2.
Pfams with statistically significant differences between butyrivibrio_bin_group and two groups of isolate Butyrivibrio and Pseudobutyrivibrio genomes. Adjusted P-value is Kruskal-Wallis test P-value with FDR controlled at 1% using Benjamini-Hochberg method
| Pfam ID | Pfam definition | Pfam functional category | Mean count, butyrivibrio_bin_group (n = 10) | Mean count enolase_positive_group (n = 46) | Mean count, enolase_negative_group (n = 32) | Adjusted P-value |
|---|---|---|---|---|---|---|
| PF03418 | Germination protease | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF03419 | Sporulation factor SpoIIGA | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF05580 | SpoIVB peptidase S55 | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF06686 | Stage III sporulation protein AC/AD protein family | Sporulation | 2 | 0.043478 | 0 | 6.91E-16 |
| PF06898 | Putative stage IV sporulation protein YqfD | Sporulation | 1.1 | 0.021739 | 0 | 6.91E-16 |
| PF07451 | Stage V sporulation protein AD (SpoVAD) | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF07454 | Stage II sporulation protein P (SpoIIP) | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF09547 | Stage IV sporulation protein A (spore_IV_A) | Sporulation | 1.1 | 0.021739 | 0 | 6.91E-16 |
| PF09548 | Stage III sporulation protein AB (spore_III_AB) | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF09551 | Stage II sporulation protein R (spore_II_R) | Sporulation | 1.2 | 0.021739 | 0 | 6.91E-16 |
| PF09578 | Spore cortex protein YabQ (Spore_YabQ) | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF12116 | Stage III sporulation protein D | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF12685 | SpoIIIAH-like protein | Sporulation | 1 | 0.021739 | 0 | 6.91E-16 |
| PF03862 | SpoVA protein | Sporulation | 2 | 0.043478 | 0 | 7.68E-16 |
| PF08486 | Stage II sporulation protein | Sporulation | 1.9 | 0.043478 | 0 | 7.68E-16 |
| PF08769 | Sporulation initiation factor Spo0A C terminal | Sporulation | 1.8 | 0.043478 | 0 | 8.61E-16 |
| PF03323 | Bacillus/Clostridium GerA spore germination protein | Sporulation | 0.9 | 0.021739 | 0 | 4.69E-14 |
| PF09581 | Stage III sporulation protein AF (Spore_III_AF) | Sporulation | 1 | 0.021739 | 0 | 4.69E-14 |
| PF11007 | Spore coat associated protein JA (CotJA) | Sporulation | 0.9 | 0.021739 | 0 | 4.69E-14 |
| PF12652 | CotJB protein | Sporulation | 0.9 | 0.021739 | 0 | 4.69E-14 |
| PF15714 | Stage V sporulation protein T C-terminal, transcription factor | Sporulation | 0.9 | 0.021739 | 0 | 4.69E-14 |
| PF12164 | Stage V sporulation protein AA | Sporulation | 0.8 | 0.021739 | 0 | 4.20E-12 |
| PF13782 | Stage V sporulation protein AB | Sporulation | 0.8 | 0.021739 | 0 | 4.20E-12 |
| PF00269 | Small, acid-soluble spore proteins, alpha/beta type | Sporulation | 0.7 | 0.021739 | 0 | 3.13E-10 |
| PF02654 | Cobalamin-5-phosphate synthase | Cobalamin biosynthesis | 0 | 0.978261 | 0.90625 | 3.48E-12 |
| PF07685 | CobB/CobQ-like glutamine amidotransferase domain | Cobalamin biosynthesis | 0 | 1.913043 | 1.75 | 6.14E-10 |
| PF01890 | Cobalamin synthesis G C-terminus | Cobalamin biosynthesis | 0 | 0.934783 | 0.84375 | 4.67E-09 |
| PF02570 | Precorrin-8X methylmutase | Cobalamin biosynthesis | 0 | 0.934783 | 0.84375 | 4.67E-09 |
| PF02571 | Precorrin-6x reductase CbiJ/CobK | Cobalamin biosynthesis | 0 | 0.934783 | 0.84375 | 4.67E-09 |
| PF06180 | Cobalt chelatase (CbiK) | Cobalamin biosynthesis | 0 | 0.934783 | 0.84375 | 4.67E-09 |
| PF11760 | Cobalamin synthesis G N-terminal | Cobalamin biosynthesis | 0 | 0.934783 | 0.84375 | 4.67E-09 |
Table 3.
Pfams with statistically significant differences between enolase-positive group and enolase-negative group. Adjusted P-value is Kruskal–Wallis test P-value with FDR controlled at 1% using Benjamini-Hochberg method
| Pfam ID | Pfam definition | Pfam functional category | Mean count, butyrivibrio_bin_group (n = 10) | Mean count enolase_positive_group (n = 46) | Mean count, enolase_negative_group (n = 32) | Adjusted P-value |
|---|---|---|---|---|---|---|
| PF03952 | Enolase, N-terminal domain | Glycolysis | 1 | 1 | 0.09375 | 3.16E-15 |
| PF00113 | Enolase, C-terminal TIM barrel domain | Glycolysis | 1 | 1 | 0.21875 | 9.74E-12 |
| PF01227 | GTP cyclohydrolase I | Coenzyme biosynthesis | 1 | 0.782609 | 0.0625 | 7.10E-10 |
| PF01729 | Quinolinate phosphoribosyl transferase, C-terminal domain | Coenzyme biosynthesis | 0 | 0.782609 | 0.125 | 1.38E-08 |
| PF02445 | Quinolinate synthetase A protein | Coenzyme biosynthesis | 0 | 0.782609 | 0.125 | 1.38E-08 |
| PF02749 | Quinolinate phosphoribosyl transferase, N-terminal domain | Coenzyme biosynthesis | 0 | 0.782609 | 0.125 | 1.38E-08 |
| PF05690 | Thiazole biosynthesis protein ThiG | Coenzyme biosynthesis | 0.4 | 0.76087 | 0.0625 | 1.37E-07 |
| PF06968 | Biotin and Thiamin Synthesis associated domain | Coenzyme biosynthesis | 0.6 | 1.5 | 0.28125 | 2.27E-06 |
| PF02548 | Ketopantoate hydroxymethyltransferase | Coenzyme biosynthesis | 0.4 | 0.695652 | 0.125 | 2.71E-05 |
| PF02569 | Pantoate-beta-alanine ligase | Coenzyme biosynthesis | 0.4 | 0.695652 | 0.125 | 2.71E-05 |
| PF08818 | Domain of unknown function (DU1801) | Unknown | 0 | 0.195652 | 0.9375 | 7.83E-07 |
| PF10670 | Domain of unknown function (DUF4198) | Unknown | 0 | 0.130435 | 0.6875 | 9.89E-06 |
| PF04402 | Protein of unknown function (DUF541) | Unknown | 0 | 0.217391 | 0.90625 | 3.29E-05 |
| PF03588 | Leucyl/phenylalanyl-tRNA protein transferase | N-degron proteolytic pathway | 0 | 0.086957 | 0.71875 | 4.18E-07 |
| PF02617 | ATP-dependent Clp protease adaptor protein ClpS | N-degron proteolytic pathway | 0 | 0.086957 | 0.65625 | 4.61E-07 |
| PF02861 | Clp amino terminal domain, pathogenicity island component | N-degron proteolytic pathway | 2 | 2.086957 | 2.65625 | 4.61E-07 |
| PF10431 | C-terminal, D2-small domain, of ClpB protein | N-degron proteolytic pathway | 4.2 | 4.108696 | 4.65625 | 1.23E-05 |
Some of the statistically significant differences between Butyrivibrio bins on one side and isolate genomes on the other side include protein families involved in sporulation (Table 2), which are present in Butyrivibrio bins, generated from human stool metagenomes, but largely absent in isolate genomes, which mostly originate from ruminal samples. On the other hand, the enzymes from cobalamin biosynthesis pathway show the opposite distribution. These results recapitulate the findings of Seshadri et al. (43), which reported that Pfams involved in cobalamin biosynthesis are overrepresented in ruminal genomes, while sporulation-specific Pfams are overrepresented in human isolates. Closer examination of the full results of statistical analysis shows that the only isolate genome with sporulation Pfams is Butyrivibrio crossotus DSM 2876 originating from human stool. The presence of sporulation genes in this genome has been reported previously. Notably, Butyrivibrio crossotus DSM 2876 is also the only strain in the enolase-positive group lacking cobalamin biosynthesis Pfams. This suggests that the presence of sporulation genes and the absence of cobalamin biosynthesis in metagenomic bins is not an artifact of metagenome assembly or binning, but a feature of Butyrivibrio crossotus-like populations inhabiting the human gut.
In addition, statistical analysis with the IMG ADG tool reveals statistically significant differences between the enolase-positive and enolase-negative groups of isolate genomes. They corroborate the findings of Seshadri et al. (43) suggesting genome evolution via gene loss in enolase-negative group. Enolase-negative genomes appear to be in the process of losing enzymes participating in biosynthesis of coenzymes including NAD, thiamin and CoA (Table 3). However, these genomes show no signs of general genome reduction, such as accumulation of pseudogenes, reduced average genome size or GC content. Furthermore, there are Pfams that are significantly more abundant in the enolase-negative group to the exclusion of enolase-positive genomes and metagenome bins. While many of these are proteins and domains of unknown function (Table 3), one unexpected finding is higher abundance of the componentry of an N-end rule pathway for degradation of proteins with specific N-terminal amino acids (44) including leucyl,phenylalanyl-tRNA-protein transferase, an adaptor protein ClpS and domains of ClpA chaperone (Table 3). The functional significance of this observation is unclear, since bacteria have other proteolytic pathways (see (45) for a review).
To summarize, the results of statistical analysis using the ADG tool highlight the differences between ruminal and human populations and isolates of Butyrivibrio and Pseudobutyrivibrio spp., point to their diverging evolutionary trajectories and suggest possible avenues for experimental studies. Combined with other IMG tools it provides a powerful framework for genomic and metagenomic data exploration. A webinar recording featuring the IMG statistical analysis tool is available in the Youtube playlist with a link from the IMG Help page (https://img.jgi.doe.gov/help.html).
IMG help and tutorials
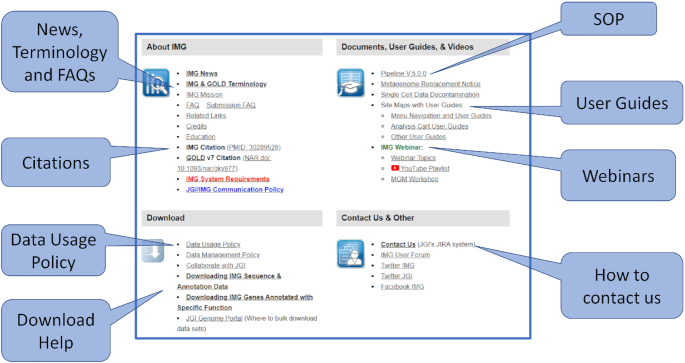
IMG currently has over 22 000 registered users from 109 countries. Many users are from academic institutions all over the world, which are also using IMG in their curriculum. Beginner users who need to learn how to perform comparative analysis on genome and metagenomes, often struggle to learn how to use the IMG system due to the complexity of data types and the tools we have provided. To further support these users, we have redesigned the IMG Help page to add more user guides and webinar recordings. The new Help page is divided into four sections: (i) About IMG, (ii) Documents, User Guides & Videos, (iii) Download, (iv) Contact Us & Other (see Figure 6).
Figure 6.
New Help Page: We have redesigned the IMG Help page to help users to better understand the system.
The About IMG section lists our mission, policy and system requirements. We have added a new IMG & GOLD Terminology guide to help users understand the special terminologies and keywords used in IMG and GOLD. There are two frequently asked questions (FAQ) links for general information and for specific questions to data submission. Users of the IMG system are encouraged to cite IMG (32) and GOLD (1) in their publications.
The Documents, User Guides & Videos section includes links to IMG user guides and our standard operating procedure (SOP) for the annotation pipeline. We have recently conducted two IMG Webinar series from April to June 2020, with a total of eight lectures. Links to the webinar recordings and relevant documents are available in this section too.
Many users are interested in downloading data from IMG to perform their own studies or analysis. The Download section includes the JGI data usage policy that needs to be followed by all users. We have also added a couple of new user guides to show how users can download certain data from IMG. The ‘Downloading IMG Sequence & Annotation Data’ guide shows how to download a large amount of genomes and metagenomes from the JGI Genome Portal. The ‘Downloading IMG Genes Annotated with Specific Function’ guide shows how to download all IMG isolate and metagenome genes annotated with a specific function (e.g., Enzyme EC:3.2.1.14).
IMG users can submit bug reports or questions to the JGI tracking system through the Contacts Us link in the Contact Us & Other section. This section also includes additional IMG social media links.
CONCLUDING REMARKS AND FUTURE PLANS
IMG continues to experience exponential data growth over the years. The growth is sustained both with regard to the number of datasets being added into IMG and the types of new data (e.g., metagenome bins) being supported. As a result, IMG constantly faces a challenge of processing, storing and querying a large amount of diverse data to serve the users with various research interests, analysis needs, and level of bioinformatics experience. In order to support this growth, we will continue improving the annotation pipeline (including the update of the reference databases), and expanding the data model and UI for analysis of metatranscriptomic datasets in IMG.
With the rapid data growth, there is also a need to provide more efficient and more diverse analysis tools in IMG. Users also request better visualization tools for data analysis. Even though we have released a new analysis tool that allows users to analyze genomes together with metagenome bins, many other tools in IMG (such as synteny viewer and other tools in the Compare Genomes) are still limited to genomes and metagenomes only. We are currently in the process of systematically reviewing all existing comparative analysis tools in IMG to determine how we can provide the necessary improvements. IMG is also forming collaborations with KBase (46) and National Microbiome Data Collaborative (NMDC) (47) for co-development of analysis tools and user interface.
IMG is currently accepting genome submissions in FASTA format only. An extension of the IMG annotation pipeline to enable submission of annotated genomes in gff (General Feature Format) format is under development. This will allow loading of annotated eukaryotic genomes, as well as prokaryotic genomes with manual user annotations, such as corrected coordinates for the frameshifted genes and potentially other predicted and experimentally identified features. The IMG functional annotations in this case will still be generated in addition to the ones available externally.
Due to the complexity of the IMG system, we are also paying more attention to the usability issues. We have conducted a few studies through user surveys and solicited feedback to improve the help page and the user guides. The IMG webinar series we conducted from April to June 2020 have enjoyed great success with our users. This will remain an area for improvements in the foreseeable future.
Contributor Information
I-Min A Chen, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Ken Chu, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Krishnaveni Palaniappan, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Anna Ratner, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Jinghua Huang, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Marcel Huntemann, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Patrick Hajek, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Stephan Ritter, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Neha Varghese, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Rekha Seshadri, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Simon Roux, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Tanja Woyke, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Emiley A Eloe-Fadrosh, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Natalia N Ivanova, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
Nikos C Kyrpides, Department of Energy Joint Genome Institute, Lawrence Berkeley National Laboratory, 1 Cyclotron Road, Berkeley, CA 94720, USA.
FUNDING
U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]; National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy. Funding for open access charge: U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]; National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.A., Kyrpides N.C., Reddy T.B.K.. Genomes OnLine Database (GOLD) v.7: updates and new features. Nucleic Acids Res. 2018; 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Field D., Sterk P., Kottmann R., Wim De Smet J., Amaral-Zettler L., Cochrane G., Cole J.R., Davies N., Dawyndt P., Garrity GM. et al.. Genomic standards consortium projects. Stand Genomic Sci. 2014; 9:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clum A., Huntemann M., Foster B., Foster B., Roux R., Hajek P., Varghese N., Mukherjee S., Reddy T.B.K., Daum C. et al.. The DOE-JGI metagenome analysis Workflow. 2020; bioRxiv doi:02 October 2020, preprint: not peer reviewed 10.1101/2020.09.30.320929. [DOI] [PMC free article] [PubMed]
- 4. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K.D., Sayers E.W.. GenBank. Nucleic Acids Res. 2018; 46:D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leinonen R., Sugawara H., Shumway M.. The Sequence Read Archive. Nucleic Acids Res. 2010; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rhoads A., Au K.F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015; 13:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nobuaki K., Kazuharu A.. Nanopore sequencing: review of potential applications in functional genomics. Dev. Growth Differ. 2019; 61:316–326. [DOI] [PubMed] [Google Scholar]
- 8. Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K., Kyrpides N.C., Hugenholtz P.. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007; 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan P.P., Lin B., Mak A.J., Lowe T.M.. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. 2019; bioRxiv doi:30 April 2019, preprint: not peer reviewed 10.1101/614032. [DOI] [PMC free article] [PubMed]
- 10. Nawrocki E.P., Eddy S.R.. Infernal 1.1: 100-fold Faster RNA Homology Searches. Bioinformatics. 2013; 29:2933–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Griffiths-Jones S., Moxon S., Marshall M., Khanna A., Eddy S.R., Bateman A.. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2015; 33:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nawrocki E.P., Kolbe D.L., Eddy S.R.. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009; 25:1335–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hyatt D., Chen G.L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J.. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010; 11:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lomsadze A., Gemayel K., Tang S., Borodovsky M.. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res. 2018; 28:1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V.. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015; 43:D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2018; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haft D.H., Selengut J.D., Richter R.A., Harkins D., Basu M.K., Beck E.. TIGRFAMs and genome properties in 2013. Nucleic Acids Res. 2013; 41:D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandurangan A.O., Stahlhacke J., Oates M.E., Smithers B., Gough J.. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 2019; 47:D490–D494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Letunic I., Bork P.. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018; 46:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sillitoe I., Dawson N., Lewis T.E., Das S., Lees J.G., Ashford P., Tolulope A., Scholes H.M., Senatorov I., Bujan A. et al.. CATH: expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res. 2019; 47:D280–D284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D.. HMMER web server: 2018 update. Nucleic Acids Res. 2018; 46:W200–W204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arndt W. Modifying HMMER3 to run efficiently on the Cori supercomputer using OpenMP tasking. IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW). 2018; 239–246. [Google Scholar]
- 23. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kielbasa S.M., Wan R., Sato K., Horton P., Frith M.C.. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011; 21:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caspi R., Billington R., Ferrer L., Foerster H., Fulcher C.A., Keseler I.M., Kothari A., Krummenacker M., Latendresse M., Mueller L.A. et al.. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016; 44:D471–D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emanuelsson O., Brunak S., von Heijne G., Nielsen H.. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2007; 2:953–971. [DOI] [PubMed] [Google Scholar]
- 27. Moller S., Croning M.D.R., Apweiler R.. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001; 17:646–653. [DOI] [PubMed] [Google Scholar]
- 28. Varghese N.J., Mukherjee S., Ivanova N., Konstantinidis K.T., Mavrommatis K., Kyrpides N.C., Pati A.. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015; 43:6761–6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palaniappan K., Chen I.A., Chu K., Ratner A., Seshadri R., Kyrpides N.C., Ivanova N.N., Mouncey N.J.. IMG-ABC v.5.0: an update to the IMG/Atlas of Biosynthetic Gene Clusters Knowledgebase. Nucleic Acids Res. 2020; 48:D422–D430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hadjithomas M., Chen I.A., Chu K., Huang J., Ratner A., Palaniappan K., Andersen E., Markowitz V., Kyrpides N.C., Ivanova N.N.. IMG-ABC: new features for bacterial secondary metabolism analysis and targeted biosynthetic gene cluster discovery in thousands of microbial genomes. Nucleic Acids Res. 2017; 45:D560–D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huntemann M., Ivanova N.N., Mavromatis K., Tripp H.J., Paez-Espino D., Palaniappan K., Szeto E., Pillay M., Chen I.A., Pati A. et al.. The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v. 4). Stand. Genomic Sci. 2015; 10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen I.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J.R., Seshadri R. et al.. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and metagenomes. Nucleic Acids Res. 2019; 47:D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paez-Espino D., Roux S., Chen I.A., Palaniappan K., Ratner A., Chu K., Huntemann M., Reddy T.B.K., Pons J.C., Llabres M.. IMG/VR v.2.0: an integrated data management and analysis system for cultured and environmental viral genomes. Nucleic Acids Res. 2016; 47:D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karp P.D., Ivanova N., Krummenacker M., Kyrpides N., Latendresse M., Midford P., Ong W.K., Paley S., Seshadri R.. A comparison of microbial genome web portals. Front. Microbiol. 2019; 10:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang D.D., Froula J., Egan R., Wang Z.. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015; 3:e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W.. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015; 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaumeil P.A., Mussig A.J., Hugenholtz P., Parks D.H.. GTDB-Tk: a tool kit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020; 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fisher R.A. Newman J.R. Mathematics of a Lady Tasting Tea. The World of Mathematics. 1956; 3:Courier Dover Publications. [Google Scholar]
- 39. Mann H.B., Whitney D.R.. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947; 18:50–60. [Google Scholar]
- 40. Welch B.L. The generalization of Student's problem when several different population variances are involved. Biometrika. 1947; 34:28–35. [DOI] [PubMed] [Google Scholar]
- 41. Fisher R.A. On the probable error of a coefficient of correlation deduced from a small sample. Metron. 1921; 1:3–32. [Google Scholar]
- 42. Field A. Discovering Statistics using SPSS. 2009; Sage Publications, Inc. [Google Scholar]
- 43. Seshadri R., Leahy S.C., Attwood G.T., Teh K.H., Lambie S.C., Cookson A.L., Eloe-Fadrosh E.A., Pavlopoulos G.A., Hadjithomas M., Varghese N.J. et al.. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018; 36:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tobias J.W., Shrader T.E., Rocap G., Varshavsky A.. The N-end rule in bacteria. Science. 1991; 254:1374–1377. [DOI] [PubMed] [Google Scholar]
- 45. Varshavsky A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:358–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S. et al.. KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 2018; 36:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wood-Charlson E.M., Anubhav, Auberry D., Blanco H., Borkum M.I., Corilo Y.E., Davenport K.W., Deshpande S. et al.. The National Microbiome Data Collaborative: enabling microbiome science. Nat. Rev. Microbiol. 2020; 18:313–314. [DOI] [PubMed] [Google Scholar]