Abstract
The mission of NASA’s GeneLab database (https://genelab.nasa.gov/) is to collect, curate, and provide access to the genomic, transcriptomic, proteomic and metabolomic (so-called ‘omics’) data from biospecimens flown in space or exposed to simulated space stressors, maximizing their utilization. This large collection of data enables the exploration of molecular network responses to space environments using a systems biology approach. We review here the various components of the GeneLab platform, including the new data repository web interface, and the GeneLab Online Data Entry (GEODE) web portal, which will support the expansion of the database in the future to include companion non-omics assay data. We discuss our design for GEODE, particularly how it promotes investigators providing more accurate metadata, reducing the curation effort required of GeneLab staff. We also introduce here a new GeneLab Application Programming Interface (API) specifically designed to support tools for the visualization of processed omics data. We review the outreach efforts by GeneLab to utilize the spaceflight data in the repository to generate novel discoveries and develop new hypotheses, including spearheading data analysis working groups, and a high school student training program. All these efforts are aimed ultimately at supporting precision risk management for human space exploration.
INTRODUCTION
Scientists are leveraging space-based platforms such as the International Space Station (ISS) and earth-based laboratories such as the NASA Space Radiation Laboratory to study the effects of space on biological organisms, generating nucleic acid, protein, and metabolite profiling data at an unprecedented rate. The 2011 National Research Council (NRC) Decadal Survey presaged this ‘wave of change’ almost a decade ago, and called for the development of more powerful informatics technologies to meet the growing needs of these researchers (1). The Survey also promulgated the use of a systems biology approach by space biosciences investigators, which led to the formation of the GeneLab project (https://genelab.nasa.gov/), a science initiative at NASA to maximize omics data collected from spaceflight and ground simulation experiments, and including the creation in 2014 of the NASA GeneLab data repository (https://genelab-data.ndc.nasa.gov/ ) (1). The mission of GeneLab is to maximize the utilization of valuable biospecimens flown in space or exposed to simulated space stressors through the collection and curation of genomic, transcriptomic, proteomic and metabolomic (so-called ‘omics’) data, in order to enable the exploration of molecular network responses to space environments using a systems biology approach. Our designs for the data repository continue to be guided by the FAIR principles (2,3). To foster open science in space biology research and to stimulate research opportunities based on an integrated multi-omics analysis approach, investigators who generate omics data are encouraged to contact GeneLab for the latest information concerning data submission requirements and procedures. All GeneLab repository data are open-access.
We review here the various components (Figure 1) of the database, then describe its new data repository web interface and the GeneLab Online Data Entry (GEODE) web portal, which provides ‘self-service’ data curation, and will support the expansion of the database in the future to include companion non-omics assay data. We discuss our design strategies for GEODE, emphasizing how they promote investigators providing more accurate metadata using controlled vocabularies, reducing the curation effort required of GeneLab staff. We also introduce here a new GeneLab Application Programming Interface (API) specifically designed to support tools for the visualization of processed omics data. Such visualization tools are essential to fulfil an ‘Open Science’ mandate, democratizing the access to data and allowing the scientific community at large to gain new knowledge on how spaceflight affects life, from humans and mice, to plants and microbes.
Figure 1.
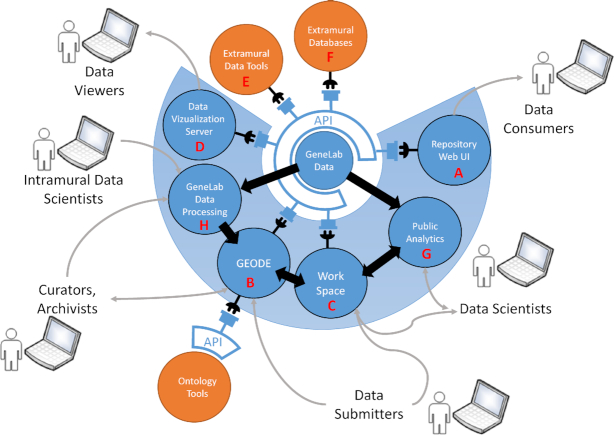
The GeneLab database and its connections to intramural and extramural systems. The database API is leveraged extensively by components of the GeneLab Data Systems: the database web browser interface (A), the GeneLab Online Data Entry tool (B), a workspace tool for private data sharing (C), and the GeneLab data visualization server (D). The database API is also being used to power various extramural data analytics and visualization tools (E, F). The GeneLab Public Analytics platform (G) and Data Processing systems (H) access the repository data files directly.
GENELAB DATABASE HOLDINGS
As of August 2020, GeneLab houses data from 271 studies representing investigations of many kinds of spaceflight experimental factors: gravity level (microgravity, hypergravity), ionizing radiation, circadian disruption, and atmospheric pressure, together with information on confounding variables such as organism diet, age, and sample preservation methods, for a variety of biological organisms (Figure 2). Approximately half the studies in the GeneLab repository are from experiments conducted in space, and the other half from ground-based (space analogue) environments (4). Mammalian (primarily rodent and human cell line) investigations account for a majority the studies. Other organisms studied include various flowering plants, (e.g. Arabidopsis thaliana), microbes, nematodes (primarily Caenorhabditis elegans), insects (Drosophila melanogaster), and bony fish. Although, the most prevalent study assay type is transcription profiling, the database also includes studies of protein expression, whole genome sequencing, metabolite profiling, metagenomics, epigenomics and epitranscriptomics. Investigators continue to submit data from new kinds of assays at an ever-increasing rate, particularly new kinds of RNA sequencing assays, such as spatial transcription profiling (5) data (6). Accommodating these new kinds of assays have not required GeneLab data systems software development, only minor changes to systems configuration files.
Figure 2.
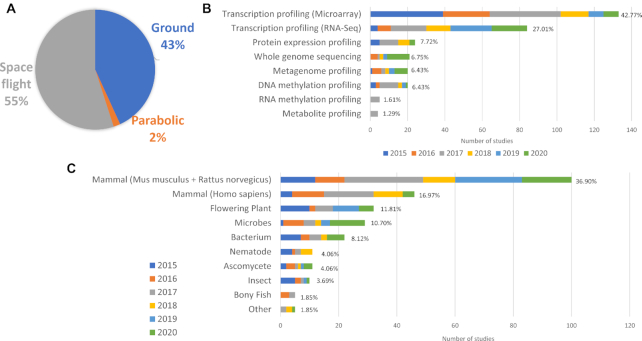
GeneLab database content by study experiment environment (A), study assay type by year of data publication (B), and organism studied by year of data publication (C).
DATA PROCESSING STANDARDIZATION
Since its inception GeneLab has focused on adopting best practices regarding the use of community-derived standards for metadata and data processing. For submitted metadata, GeneLab curators encourage the use of domain ontologies, and meticulously, manually harmonize submitted metadata with those of existing studies (1). To support consistent interpretation of submitted data, GeneLab curators augment submitted metadata for spaceflight studies with spacecraft environmental metadata. For example, for experiments conducted on board the U.S. Space Transportation System (the ‘Space Shuttle’), on the ISS, and on board various other spacecraft, GeneLab has gathered, summarized, and attached radiation exposure metadata to the other experiment metadata submitted by investigators (4). Ambient CO2, temperature, and humidity data are also gathered, summarized, and associated with each sample for metadata submitted for ISS experiments.
GeneLab has also begun to process submitted transcriptomic data using consensus pipelines developed by representatives of the space biology community (https://github.com/nasa/GeneLab_Data_Processing). This yields gene expression analysis data that are then made available in the database together with the raw data, and all intermediate data processing files. At the time of this writing, 103 GeneLab transcription profiling studies in the database have processed data available.
KEY DATABASE FEATURES
When originally designed, the GeneLab data systems sought merely to replicate features of existing, publicly-available life sciences data repositories that have long been recognized as valuable to end-users. These included the use of persistent, immutable accession numbers for submitted data, completely open access, and an API to allow for data file and metadata retrieval. Since this original design several years ago, GeneLab has sought to improve on this set of features, and implement new ones in line with advancing end-user needs and new, community-derived design principles for data repositories.
User interface
The initial user interface for the data repository organized study metadata using a series of tabs, one each for administrative, protocol, sample, and assay metadata. However, this layout makes it difficult for users to see more than one active tab's content at the same time. We have since revised the layout to use navigable, movable, and collapsible sections for each kind of metadata (Figure 3) so that users can create custom displays to organize and view the exact kind of metadata they want.
Figure 3.
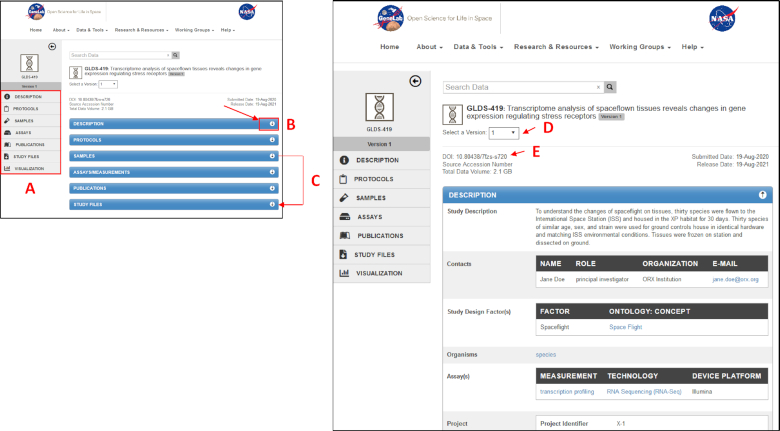
Redesigned ‘individual study page’ of the GeneLab data repository. Users can click on the section links (A) to navigate to each panel of metadata, open/close (collapse) each panel of metadata using the arrow on the panel header (B), and/or drag panels to re-order metadata displays (C). For each study dataset, a DOI is generated (D), and every submitted version of the dataset can be displayed using a drop-down menu (E).
Increased FAIRness
Digital object identifiers
From its inception, GeneLab has sought to implement community-derived (i.e. open) standards for the data and metadata it curates, and for the operation of the database (end-user interactions, curation by GeneLab, etc.). Largely concurrent with the design of the GeneLab Data Repository, the global scientific community converged on a set of optimal design features for data repositories, now termed the FAIR (Findability, Accessibility, Interoperability, and Reusability) principles. In the area of Findability, the data repository now provides Digital Object Identifiers (DOIs) for all submitted datasets. DOIs have guaranteed persistence, and resolution control mechanisms that are centralized (independent of the data source), and provide users of the GeneLab datasets with a way to cite them easily and uniformly. In addition, each version of a dataset (distinguished by changes in data files or in metadata) is tracked in the data repository, and users can opt to see previous versions through the web interface (Figure 3). This ensures persistence of the specific version of data as cited by others. Finally, NASA is currently developing processes for the automated submission to other databases of data submitted first to GeneLab, with the twin goals of making the data more findable from extramural database systems, and reducing the burden to data providers of generating multiple data submissions.
Accessibility
All data in the GeneLab data repository continues to be accessible to all users without user authentications. To provide the greatest accessibility to its data analysis tools and collaborative workspace, the GeneLab analysis and collaboration platforms have integrated the widely-available Google credentialing system into its authentication systems. Users can opt to use their Google-managed credentials or, if they have them, use their NASA credentials to access these subsystems. It is our intention that this development leads to increased access to and usage of these tools for more investigators.
Linked data
One the biggest areas where many data repositories fall short using FAIR-based metrics is interoperability (2), which often presents a barrier to data reusability. In order to support systems interoperation and increase data reuse, the GeneLab repository data uses the widely known ISA experiment model (7) for tagging data files with metadata. The ISA model leverages community-developed concept vocabularies (ontologies), so that there is agreement among users on metadata meaning, and therefore data characterization. Furthermore these models support linkage of data values (‘linked data’) so that, for example, biological samples, personnel or resources reused across experiments need only be described once by the first experiment to refer to them. Subsequent investigations that also use these objects then can quickly search for and refer to them in their investigation descriptions.
In the specific domain of space biology, it is common to re-use samples across different investigations, given the high cost of generating biological sample data in space environments. In many other application areas, it is common to re-use parts of experimental protocols, and these too can be similar referred to by data submitters using the ISA model, rather than duplicated semantically in the data repository.
Web-browser data submissions
The most recent addition to the GeneLab Data Systems is GEODE, the GeneLab Environment for Online Data Entry. This interface supports the creation of complex metadata descriptions of datasets, and was built in collaboration with, and by extending an interface originally developed by, the Metabolights (8) database at EBI. This web browser interface (Figure 4) allows users to develop ISA-tab sets of metadata through either a guided submission ‘wizard’, in which users create metadata step-by-step, or ‘Study Overview’ method, in which metadata can be arbitrarily edited. Data submitters and GeneLab curation staff can both create metadata using GEODE; submitters are limited to editing only their own submitted metadata, while GeneLab curation staff can access and edit all metadata. The web interface provides the capability for submitters (or curators) to look up metadata concepts in online ontologies dynamically, which reduces metadata normalization effort of the curation staff. Furthermore, GEODE provides continuously available data set editing for submitters (or curators), so that they can rapidly and efficiently update and correct their data; any data set owner (or curator) can change the status of their published data sets to ‘In Review’, make changes to the data/metadata, and re-submit it for release at any time.
Figure 4.
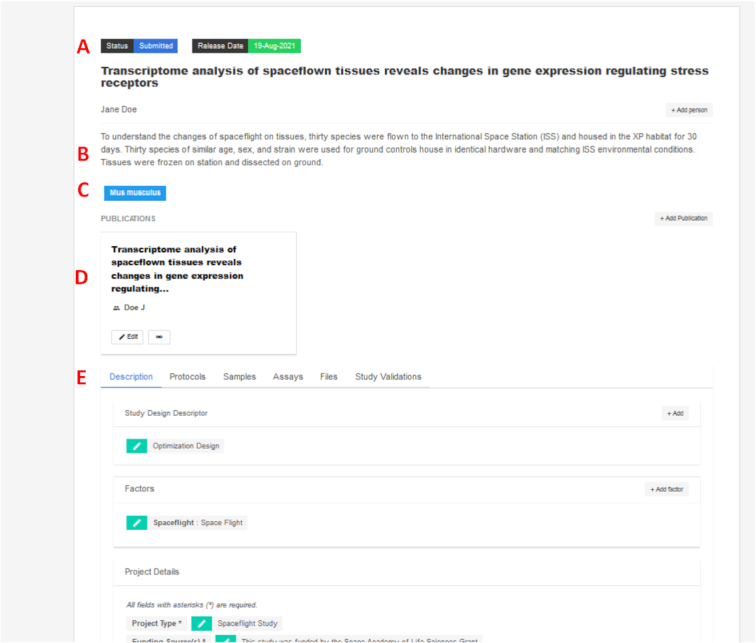
The GEODE web browser interface used by both data submitters and GeneLab data curation staff. Below the data set ‘Status’ and desired release date fields (A) are the user-entered title, contact(s) and brief study description fields (B), the user-selected chosen taxon(s) studied (C), relevant publication objects (D) and subsections for users to specify further details of the study's description, protocols, samples, assays and study files (E), all validated against the ISA model.
Enhanced API
GeneLab has always provided an Application Programming Interface (API) for the retrieval of individual experiment data and metadata files. Improvements in this API now enables users to query the database for samples, potentially across multiple datasets and assays, by combinations of values of metadata fields, and then to retrieve sample-associated experiment data. Both the data and the accompanying annotations are returned in a tabular format suitable for downstream processing in a variety of languages and environments commonly used by data scientists. This, in turn, empowers researchers to conduct multi-dataset and multi-omics analyses, such as investigating the impact of single experiment factors on a variety of organisms during space flight. For example, the API can be leveraged to interrogate the entirety of the GeneLab database for RNA-Seq assay gene expression counts from all samples from Mus musculus that experienced a specified level of ionizing radiation; such a request returns gene counts from multiple datasets as well as all other metadata values associated with the samples (status of space flight, radiation dose, diet, age, etc.), immediately providing the end-user with data and annotations suitable to build a complex model that accounts for all main and confounding factors known to the database. The API also supports queries for multiple assay types, for example, all transcriptomic and epigenomic assays, facilitating multi-omics data analyses. This enhanced API layer is currently being leveraged within GeneLab to provide advanced data visualizations of GeneLab processed data (9). These include a data table browser for sample data and annotations, and common statistical data visualizations such as principal component analysis, in a web-browser interface. Finally, we should note that, as the data API leverages the versatile ISA-Tab format to bring annotations to data files, this API could be easily extended to numerous types of assay data, not just omics.
COMMUNITY
Scientific outreach
A key goal of the GeneLab project is to bring together a community of scientists to generate novel hypotheses and make valuable scientific discoveries using the data in the GeneLab database. GeneLab has sought to engage working groups of students, bioinformaticians, space biologists, researchers and citizen scientists by providing freely-available data analysis tools and support for their use. Users can access user guides and tutorial videos on how to use the various GeneLab applications, including how to analyze RNA-seq datasets using the GeneLab Analysis Platform (https://genelab.nasa.gov/help/tutorials). Furthermore, NASA has developed the GeneLab 4 High School (GL4HS) training program, in which fifteen high school students learn how to use the GeneLab tools over four weeks to analyze spaceflight microarray and RNA-seq data, how to generate follow-on hypotheses based on these analyses, how to design a ground-based experiment to test these hypotheses, and ultimately write research proposals for the experiments (https://www.nasa.gov/ames/genelab-for-high-schools). GeneLab is also launching an Education Working Group, a group of educators and scientists, who will develop educational content that promotes the use of the GeneLab database, and its visualization and analytical tools.
Analysis working groups
In 2018 GeneLab formed four Analysis Working Groups (AWGs) comprised of over 100 scientists and students from multiple space agencies, academia, and industry, for the purpose of optimizing and standardizing the processing of raw omics data from the GeneLab repository. The AWGs work in four major areas: animal, plant, microbial, and multi-omics data processing. The consensus data processing pipelines developed in each group are then used by GeneLab to process the data in the repository (see Data Processing Standardization above). The AWGs have begun to utilize the processed data to publish insights in important areas of space biology (10–14).
Data re-use
The number of GeneLab studies continue to grow rapidly, providing more opportunities for meta-analysis. Currently there are 18 publications of this type linked to repository datasets that together leveraged 78 repository datasets (see https://genelab.nasa.gov/publications/derived). In addition, an increasing number of extramural visualization tools are consuming the processed data in the repository through its API. For example, the Test of Arabidopsis Space Transcriptome (TOAST) tool has a graphical user interface through which users can quickly compare the data from plant spaceflight experiments using real-time, gene-specific queries, or explore gene networks of interest (15). Likewise, Project MANGO (https://www.ebi.ac.uk/metagenomics/super-studies/3) users can generate and visualize taxonomic analyses of GeneLab processed data to explore the microbiome of the ISS.
FUTURE DIRECTIONS
Astronaut data
In 2011 the National Research Council of the U.S. recognized the importance of collecting astronaut data, in addition to model organism data in the Decadal Survey (16): ‘because of the limited number of humans who undergo exposure to the space environment, maintaining an extensive and well-organized database and managing it as a resource to be shared with the scientific community has long been viewed as an essential step for scientific discovery’. However, anonymizing astronaut data presents a unique challenge as the cohort is indeed extremely small, and very few metadata (e.g. just gender and flight dates) may be used to identify individuals. Nevertheless, to achieve greater reuse of these data, they too should be made part of an Open Science framework using FAIR principles. They should be stored in non-proprietary formats based on open (community-derived) standards, with open access and with linkages to other data that provide important context (17). This goal was nicely summarized by Higman et al. (17), ‘data should be as open as possible, as closed as necessary’. More effort should be put into identifying what types of data can remain open, even if one can identify the individuals to which they belong. For example, gene expression levels do not present any sensitive information in the context of the response to stresses like microgravity and radiation. Such information should therefore be made available through open science frameworks as soon as these data are released for publication by investigators. Any information revealing sensitive and personal information, such as that which can be derived from DNA or RNA sequences, would remain access-controlled.
Enabling precision space risk management
Although valuable knowledge has been gained from experiments conducted in space or from space simulations on the ground, little progress has been made in terms of modelling the impact of spaceflight on humans. Most risk models have been focusing on space radiation and cancer (18–20). The strong epidemiological data on cancer risk from ionizing radiation (21) and the existence of accelerators dedicated to simulating space radiation have been the main thrust behind these risk models. Ionizing radiation is an easy-to-use experimental factor, with good quantitative input and output, enabling mathematical models. Briefly, scientists control precisely the dose, the dose rate and the type of radiation delivered and they can measure precisely short-term endpoints such as DNA double strand breaks (22,23). Interestingly, such responses have been shown to be modulated by the genetic background of an individual (24–26), eventually leading to DNA mutations and cancer (27–29). Because modelling requires quantitative data, omics data are very well suited to informing risk models and to tackling more challenging stressors such as microgravity. The growing amount of omics data generated by NASA principal investigators and the recent guidelines from NASA emphasizing the importance of submitting all omics data to GeneLab are therefore exciting developments for modellers. Indeed, the software architecture designed by GeneLab and its strategies around data and metadata harmonization will enable the development of the next generation of space risk models. By classifying experimental factors into digital objects and by transforming raw data into comparable quantitative endpoints using a workflow approved by the community, GeneLab lets users see how space factors affect a large array of biological processes (13,30–32). By establishing mathematical laws describing the relationship between factors’ values and biological endpoints, one can start building more elaborate computer models. By integrating information from other NASA databases with more qualitative data (e.g. immunocytochemistry, behavioural data) using the same metadata schema and harmonization strategies, GeneLab can continue to evolve as a knowledge-based system, fully leveraging advances in machine learning, providing insights on enabling human life in space, and enabling individualized space risk models for precision space medicine.
GeneLab plans to utilize machine learning algorithms and artificial intelligence (AI) to maximize the usage of its data, especially in the context of modelling and extrapolation to human risks. Such effort may start by providing a portal proposing mechanistic models describing known health risks (cancer, muscle atrophy, bone loss, etc.). Such a tool would allow users to test how well GeneLab omics data and data from other databases support these models and test alternative models. Such approach will involve collaborating with the NASA Ames Life Sciences Data Archive (ALSDA), which archives biospecimens and non-omics data from mammalian and microbial NASA experiments.
DATA AVAILABILITY
NASA GeneLab is an open-access data repository (https://genelab.nasa.gov).
ACKNOWLEDGEMENTS
We would like to thank all members of the GeneLab team and the data systems team in particular: Ms Jamie Bales, Mr Peter O’Donnell, Ms Sonja Caldwell and Mr Matias Fernandez, for developing the database and web applications.
Contributor Information
Daniel C Berrios, USRA/NASA Ames Research Center, Moffett Field, CA 94035, USA.
Jonathan Galazka, NASA Ames Research Center, Moffett Field, CA 94035, USA.
Kirill Grigorev, Weill Cornell Medicine, New York, NY 10065, USA.
Samrawit Gebre, KBR/NASA Ames Research Center, Moffett Field, CA 94035, USA.
Sylvain V Costes, NASA Ames Research Center, Moffett Field, CA 94035, USA.
FUNDING
Space Biology Program (Science Mission Directorate, Biological and Physical Sciences Division) of the National Aeronautics and Space Administration. Funding for open access charge: NASA.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ray S., Gebre S., Fogle H., Berrios D.C., Tran P.B., Galazka J.M., Costes S.V.. GeneLab: Omics database for spaceflight experiments. Bioinformatics. 2019; 35:1753–1759. [DOI] [PubMed] [Google Scholar]
- 2. Berrios D.C., Beheshti A., Costes S.V.. FAIRness and usability for open-access omics data systems. Proceedings of the 2018 AMIA Annual Symposium. 2018; AMIA; 232–241. [PMC free article] [PubMed] [Google Scholar]
- 3. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E. et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beheshti A., Miller J., Kidane Y., Berrios D., Gebre S.G., Costes S.V.. NASA GeneLab project: bridging space radiation omics with ground studies. Radiat. Res. 2018; 189:553–559. [DOI] [PubMed] [Google Scholar]
- 5. Ståhl P.L., Salmén F., Vickovic S., Lundmark A., Navarro J.F., Magnusson J., Giacomello S., Asp M., Westholm J.O., Huss M. et al.. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016; 353:78–82. [DOI] [PubMed] [Google Scholar]
- 6. Galazka J., Giacomello S., Lai Polo S.-H., Andrusivova Z., Saravia-Butler A., Fogle H., Andrusivova Z., Giacomello S., Andrusivova Z., Boyko V. et al.. Spatially resolved transcriptional analysis of hearts from mice flown on the RR-3 mission. GeneLab. 2020; 10.26030/09s2-9×11 version 2. [Google Scholar]
- 7. González-Beltrán A., Maguire E., Sansone S.A., Rocca-Serra P.. linkedISA: semantic representation of ISA-Tab experimental metadata. BMC Bioinformatics. 2014; 15:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O’Donovan C.. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020; 48:D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berrios D., Weitz E., Grigorev K., Costes S.V., Gebre S., Beheshti A.. Ding Q., Eulenstein O., Al-Mubaid H.. Proceedings of the 12th International Conference on Bioinformatics and Computational Biology. 2020; 70:EasyChair; 89–98. [Google Scholar]
- 10. Bailey S., Mason C.E., Beheshti A., Hassane D., Costes S.V., Grabham P., Scott R., Keune J., Bristow T., Klotz R. et al.. Hallmarks of space biology: the next stage of exploration. Cell. 2020; In press. [Google Scholar]
- 11. Beheshti A., Chakravarty K., Fogle H., Fazelinia H., Silveira W.A.D., Boyko V., Polo S.-H.L., Saravia-Butler A.M., Hardiman G., Taylor D. et al.. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019; 9:19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beheshti A., McDonald J.T., Miller J., Grabham P., Costes S.V.. GeneLab database analyses suggest long-term impact of space radiation on the cardiovascular system by the activation of FYN through reactive oxygen species. Int. J. Mol. Sci. 2019; 20:661–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McDonald J.T., Stainforth R., Miller J., Cahill T., da Silveira W.A., Rathi K.S., Hardiman G., Taylor D., Costes S.V., Chauhan V. et al.. NASA genelab platform utilized for biological response to space radiation in animal models. Cancers (Basel). 2020; 12:318–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Overbey E.G., Paul A.M., da Silveira W.A., Tahimic C.G.T., Reinsch S.S., Szewczyk N., Stanbouly S., Wang C., Galazka J.M., Mao X.W.. Mice exposed to combined chronic low-dose irradiation and modeled microgravity develop long-term neurological sequelae. Int. J. Mol. Sci. 2019; 20:4094–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barker R., Lombardino J., Rasmussen K., Gilroy S.. Test of arabidopsis space transcriptome: a discovery environment to explore multiple plant biology spaceflight experiments. Front. Plant Sci. 2020; 11:147–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. National Academies of Sciences Engineering and Medicine (U.S.). Committee on a Midterm Assessment of Implementation of the Decadal Survey on Life and Physical Sciences Research at NASA, National Academies of Sciences Engineering and Medicine (U.S.). Aeronautics and Space Engineering Board., National Academies of Sciences Engineering and Medicine (U.S.). Space Studies Board. and National Academies of Sciences Engineering and Medicine (U.S.). Division on Engineering and Physical Sciences A Midterm Assessment of Implementation of the Decadal Survey on Life and Physical Sciences Research at NASA. 2018; Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- 17. Rosie H., Bangert D., Jones S.. Three camps, one destination: the intersections of research data management, FAIR and open. Insights. 2019; 32:18–27. [Google Scholar]
- 18. Schimmerling W., Cucinotta F.A., Wilson J.W.. Radiation risk and human space exploration. Adv. Space Res. 2003; 31:27–34. [DOI] [PubMed] [Google Scholar]
- 19. Cucinotta F.A., Durante M.. Cancer risk from exposure to galactic cosmic rays: implications for space exploration by human beings. Lancet Oncol. 2006; 7:431–435. [DOI] [PubMed] [Google Scholar]
- 20. Heuskin A.C., Osseiran A.I., Tang J., Costes S.V.. Simulating space radiation-Induced breast tumor incidence using automata. Radiat. Res. 2016; 186:27–38. [DOI] [PubMed] [Google Scholar]
- 21. Preston D.L., Ron E., Tokuoka S., Funamoto S., Nishi N., Soda M., Mabuchi K., Kodama K.. Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat. Res. 2007; 168:1–64. [DOI] [PubMed] [Google Scholar]
- 22. Costes S.V., Boissiere A., Ravani S., Romano R., Parvin B., Barcellos-Hoff M.H.. Imaging features that discriminate between foci induced by high- and low-LET radiation in human fibroblasts. Radiat. Res. 2006; 165:505–515. [DOI] [PubMed] [Google Scholar]
- 23. Georgescu W., Osseiran A., Rojec M., Liu Y., Bombrun M., Tang J., Costes S.V.. Characterizing the DNA damage response by cell tracking algorithms and cell features classification using high-content time-lapse analysis. PLoS One. 2015; 10:e0129438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ochola D.O., Sharif R., Bedford J.S., Keefe T.J., Kato T.A., Fallgren C.M., Demant P., Costes S.V., Weil M.M.. Persistence of gamma-H2AX foci in bronchial cells correlates with susceptibility to radiation associated lung cancer in mice. Radiat. Res. 2019; 191:67–75. [DOI] [PubMed] [Google Scholar]
- 25. Penninckx S., Cekanaviciute E., Degorre C., Guiet E., Viger L., Lucas S., Costes S.V.. Dose, LET and strain dependence of radiation-induced 53BP1 foci in 15 mouse strains ex vivo introducing novel DNA damage metrics. Radiat. Res. 2019; 192:1–12. [DOI] [PubMed] [Google Scholar]
- 26. Pariset E., Penninckx S., Degorre C., Guiet E., Lopez Macha A., Cekanaviciute E., Snijders A.M., Mao J.H., Paris F., Costes S.V.. 53BP1 repair kinetics for prediction of in vivo radiation susceptibility in 15 mouse strains. Radiat. Res. 2020; doi:10.1667/RADE-20-00122.1. [DOI] [PubMed] [Google Scholar]
- 27. Costes S., Sachs R., Hlatky L., Vannais D., Waldren C., Fouladi B.. Large-mutation spectra induced at hemizygous loci by low-LET radiation: evidence for intrachromosomal proximity effects. Radiat. Res. 2001; 156:545–557. [DOI] [PubMed] [Google Scholar]
- 28. Tang j., Georgescu W., Deschamps T., Yannone S.M., Costes S.V.. Genomic Instability and Cancer Metastasis. 2015; Springer Science & Business Media; 75–93. [Google Scholar]
- 29. Heuskin A.C., Osseiran A.I., Tang J., Costes S.V.. Simulating space radiation-induced breast tumor incidence using automata. Radiat. Res. 2016; 186:27–38. [DOI] [PubMed] [Google Scholar]
- 30. Beheshti A., Shirazi-Fard Y., Choi S., Berrios D., Gebre S.G., Galazka J.M., Costes S.V.. Exploring the effects of spaceflight on mouse physiology using the open access NASA GeneLab platform. J. Vis. Exp. 2019; 143:e58447. [DOI] [PubMed] [Google Scholar]
- 31. Beheshti A., Chakravarty K., Fogle H., Fazelinia H., Silveira W.A.D., Boyko V., Polo S.L., Saravia-Butler A.M., Hardiman G., Taylor D. et al.. Multi-omics analysis of multiple missions to space reveal a theme of lipid dysregulation in mouse liver. Sci. Rep. 2019; 9:19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beheshti A., Cekanaviciute E., Smith D.J., Costes S.V.. Global transcriptomic analysis suggests carbon dioxide as an environmental stressor in spaceflight: a systems biology GeneLab case study. Sci. Rep. 2018; 8:4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NASA GeneLab is an open-access data repository (https://genelab.nasa.gov).