Abstract
PubChem (https://pubchem.ncbi.nlm.nih.gov) is a popular chemical information resource that serves the scientific community as well as the general public, with millions of unique users per month. In the past two years, PubChem made substantial improvements. Data from more than 100 new data sources were added to PubChem, including chemical-literature links from Thieme Chemistry, chemical and physical property links from SpringerMaterials, and patent links from the World Intellectual Properties Organization (WIPO). PubChem's homepage and individual record pages were updated to help users find desired information faster. This update involved a data model change for the data objects used by these pages as well as by programmatic users. Several new services were introduced, including the PubChem Periodic Table and Element pages, Pathway pages, and Knowledge panels. Additionally, in response to the coronavirus disease 2019 (COVID-19) outbreak, PubChem created a special data collection that contains PubChem data related to COVID-19 and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
INTRODUCTION
PubChem (https://pubchem.ncbi.nlm.nih.gov) (1–4) is a public chemical database at the National Library of Medicine (NLM), an institute within the U.S. National Institutes of Health (NIH). It collects chemical information from more than 750 data sources and disseminates it to the public free of charge. With more than four million unique interactive users per month at peak time (see Figure 1), PubChem is one of the most visited chemistry web sites in the world. PubChem serves as a key chemical information resource for biomedical research communities in many areas such as cheminformatics, chemical biology, medicinal chemistry and drug discovery. Importantly, PubChem is widely used as a ‘big data’ source in machine learning and data science studies for virtual screening (5–9), drug repurposing (10–13), chemical toxicity prediction (14–16), drug side effect prediction (17,18) and metabolite identification (19–22) and so on.
Figure 1.
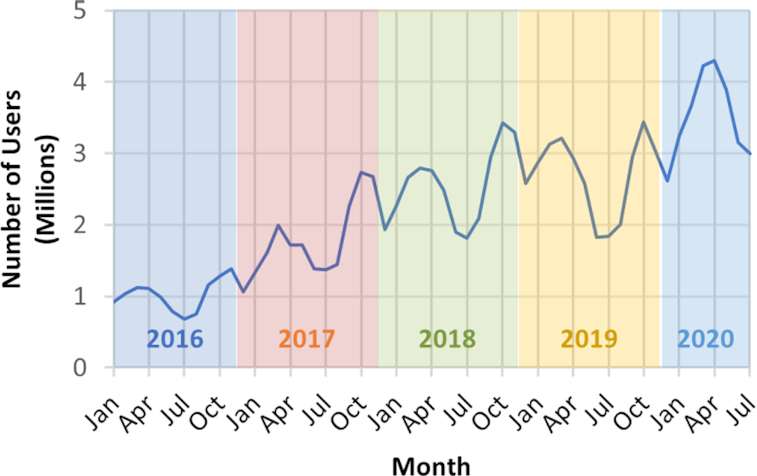
Growth of the number of unique PubChem users per month, as tracked by Google Analytics. The data shown here are for interactive users who access PubChem through web browsers only and does not include programmatic users.
PubChem organizes its data into three databases (2): Substance, Compound and BioAssay. The Substance database is a repository where depositor-provided chemical data are archived. The Compound database stores unique chemical structures extracted from the Substance database. The BioAssay database stores biological assay descriptions and test results. For each record in these databases, PubChem provides a dedicated web page that presents its available data. In addition, PubChem also provides alternative views through web pages that present the data related to a specific gene, protein, pathway, and patent (examples of these pages are listed in Table 1). PubChem data can be downloaded interactively through web interfaces or programmatically through several web service access routes including PUG-REST (23,24) and PUG-View (25).
Table 1.
Links to examples of the pages that present data for various types of PubChem records
| Record type | Example ID | Link |
|---|---|---|
| Compound | CID 1983 | https://pubchem.ncbi.nlm.nih.gov/compound/1983 |
| Chemical name ‘ibuprofen’ | https://pubchem.ncbi.nlm.nih.gov/compound/ibuprofen | |
| Substance | SID 138460 | https://pubchem.ncbi.nlm.nih.gov/substance/138460 |
| BioAssay | AID 248 | https://pubchem.ncbi.nlm.nih.gov/bioassay/248 |
| Gene | Gene ID 1956 | https://pubchem.ncbi.nlm.nih.gov/gene/1956 |
| Gene name ‘EGFR’ | https://pubchem.ncbi.nlm.nih.gov/gene/egfr | |
| Protein | Accession P00533 | https://pubchem.ncbi.nlm.nih.gov/protein/P00533 |
| PDB Chain ID 1XKK_A | https://pubchem.ncbi.nlm.nih.gov/protein/1XKK_A | |
| Pathway | Reactome ID R-BTA-177929 | https://pubchem.ncbi.nlm.nih.gov/pathway/Reactome:R-BTA-177929 |
| Patent | US7651687 | https://pubchem.ncbi.nlm.nih.gov/patent/US7651687 |
| Bioactivity | AID 248 & SID 553777 | https://pubchem.ncbi.nlm.nih.gov/bioassay/248#sid=553777 |
| Accession P00533 & CID 5328245 | https://pubchem.ncbi.nlm.nih.gov/protein/P00533#cid=5328245 | |
| Gene ID 1956 & CID 5328245 | https://pubchem.ncbi.nlm.nih.gov/gene/1956#cid=5328245 |
As a public resource used by millions of users with diverse backgrounds, it is challenging for PubChem to satisfy the data needs of all users. For example, PubChem needs to efficiently handle large amounts of heterogeneous data collected from hundreds of data sources in various scientific domains, while simultaneously striving to limit data errors and handle important scientific data representation nuances. An increase in mobile users raises the need for responsive webpages that are rendered appropriately on both large screens (e.g. desktops) and small screens (e.g. tablets and smartphones). To address these and other challenges, many changes have been made to PubChem since our latest update paper published in this journal (1).
DATA CONTENTS
With data integration from over 100 new sources for the past two years, PubChem now contains more than 293 million depositor-provided substance descriptions, 111 million unique chemical structures, and 271 million bioactivity data points from 1.2 million biological assays experiments (as of August 2020). This corresponds to an increase in substances, compounds and bioactivities by 19%, 14% and 14%, respectively, compared to the statistics reported in our previous paper two years ago (1). This section highlights some of the notable new data added to PubChem.
COVID-19 data set
The coronavirus disease 2019 (COVID-19) outbreak (26–28), which began in early 2020, caused a serious global health crisis and scientific communities initiated various efforts to stop this deadly disease. This created a huge demand for convenient access to data pertinent to COVID-19 and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In response to this urgent demand, PubChem created a special data collection, which contains PubChem data related to COVID-19 and SARS-CoV-2. This COVID-19 data collection can be accessed either from the PubChem homepage or via the following URL:
https://pubchem.ncbi.nlm.nih.gov/#query=covid-19.
The data in this collection are collected from various authoritative sources, including several NCBI databases (RefSeq, Gene, Protein, Structure, GenBank, ClinicalTrials.gov) (4,29,30) as well as other external resources (UniProt (31), RCSB Protein Databank (PDB) (32), IUPHAR/BPS Guide to PHARMACOLOGY (33), WikiPathways (34), DrugBank (35), COVID-19 Disease Map (https://fairdomhub.org/projects/190), CAS COVID-19 Protein Target Thesaurus (https://www.cas.org/covid-19-protein-target-thesaurus), European Clinical Trials Register (https://www.clinicaltrialsregister.eu/ctr-search/search?query=covid-19)). This data collection is updated on a weekly basis and PubChem is actively seeking new relevant data to add to it.
Chemical-literature links
PubChem added more than 1.35 million links between chemicals and scientific articles that mention them, thanks to data contributed by the publisher Thieme Chemistry (https://go.usa.gov/xEDCA). These data contain about 745 000 chemicals, along with relevant articles and their metadata (including the digital object identifier (DOI), publication title, name of the journal or book, publication type, language, and publication year). The summary page of each compound contributed by Thieme Chemistry includes a table containing document links from Thieme Chemistry, as shown in the following example (for CID 9568614, esomeprazole):
https://pubchem.ncbi.nlm.nih.gov/compound/9568614#section=Thieme-References
The addition of Thieme Chemistry information drastically increased the number of chemical structures in PubChem with links to the scientific literature. Of the 750 000 chemicals, >40% were new to PubChem and 90% previously lacked literature links. Importantly, because many of these articles are about chemical synthesis, they are often not found in PubMed, which focuses on literature in biomedical and life sciences. Therefore, these chemical-literature links dramatically expand the findability, accessibility, interoperability, and reusability (FAIR) of synthesis-related chemical information.
Links to molecular property data in SpringerMaterials
For >32 000 compounds, PubChem now provides links to hundreds of chemical and physical properties pertinent to material science and related fields, available from SpringerMaterials (https://go.usa.gov/xvqfq). The summary page of a compound with the SpringerMaterials link has a list of properties available at SpringerMaterials for that compound in the ‘SpringerMaterials Properties’ subsection in the ‘Chemical and Physical Properties’ section. For example, the following link shows the list of the material properties for CID 702 (ethanol):
https://pubchem.ncbi.nlm.nih.gov/compound/702#section=SpringerMaterials-Properties
Clicking on one of the properties in this list directs to the SpringerMaterials web page showing a list of articles containing detailed information on that property. The SpringerMaterials links help users to locate property data and related literature available for chemicals.
Links to PATENTSCOPE at WIPO
The World Intellectual Property Organization (WIPO) provided PubChem with >16 million chemical structures searchable in its patent database called PATENTSCOPE (https://go.usa.gov/xdhfK). For each chemical structure contributed by WIPO, PubChem provides a direct link to PATENTSCOPE, enabling PubChem users to search PATENTSCOPE for patent documents relevant to that chemical structure. For example, the following URL directs users to the ‘WIPO PATENTSCOPE’ section of CID 3672 (ibuprofen):
https://pubchem.ncbi.nlm.nih.gov/compound/3672#section=WIPO-PATENTSCOPE.
The link presented in this section allows users to search PATENTSCOPE for patent documents relevant to ibuprofen and further analyze returned hits using the tools available at PATENTSCOPE. The integration of WIPO’s chemical information with PubChem makes it easier for PubChem users to find pertinent patent information about chemicals.
ToxNet migration
ToxNet (36) was a collection of NLM databases that provided a wide range of toxicological information. As part of a broad reorganization at NLM, ToxNet was retired in December 2019 and PubChem is now hosting chemical toxicology information for several ToxNet databases, including the Chemical Carcinogenesis Research Information System (CCRIS), Genetic Toxicology Data Bank (Gene-Tox) (37,38), and Hazardous Substances Data Bank (HSDB) (39,40). Text data from HSDB, ChemIDplus (41), LactMed (42,43) and LiverTox (44) are integrated as annotations into compound records in PubChem. Chemical toxicity test results of chemicals from CCRIS and Gene-Tox are archived as bioassay records and substances tested in them. More detailed information on the ToxNet integration into PubChem can be found at: https://pubchemdocs.ncbi.nlm.nih.gov/toxnet.
WEB INTERFACES
Redesigned webpages
To provide users with easier access to its content, we redesigned the PubChem homepage and the pages dedicated to individual records (e.g. compound, substance, bioassay, gene, protein and patent) (Table 1). The new pages use responsive design that works on both desktop computers and mobile devices like smartphones and tablets. In addition, color-themes were introduced for different collections. For example, compound pages have a light blue theme, while substance pages use a yellow theme.
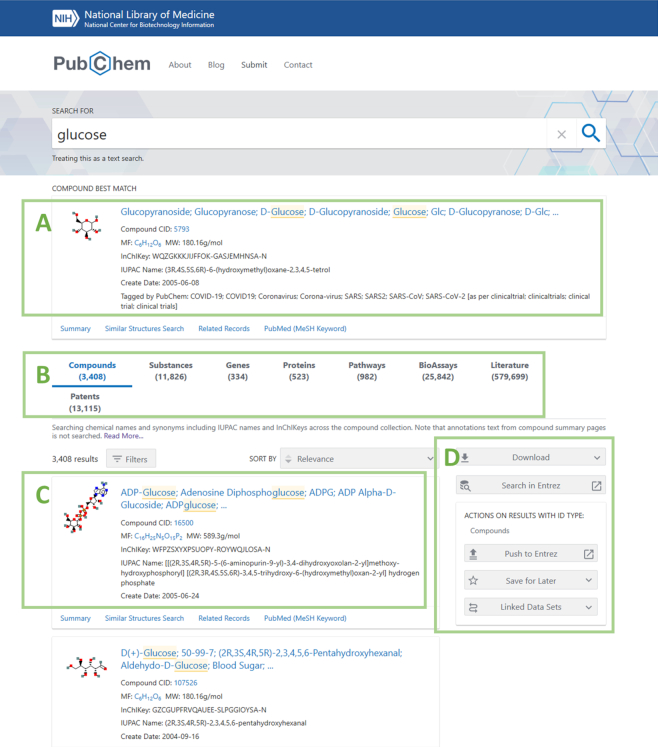
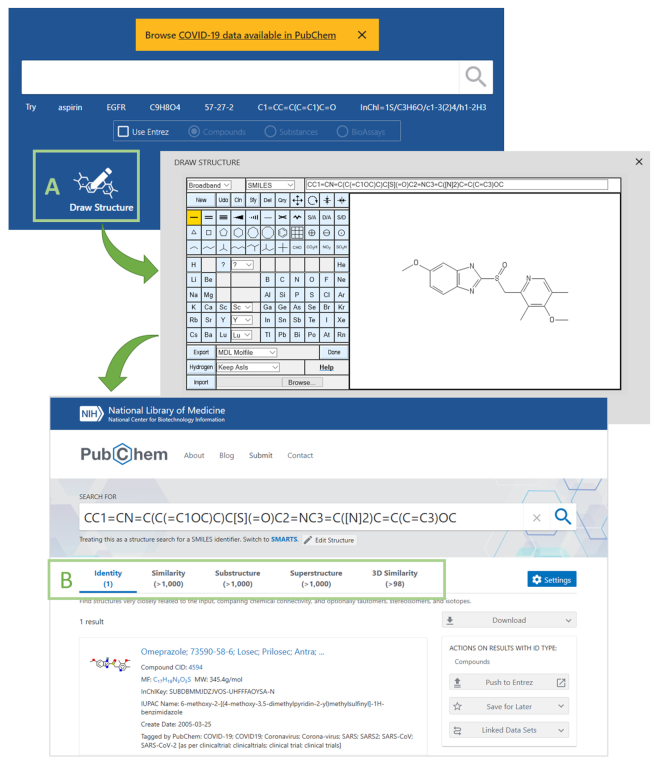
Importantly, the new PubChem homepage provides improved search capabilities. The new homepage has a single search box that allows users to search multiple data collections simultaneously using a single query (Figure 2). For example, a text query ‘glucose’ in the search box returns not only hit chemicals from the Compound and Substance databases, but also hits from other collections (e.g., proteins, genes, pathways, assays, scientific articles, patents). In addition, the new search box accepts chemical structure queries, allowing users to perform various types of structure search, including identity, similarity, substructure and superstructure searches (Figure 3). The query structure can be provided as SMILES (45–47) or InChI strings (48) or drawn using the PubChem Sketcher (49).
Figure 2.
Partial screen shot of the page returned from a text query ‘glucose’ (https://pubchem.ncbi.nlm.nih.gov/#query=glucose). PubChem interprets the query and suggest the best record pertinent to the query (Box A). The new search interface allows the user to simultaneously search multiple collections and hits returned from different collections can be viewed by click the corresponding tabs (Box B). Clicking one of the returned hits directs the user to the page dedicated to the record (Box C). The user can perform additional tasks using the buttons available on the right column of the page (Box D).
Figure 3.
Searching PubChem using a chemical structure input. The user can provide a chemical structure query by either using a line notation (e.g. SMILES or InChI) or by drawing the input molecule with the PubChem Sketcher (Box A). The new search interface performs different types of structure searches, including identity, similarity, substructure and superstructure searches (Box B).
Knowledge panels
To help users quickly find important relationships among chemicals, genes, and diseases, we introduced knowledge panels. They show a list of a few selected chemicals, genes, or diseases that are most commonly mentioned together with a given chemical or gene in scientific articles. For example, the Compound summary page of CID 1983 (acetaminophen) has the following three knowledge panels:
Chemical–chemical co-occurrences: https://pubchem.ncbi.nlm.nih.gov/compound/1983#section=Chemical-Co-Occurrences-in-Literature
Chemical–gene co-occurrences: https://pubchem.ncbi.nlm.nih.gov/compound/1983#section=Chemical-Gene-Co-Occurrences-in-Literature
Chemical–disease co-occurrences: https://pubchem.ncbi.nlm.nih.gov/compound/1983#section=Chemical-Disease-Co-Occurrences-in-Literature
For each co-occurring entity listed on the knowledge panel, a sample of relevant PubMed records is displayed as evidence (with a link to the full set). The content displayed in knowledge panels is generated from statistical analysis of mentions of chemicals, genes, and diseases in PubMed records. This analysis involves annotating PubMed records using natural-language processing (NLP) software, called LeadMine (50), and matching the annotations with information in PubChem and other databases. More detailed information on this co-occurrence analysis can be found at: https://pubchemdocs.ncbi.nlm.nih.gov/knowledge-panels.
PubChem Pathway View page
The PubChem Pathway page for a given biological pathway provides information on chemicals, proteins, genes and diseases involved in or associated with that pathway, and can be very important to provide a context to observed biological activity. All pathway records were integrated from existing pathway resources (34,51–59) without any attempt to merge or combine them, meaning that a pathway may have multiple records within PubChem, with each originating from a different source. Each page for a given pathway can be accessed via an URL of the form:
https://pubchem.ncbi.nlm.nih.gov/pathway/SOURCE:PATHID
where SOURCE is the information source for the pathway and PATHID is the record identifier used by that source. For instance, the following URL directs to the pathway page for the human glycolysis pathway from PathBank (ID: SMP0000040):
https://pubchem.ncbi.nlm.nih.gov/pathway/PathBank:SMP0000040
PubChem Pathways resource supersedes the NCBI BioSystems database (60), which is no longer being updated. Many records in the BioSystems database are also available via PubChem Pathways. The Pathway pages for those records can be accessed via an URL containing NCBI BioSystems identifiers (BSIDs), as shown in the following example (for BSID 1458456):
https://pubchem.ncbi.nlm.nih.gov/pathway/BSID:1458456
Chemicals, proteins, and genes presented on PubChem Pathway pages are linked to the corresponding PubChem pages, providing quick access to more detailed information on these entities. In addition, the Pathway pages provide information on the interactions or reactions among these entities. The Pathway pages are searchable within PubChem Search.
PubChem Periodic Table and Element pages
The PubChem Periodic Table and Element pages (61) were introduced to help users navigate the abundant chemical elemental data within PubChem, while providing a convenient entry point to explore additional chemical content, such as biological activities and health and safety data available in PubChem Compound pages for specific elements and their isotopes. The Periodic Table can be accessed from the PubChem homepage, from which Element pages for individual elements can be reached. The Periodic Table and Element pages are also available as widgets, enabling one to display PubChem's element data on external web pages. Because the widgets load data directly from PubChem, it always shows the most current information in PubChem.
The data presented on the Periodic Table and Element pages are integrated from scientific articles and authoritative data sources. The elemental data can be downloaded in common file formats and imported into data analysis programs (spreadsheet software like Microsoft Excel and Google Sheets or computer scripts in python or R). In addition, these data are not only available through the web interfaces but also programmatically through PUG-REST (23,24) and PUG-View (25).
DATA MODEL CHANGE
Among the multiple programmatic access routes to PubChem data is PUG-View (25), which is a Representational State Transfer (REST)-style interface (62,63). It serves information necessary to render interactive web pages but also allows one to programmatically access the chemical annotations and summary information in PubChem. In 2019, major changes were made to the data model for JSON/XML objects returned by the PUG-View server. More detailed information on these changes can be found at https://go.usa.gov/xEHXj. While these changes do not directly affect web users, programmatic users need to update the programs that retrieve and interpret data from PUG View.
SUMMARY
The present paper describes the updates made to PubChem over the past two years. The data integration from more than 100 new sources to PubChem has greatly broadened the scope of its information content. Notable additions include the chemical-literature links from Thieme Chemistry, links to property data available from SpringerMaterials and links to PATENTSCOPE at WIPO. In addition, a substantial amount of chemical toxicological information from several ToxNet databases has been integrated into PubChem. These new additions significantly improved the FAIR-ness of chemical information. It is also noteworthy that the special data collection for COVID-19 was created to provide scientists with quick access to data critical to their efforts to stop this deadly disease.
There were major changes to PubChem's web interfaces. The PubChem homepage and pages dedicated to individual records were redesigned to provide users with an easier, mobile-friendly access to PubChem data. These changes involve a data model change for the objects used by the new web pages as well as for programmatic users. Therefore, the programs that retrieve and interpret data from PUG-View should be updated accordingly.
In addition, several new services were introduced. The new search interface allows users to search multiple data collections within PubChem simultaneously. It also accepts structure queries and supports different types of structure search (i.e. identity, similarity, substructure and superstructure searches). The knowledge panels, which display chemicals, genes, or diseases most-frequently mentioned in PubMed articles together with a given chemical or gene, were introduced to help users quickly identify important relationships between these entities. The Pathway pages provide information on chemicals, proteins, genes, and diseases involved in each pathway. The PubChem Periodic Table and Element pages assist users in exploring the abundant chemical element data.
DATA AVAILABILITY
All PubChem data, tools, and services are provided to the public free of charge.
ACKNOWLEDGEMENTS
We appreciate the hundreds of data contributors for making their data openly accessible within PubChem. Special thanks go to the entire NCBI staff (especially to the help desk and systems support teams).
Contributor Information
Sunghwan Kim, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jie Chen, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Tiejun Cheng, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Asta Gindulyte, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jia He, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Siqian He, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Qingliang Li, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Benjamin A Shoemaker, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Paul A Thiessen, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Bo Yu, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Leonid Zaslavsky, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Jian Zhang, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
Evan E Bolton, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Department of Health and Human Services, Bethesda, MD, 20894, USA.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim S. Getting the most out of PubChem for virtual screening. Expert. Opin. Drug Discov. 2016; 11:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A. et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2020; 48:D9–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh N., Chaput L., Villoutreix B.O.. Fast rescoring protocols to improve the performance of structure-based virtual screening performed on protein-protein interfaces. J. Chem. Inf. Model. 2020; 60:3910–3934. [DOI] [PubMed] [Google Scholar]
- 6. Xiao T., Qi X.X., Chen Y.Z., Jiang Y.Y.. Development of ligand-based big data deep neural network models for virtual screening of large compound libraries. Mol. Inf. 2018; 37:1800031. [DOI] [PubMed] [Google Scholar]
- 7. Pasupa K., Kudisthalert W.. Virtual screening by a new clustering-based weighted similarity extreme learning machine approach. PLoS One. 2018; 13:e0195478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J.J., Schmucker L.N., Visco D.P.. Pharmaceutical machine learning: virtual high-throughput screens identifying promising and economical small molecule inhibitors of complement factor C1s. Biomolecules. 2018; 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deshmukh A.L., Chandra S., Singh D.K., Siddiqi M.I., Banerjee D.. Identification of human flap endonuclease 1 (FEN1) inhibitors using a machine learning based consensus virtual screening. Mol. Biosyst. 2017; 13:1630–1639. [DOI] [PubMed] [Google Scholar]
- 10. Huang H., Nguyen T., Ibrahim S., Shantharam S., Yue Z.L., Chen J.Y.. DMAP: a connectivity map database to enable identification of novel drug repositioning candidates. BMC Bioinformatics. 2015; 16:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Crisan L., Avram S., Pacureanu L.. Pharmacophore-based screening and drug repurposing exemplified on glycogen synthase kinase-3 inhibitors. Mol. Divers. 2017; 21:385–405. [DOI] [PubMed] [Google Scholar]
- 12. Gad A., Manuel A.T., Jinuraj K.R., John L., Sajeev R., Priya V.G.S., Jaleel U.C.A.. Virtual screening and repositioning of inconclusive molecules of beta-lactamase Bioassays-A data mining approach. Comput. Biol. Chem. 2017; 70:65–88. [DOI] [PubMed] [Google Scholar]
- 13. Wang J.M. Fast identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020; 60:3277–3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee J.H., Basith S., Cui M., Kim B., Choi S.. In silico prediction of multiple-category classification model for cytochrome P450 inhibitors and non-inhibitors using machine-learning method. SAR QSAR Environ. Res. 2017; 28:863–874. [DOI] [PubMed] [Google Scholar]
- 15. Ciallella H.L., Zhu H.. Advancing computational toxicology in the big data era by artificial intelligence: data-driven and mechanism-driven modeling for chemical toxicity. Chem. Res. Toxicol. 2019; 32:536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu X.Z., Ho C.H., Wang X.N.. Application of life cycle assessment and machine learning for high-throughput screening of green chemical substitutes. ACS Sustain. Chem. Eng. 2020; 8:11141–11151. [Google Scholar]
- 17. Zhang W., Zou H., Luo L.Q., Liu Q.C., Wu W.J., Xiao W.Y.. Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing. 2016; 173:979–987. [Google Scholar]
- 18. Zhang W., Liu X.R., Chen Y.L., Wu W.J., Wang W., Li X.H.. Feature-derived graph regularized matrix factorization for predicting drug side effects. Neurocomputing. 2018; 287:154–162. [Google Scholar]
- 19. Ludwig M., Duhrkop K., Bocker S.. Bayesian networks for mass spectrometric metabolite identification via molecular fingerprints. Bioinformatics. 2018; 34:333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Allen F., Greiner R., Wishart D.. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics. 2015; 11:98–110. [Google Scholar]
- 21. Shen H.B., Zamboni N., Heinonen M., Rousu J.. Metabolite identification through machine learning - tackling CASMI challenge using FingerID. Metabolites. 2013; 3:484–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heinonen M., Shen H.B., Zamboni N., Rousu J.. Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics. 2012; 28:2333–2341. [DOI] [PubMed] [Google Scholar]
- 23. Kim S., Thiessen P.A., Bolton E.E., Bryant S.H.. PUG-SOAP and PUG-REST: web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015; 43:W605–W611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S., Thiessen P.A., Cheng T., Yu B., Bolton E.E.. An update on PUG-REST: RESTful interface for programmatic access to PubChem. Nucleic Acids Res. 2018; 46:W563–W570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S., Thiessen P.A., Cheng T., Zhang J., Gindulyte A., Bolton E.E.. PUG-View: programmatic access to chemical annotations integrated in PubChem. J. Cheminform. 2019; 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C. et al.. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020; 382:1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Northwell C.-R.C.. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA-J. Am. Med. Assoc. 2020; 323:2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Spinelli A., Pellino G.. COVID-19 pandemic: perspectives on an unfolding crisis. Br. J. Surg. 2020; 107:785–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Madej T., Lanczycki C.J., Zhang D.C., Thiessen P.A., Geer R.C., Marchler-Bauer A., Bryant S.H.. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014; 42:D297–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2020; 48:D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bateman A., Martin M.J., Orchard S., Magrane M., Alpi E., Bely B., Bingley M., Britto R., Bursteinas B., Busiello G. et al.. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burley S.K., Berman H.M., Bhikadiya C., Bi C.X., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S. et al.. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019; 47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Southan C., Sharman J.L., Campo B., Cavanagh D.R., Alexander S.P.H., Davenport A.P. et al.. The IUPHAR/BPS Guide to PHARMACOLOGY in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV Guide to MALARIA PHARMACOLOGY. Nucleic Acids Res. 2020; 48:D1006–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slenter D.N., Kutmon M., Hanspers K., Riutta A., Windsor J., Nunes N., Melius J., Cirillo E., Coort S.L., Digles D. et al.. WikiPathways: a multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018; 46:D661–D667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wexler P. TOXNET: An evolving web resource for toxicology and environmental health information. Toxicology. 2001; 157:3–10. [DOI] [PubMed] [Google Scholar]
- 37. Auletta A.E., Brown M., Wassom J.S., Cimino M.C.. Current status of the gene-tox program. Environ. Health Perspect. 1991; 96:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cimino M.C., Auletta A.E.. Draper WM. The gene-tox program - data evaluation of chemically-induced mutagenicity. Environmental Epidemiology: Effects of Environmental Chemicals on Human Health. 1994; 241:Washington: Amer Chemical Soc; 89–104. [Google Scholar]
- 39. Fonger G.C. Hazardous substances data bank (HSDB) as a source of environmental fate information on chemicals. Toxicology. 1995; 103:137–145. [DOI] [PubMed] [Google Scholar]
- 40. Fonger G.C., Hakkinen P., Jordan S., Publicker S.. The National Library of Medicine's (NLM) Hazardous Substances Data Bank (HSDB): background, recent enhancements and future plans. Toxicology. 2014; 325:209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tomasulo P. ChemIDplus-Super source for chemical and drug information. Med. Ref. Serv. Q. 2002; 21:53–59. [DOI] [PubMed] [Google Scholar]
- 42. Tomasulo P. LactMed-new NLM database on drugs and lactation. Med. Ref. Serv. Q. 2007; 26:51–58. [DOI] [PubMed] [Google Scholar]
- 43. Anderson P.O. LactMed update—an introduction. Breastfeed. Med. 2016; 11:54–55. [DOI] [PubMed] [Google Scholar]
- 44. Hoofnagle J.H., Serrano J., Knoben J.E., Navarro V.J.. LiverTox: a website on drug-induced liver injury. Hepatology. 2013; 57:873–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Weininger D. SMILES, a chemical language and information-system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988; 28:31–36. [Google Scholar]
- 46. Weininger D., Weininger A., Weininger J.L.. SMILES. 2. Algorithm for generation of unique smiles notation. J. Chem. Inf. Comput. Sci. 1989; 29:97–101. [Google Scholar]
- 47. Weininger D. SMILES. 3. Depict - graphical depiction of chemical structures. J. Chem. Inf. Comput. Sci. 1990; 30:237–243. [Google Scholar]
- 48. Heller S.R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D.. InChI, the IUPAC international chemical identifier. J. Cheminform. 2015; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ihlenfeldt W.D., Bolton E.E., Bryant S.H.. The PubChem chemical structure sketcher. J. Cheminform. 2009; 1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lowe D.M., Sayle R.A.. LeadMine: a grammar and dictionary driven approach to entity recognition. J. Cheminform. 2015; 7:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jassal B., Matthews L., Viteri G., Gong C.Q., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R. et al.. The reactome pathway knowledgebase. Nucleic Acids Res. 2020; 48:D498–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. O’Donnell V.B., Dennis E.A., Wakelam M.J.O., Subramaniam S.. LIPID MAPS: Serving the next generation of lipid researchers with tools, resources, data, and training. Sci. Signal. 2019; 12:eaaw2964. [DOI] [PubMed] [Google Scholar]
- 53. Schläpfer P., Zhang P.F., Wang C.A., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T. et al.. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017; 173:2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Naithani S., Gupta P., Preece J., D’Eustachio P., Elser J.L., Garg P., Dikeman D.A., Kiff J., Cook J., Olson A. et al.. Plant Reactome: a knowledgebase and resource for comparative pathway analysis. Nucleic Acids Res. 2020; 48:D1093–D1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E.. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012; 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schaefer C.F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K.H.. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009; 37:D674–D679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wishart D.S., Li C., Marcu A., Badran H., Pon A., Budinski Z., Patron J., Lipton D., Cao X., Oler E. et al.. PathBank: a comprehensive pathway database for model organisms. Nucleic Acids Res. 2020; 48:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yamamoto S., Sakai N., Nakamura H., Fukagawa H., Fukuda K., Takagi T.. INOH: ontology-based highly structured database of signal transduction pathways. Database. 2011; 2011:bar052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q. et al.. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019; 20:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Geer L.Y., Marchler-Bauer A., Geer R.C., Han L.Y., He J., He S.Q., Liu C.L., Shi W.Y., Bryant S.H.. The NCBI BioSystems database. Nucleic Acids Res. 2010; 38:D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim S., Gindulyte A., Zhang J., Thiessen P.A., Bolton E.E.. PubChem Periodic Table and Element pages: improving access to information on chemical elements from authoritative sources. Chem. Teacher Int. 2020; 2:20200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fielding R.T. Representational State Transfer (REST). Architectural Styles and the Design of Network-based Software Architectures. 2000; Irvine: University of California. [Google Scholar]
- 63. Fielding R.T., Taylor R.N.. Principled design of the modern Web architecture. Proceedings of the 22nd International Conference on Software Engineering. 2000; 407–416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All PubChem data, tools, and services are provided to the public free of charge.