Abstract
Since the outbreak of the current pandemic in 2020, there has been a rapid growth of published articles on COVID-19 and SARS-CoV-2, with about 10 000 new articles added each month. This is causing an increasingly serious information overload, making it difficult for scientists, healthcare professionals and the general public to remain up to date on the latest SARS-CoV-2 and COVID-19 research. Hence, we developed LitCovid (https://www.ncbi.nlm.nih.gov/research/coronavirus/), a curated literature hub, to track up-to-date scientific information in PubMed. LitCovid is updated daily with newly identified relevant articles organized into curated categories. To support manual curation, advanced machine-learning and deep-learning algorithms have been developed, evaluated and integrated into the curation workflow. To the best of our knowledge, LitCovid is the first-of-its-kind COVID-19-specific literature resource, with all of its collected articles and curated data freely available. Since its release, LitCovid has been widely used, with millions of accesses by users worldwide for various information needs, such as evidence synthesis, drug discovery and text and data mining, among others.
INTRODUCTION
Since the outbreak of the COVID-19 pandemic, researchers from all over the world have been rapidly gearing up to study the disease and racing toward safe and effective treatments and vaccines. As a result, there is an explosion of new scientific literature about the disease and the virus that causes it, totaling over 55 000 articles in PubMed alone (as of September 2020). This is causing an increasingly serious information overload, making it difficult for scientists, healthcare professionals, and the general public to keep pace with the latest SARS-CoV-2 and COVID-19 research.
In response, we developed LitCovid (https://www.ncbi.nlm.nih.gov/research/coronavirus/), a curated literature hub, to track up-to-date published research on COVID-19 and SARS-CoV-2 in the biomedical literature (1). LitCovid aims to help users to follow the latest SARS-CoV-2 and COVID-19 literature, which is growing rapidly with ∼10,000 new articles added each month. A recent study shows that the median time to acceptance for COVID-19 related papers were 6 days, whereas the counterparts for Ebola-related and cardiovascular disease-related papers were 15 and 102 days, respectively (2).
LitCovid is updated daily with new COVID-19-relevant articles identified from PubMed (3,4) and organized into curated categories, such as treatment, diagnosis, prevention or transmission. Initially, all data collection and literature curation was done manually with little machine assistance. As the outbreak evolved, however, we developed automated approaches to support manual curation and to maximize curation productivity in order to keep up with the rapid literature growth.
In addition to being the first of its kind, LitCovid provides several unique features that enhance literature discoverability and interpretability (5) and that distinguish it from other tools, such as BIP4COVID19 (6), covidscholar (https://covidscholar.org/), iSearch COVID-19 Portfolio (https://icite.od.nih.gov/covid19/search/), CORD-19 (7) and WHO Global literature on coronavirus disease (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov). Specifically, we focus on identifying published articles on COVID-19 in PubMed (i.e. articles on other coronaviruses, such as SARS or MERS, are beyond our scope) in a comprehensive and precise manner.
The articles are curated daily. Through curated information, we allow users to quickly navigate the body of COVID-19 research with higher-level topic, geolocation and related organizers. The curated information also bridges the gap between data and knowledge (8,9), enabling knowledge discovery in downstream applications such as evidence synthesis and drug repurposing (10,11). Moreover, it facilitates information discovery through advanced search functions (e.g. relevance ranking, phrase searches) and personalized RSS feed subscriptions. Finally, LitCovid is an open database: all its articles and associated curated data can be freely downloaded for both research discovery and for machine processing.
Since its release, LitCovid has been referenced by hundreds of institutions in academia, government, and health organizations. Users worldwide access LitCovid millions of times each month for various information needs, such as evidence synthesis, drug discovery and literature review for diagnosis and treatment guidelines. LitCovid was briefly introduced in (1). To enhance transparency and allow for more effective use, here we detail its main features, curation methods and search functions, along with the many new developments and significant improvements since its inception. We also identify its limitations and future directions and welcome user feedback for its further enhancement.
MATERIALS AND METHODS
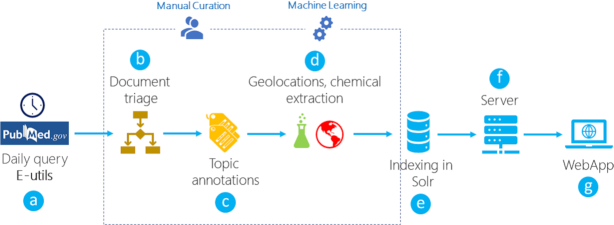
The overall curation workflow of LitCovid is illustrated in Figure 1. Each day, candidate articles are first retrieved using a query set of keywords from PubMed via NCBI’s E-utils tool (see below). The retrieved articles are then examined and classified as relevant or irrelevant. The set of COVID-19-relevant articles are further curated in depth: (i) they are assigned one or more of eight broad topics when applicable, and (ii) geolocation and drug/chemical mentions in the title and abstract are extracted. Finally, relevant articles with their curated information are indexed by Solr, a standalone open-source enterprise search platform. As noted earlier, initially, all data collection and curation was done manually with little machine assistance by two (part-time) human curators with training background in biomedical data sciences. As the outbreak evolved, automated approaches were developed to support manual curation and maximize curation productivity to keep up with the rapid literature growth. The curation details are summarized below the figure. The evaluation results of our automation tools are provided in the ‘Results’ section.
Figure 1.
An overview of the LitCovid daily workflow (A–G). The PubMed publications are retrieved using a query via NCBI’s E-utils. The publications are classified as to whether they are relevant to COVID-19. The relevant publications are curated further by topic categorization as well as geolocation and chemical extraction. The curated publications are then imported into our Solr database.
Document triage
The first step of our curation pipeline is to triage publications and to identify those relevant to COVID-19. Given the high variability of the English language and the ambiguity of terms to describe COVID-19 and SARS-Cov-2 in the literature, we decided to apply a two-step approach to maximize both the coverage and precision of retrieved publications.
Specifically, we first use a broad query ‘coronavirus’[All Fields] OR ‘ncov’[All Fields] OR ‘cov’[All Fields] OR ‘2019-nCoV’[All Fields] OR ‘COVID-19’[All Fields] OR ‘SARS-CoV-2’[All Fields] to retrieve candidate articles. The aim of this step is to include all possible relevant articles. Then, for each article retrieved, we review its relevance to COVID-19 and remove those that are irrelevant. To support human review, we developed an ensemble of automated document classifiers that include both traditional machine-learning models, such as support vector machines, using bag-of-words features and recent deep-learning models, such as convolutional neural networks, using word embeddings (12–14). Given a target article, each model predicts a score that indicates its likelihood of relevance to COVID-19. The average of all model prediction scores is then used as a reference during human review. The entire curation process is streamlined via a newly developed online system called LitSuggest (https://www.ncbi.nlm.nih.gov/research/litsuggest/), which provides a user-friendly interface for manual literature review and curation.
Document annotation
Relevant articles selected from the previous triage step are further annotated for applicable topics, geolocations and chemicals/drugs. For topic annotation, each article is considered for one or more of eight broad topics (general information, mechanism, transmission, diagnosis, treatment, prevention, case report, or epidemic forecasting) when applicable. To support manual curation on topic classification, we developed a deep learning model that integrates the embeddings produced by BioBERT (a contextualized language model trained in PubMed) (15) with manually crafted features (e.g. publication types such as journal article or case report) to predict the topic probability score. In total, eight topic classification models are developed (one per topic). The predicted topics are then manually reviewed by examining the article content (abstract and/or full text), in which irrelevant topic labels are removed and missing ones are added.
For entity extraction, the geolocations mentioned in the article title/abstract are first extracted using an advanced named entity recognition tool provided in spaCy (https://spacy.io/), a Python library. Extracted entities of the type of country, city, state and nationality are then mapped into countries, using dictionaries. Extracted countries are then manually reviewed to improve accuracy. Chemical and drug mentions are retrieved from PubTator API (16,17), which provides state-of-the-art concept annotations of the biomedical literature.
Article indexing
Annotated articles are subsequently indexed by Solr (https://lucene.apache.org/solr/) in following steps:
The raw text is lowercased, transformed into ASCII, tokenized and stemmed.
Stopwords and English possessives are removed.
Synonymous tokens are mapped to each other when applicable using those extracted from Medical Subject Headings (MeSH).
System implementation details
The LitCovid backend is implemented as a pure JSON API, based on the Django and Django REST framework, and communicates with a front-end app built with Angular. To optimize its performance and reliability, LitCovid uses multiple Solr instances, front-end and back-end server instances.
User interface and system features
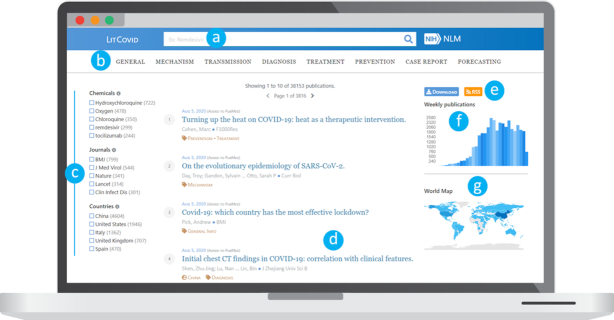
LitCovid user interface is illustrated in Figure 2. It provides central access for users to navigate through topics, perform searches, subscribe to daily alerts on a topic of interest and download relevant data. The primary functions are summarized below.
Figure 2.
LitCovid user interface: (A) free-text search box; (B) topic navigation; (C) filter facets by chemical, journal title, or country; (D) document summary with article meta-information; (E) download and RSS options; (F) growth of articles per week; and (G) a visualization on geolocations mentioned in titles and abstracts.
Key search functions
LitCovid provides free-text search (Figure 2A). By default, it performs OR searches; i.e. a search for SARS-CoV-2 RNA will return all papers that contain either SARS-CoV-2, RNA, or both. In addition, LitCovid supports AND searches (the query SARS-CoV-2 AND RNA will return articles that contain both words), NOT searches (the query SARS-CoV-2 NOT RNA will return articles that contain SARS-CoV-2 but not RNA) and phrase searches that use double quotes (the query ‘SARS-CoV-2 RNA’ will return only articles that contain this exact phrase). Note that AND, OR and NOT operators are case sensitive.
In addition, users can perform field-based searches using field names with the format field:query. Available searchable fields include title (for titles), abstract (for abstracts), journal (for journals), countries (for countries), authors (for authors), topics (for topics) and e_drugs (for chemicals). Parentheses are used for more complex searches such as combining both free-text and field-based searches. Table 1 illustrates representative query examples.
Table 1.
Representative query examples and their corresponding explanations
| Query example | Query interpretation |
|---|---|
| fever AND rash | It retrieves the papers that contain both terms ‘fever’ and ‘rash’. |
| treatment NOT cancer | It retrieves the papers that contain the term ‘treatment’ and do not contain the term ‘cancer’. |
| “corticosteroid therapy" | It retrieves the papers that contain the phrase ‘corticosteroid therapy’ |
| 32744591 | It retrieves the paper with the PMID:32744591 |
| topics:Treatment | It retrieves the papers annotated with the topic treatment |
| topics:(Treatment AND Diagnosis NOT Prevention) | It retrieves the papers annotated with the topics treatment and diagnosis, but without the topic prevention |
| journal:Nature AND abstract:“vaccine" | It retrieves the papers from the Journal Nature and contain the term ‘vaccine’ in the abstract. |
Furthermore, search results can be sorted by either recency or relevance (based on the widely used BM25 ranking function (18). The matched terms are highlighted in passages for user convenience using Solr's built-in unified highlighter.
Search filters
After performing a search, users can view top journals, geolocations, and chemicals associated with the matching articles as well as weekly trends of these publications. They also can further filter their search by journal, country, chemical, or week. Two of the filters, a weekly histogram and a world map of all countries mentioned in the abstracts, are also available on the home page for convenient access.
Abstract page
Users can click on a publication title to display its abstract on a dedicated page along with a world map of related countries and a list of similar publications (article title, abstract, author-provided keywords and curated topics are used to compute similar articles). On the abstract page, each article in LitCovid can be readily shared to Facebook or Twitter with a single click.
RSS feeds
LitCovid offers users an opportunity to subscribe to personalized RSS feeds (for all publications, topic-related publications, or query-related publications) with their favorite RSS clients.
Customized download options
The entire set of articles and curated data in LitCovid are publicly available. There are three download options for different downstream use cases. Publications can be downloaded in (i) TSV format to be processed by automated software, (ii) RIS format to be imported into bibliography reference software and (iii) JSON/XML format for data and text mining.
RESULTS
Rapid growth of COVID-19 literature
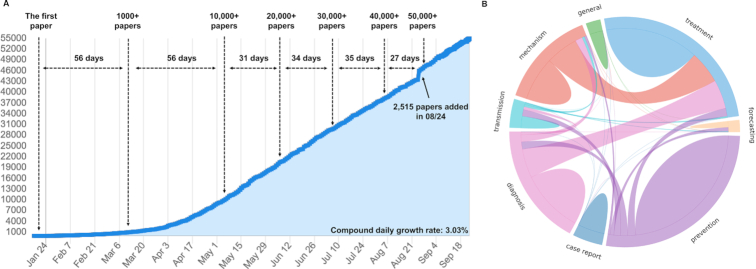
Figure 3A demonstrates the cumulative growth of LitCovid (retrieved on 21 September 2020). Although it took almost two months (from January to mid-March) for the first 1000 articles to appear, the database had a 10-fold increase during the next 2 months and has continued to increase rapidly since then (at a rate of ∼10 000 per month), with over 50 000 articles to date. On 24 August, we noted the largest single day increase of 2515 new articles from PubMed.
Figure 3.
(A) cumulative growth of LitCovid, accessed 21 September 2020. The compound daily growth rate was calculated based on the number of articles added daily; (B) topic distributions and co-occurrences among the topics.
LitCovid topic distribution
Figure 3B shows the topic distributions and co-occurrences. The majority of the articles are associated with the topics prevention and treatment, followed by diagnosis and mechanism. In addition, ∼20% of the articles are assigned more than one topic, and we observed three representative topic co-occurrence patterns: (i) diagnosis-treatment, e.g. papers that describe both symptoms and treatment outcomes (PMID:32449128); (ii) mechanism-treatment, e.g. papers that describe both underlying biological pathways and potential drug efficacy (PMID: 32503821); and (iii) transmission-prevention, e.g. papers that describe both potential transmission routes and the necessary prevention procedures (PMID: 32498142).
Evaluation performance of automated methods in curation assistance
All the automated methods described above were evaluated before their first use and have been improved continuously. Current performance is summarized in Table 2, based on the papers in LitCovid as of 12 August 2020. Human annotations are used as the ground truth for model training and evaluation. For document classification, papers judged as relevant for LitCovid are considered positive instances, while papers rejected during manual review and a random sample of covid19-unrelated papers are considered negative instances. The entire dataset is randomly split for training (80%) and testing (20%). For topic assignment, we used the subset of articles in LitCovid with assigned topics and randomly split them for both training (80%) and testing (20%). For evaluating geolocation tagging, a test set was manually annotated, with no training data, as the model uses an unsupervised method. The evaluation datasets are available via ftp://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/litcovid_evaluation_datasets.zip.
Table 2.
Training and testing dataset split and evaluation results of automation tools
| Task | Document classification | Topic assignment | Geolocation tagging |
|---|---|---|---|
| Training/test | 63 998/16 000 articles | 32 105/8026 articles | N.A./ 361 articles |
| Evaluation (P/R/F) | 0.99/0.99/0.99 | 0.80/0.82/0.81 | 0.96/0.93/0.94 |
Note: P: Precision, R: Recall, and F: micro F1-score. The geolocation extraction model was directly applied, so no training instances were required.
As shown in Table 2, both automatic document classification and geolocation tagging methods achieved exceptionally high performance. While achieving an average micro F1-score of 0.81, the overall performance was relatively lower for predicting relevant topics, particularly in recall. This is mainly due to the complexity of the task, requiring the assignment of up to eight topics, and the lack of abstracts and full-texts available for text mining in many articles. For ∼40% of articles in LitCovid, the only available information is the title (i.e. no abstract or full text), thus increasing the difficulty for automatic text processing. When abstract information is available, the performance of our automatic topic assignment is indeed significantly higher, with a micro F1-score of 0.89.
Comparison of LitCovid versus keyword query
To demonstrate that our method is superior to conventional keyword searches in collecting relevant articles with higher precision and better coverage, we performed a direct comparison of the articles collected in LitCovid with the ones directly retrieved from the query used in PubMed [data accessed at the beginning of March, 2020]: ((wuhan[all fields] and (‘coronavirus’[mesh terms] or ‘coronavirus’[all fields])) and 2019/12[pdat]: 2030[pdat]) or 2019-ncov[all fields] or 2019ncov[all fields] or covid-19[all fields] or sars-cov-2[all fields]. At that time, 593 papers were collected in LitCovid in total, whereas the PubMed query retrieved 457 papers; thus, our method yielded approximately 30% more papers on COVID-19.
Figure 4 provides a detailed comparison of the overlap and differences in the articles retrieved by the two methods. Of the articles retrieved via the PubMed query, 97% (446 out of 457) were found in LitCovid. Further, we manually checked the 11 missing articles in LitCovid and found that none were related to COVID-19. For example, PMID: 31670218 (https://pubmed.ncbi.nlm.nih.gov/31670218/) is about the Porcine deltacoronavirus (PDCoV) and was published in August 2019. Similarly, PMID: 31896597 (https://pubmed.ncbi.nlm.nih.gov/31896597/) is about Baculovirus and was published in September 2019. The main topics of these papers are not related to COVID-19 and, thus, not collected in LitCovid. In contrast, 147 distinct papers were found only in LitCovid and are truly COVID-19 relevant (e.g. PMID: 32060619 (https://pubmed.ncbi.nlm.nih.gov/32060619/), PMID: 32061312 (https://pubmed.ncbi.nlm.nih.gov/32061312/) and PMID: 32061333 (https://pubmed.ncbi.nlm.nih.gov/32061333/)). These articles were missed by the PubMed query due to its keyword limitations. Overall, the comparison shows that our two-step approach achieves both higher coverage and greater precision than the conventional keyword-based search.
Figure 4.
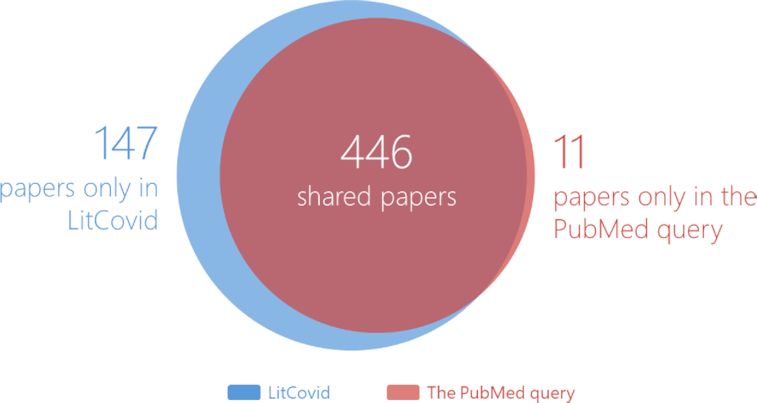
Comparative analysis of the database coverage between LitCovid and PubMed queries.
Usage of different features
Since the release of LitCovid, the database has been accessed over ten million times by users from ∼200 countries. Figure 5 presents the usage breakdown of different LitCovid functions and search features in July 2020. Notably, 58% of uses are related to clicking and reading the abstract page, suggesting that users find the papers of interest and read in more detail. The high access of the abstract page is primarily contributed by article navigation (for example, users browse through the topic treatment and click the abstract page of interesting articles) which accounts for 18% of total uses and keyword search (for example, users search a query and click the abstract page of relevant articles) which accounts for 15%. The RSS feature also accounts for 7% of total usage. Taken together, we can see that the features employed in LitCovid are widely used among its users.
Figure 5.
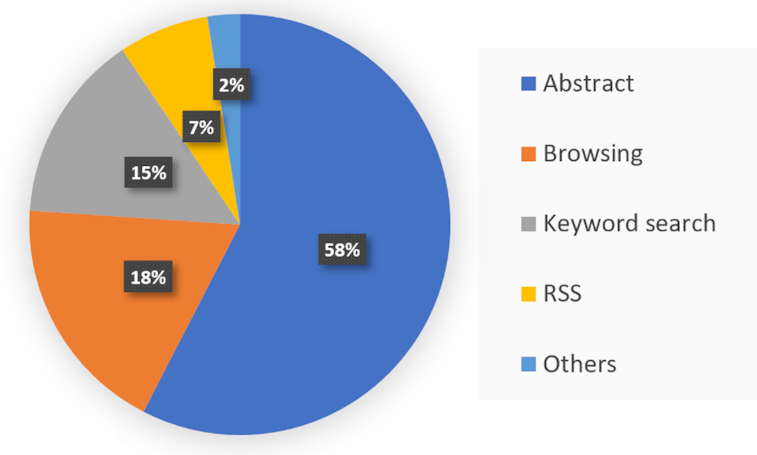
Proportion of users accessing different features of LitCovid, based on the data in July 2020.
DISCUSSION
In this work, we summarized the curation workflow, system design, and primary functions of LitCovid in detail. The significant changes in the curation workflow and the development of recent features are driven directly by the rapid growth of COVID-19 literature and the increasing needs of LitCovid users. Here we discuss some of these opportunities and challenges.
As stated earlier, LitCovid has been widely used to fill various information needs, such as evidence synthesis (11), drug repurposing (10) and guidelines for diagnosis (19). With the rapid growth of COVID-19 literature, these three primary use cases of LitCovid have been increasingly popular. First and foremost, LitCovid is used by researchers, healthcare professionals, and the general public to keep up with the latest COVID-19 research in the literature. The recently introduced RSS feed is a popular feature with over a half million uses per month—along with several other features that keep users afloat in a sea of literature.
LitCovid provides timely and central access for systematic literature reviews, which are greatly needed for knowledge synthesis in evidence-based medicine. LitCovid has been used for several such studies. For instance, it has been used as a primary resource in the LitCOVID Systematic Review Daily Report (http://covidlit.spectrumhealth.org/), where it provides rapid systematic reviews on representative topics via community efforts.
LitCovid enables large-scale data-driven discovery. All of the articles and associated curation data in LitCovid are freely available, and were downloaded and used in other computational tools and systems (6,20–21). In addition, we have recently performed a computer annotation of key semantic concepts of COVID-19 literature (22), using PubTator (16), a state-of-the-art natural language processing and concept annotation system. The LitCovid data and their associated annotations play critical roles in the analysis of the COVID-19 research landscape, and the building of a COVID-19 knowledge graph.
The previous curation workflow of LitCovid was mostly manual. However, it is not sufficient and scalable to the ever-increasing COVID-19 literature. Through large-scale evaluation, we have demonstrated that automated methods can achieve exceptionally high performance for classifying publications and tagging geolocations in the title and abstract. The performance of the automatic topic assignment is also expected to be continuously improved with additional training data and the use of passage-level and entity-level information in addition to the current deep learning model (14,23). According to (2), many published COVID-19 articles without abstract information in PubMed are not descriptions of formal research studies but rather commentary, perspective, or news articles. Thus, in order to make the best use of human curation efforts (24), we have prioritized annotating topics for those articles with abstract available in PubMed since late August, when the number of daily new articles reached a record high of over 2500. For such articles, the automatic text-mining results are also more accurate, thus further maximizing curation efficiency. Overall, the current curation workflow uses a combination of manual and automatic curation, which scales up manual curation with automated methods while ensuring the usability of our database.
There are several additional research directions for LitCovid development. For example, relevant preprints might be considered in addition to PubMed articles, as shown in similar tools such as the WHO COVID-19 Global literature on coronavirus disease. Further, when more full-length articles become available for text and data mining, finer-grained search topics and full-text search functions are worth considering. For instance, several recent AI studies have shown early results for more advanced question answering and topic analysis of the COVID-19 literature (22,25). Finally, a particular challenge is to validate the findings of COVID-19 papers especially given the rapid paper acceptance time. A potential solution might be to adapt existing evidence attribution tools to measure the quality and validity of these papers (26,27). We also look forward to user feedback to guide further enhancements.
DATA AVAILABILITY
LitCovid is free and open to all users and there is no login requirement. LitCovid can be accessed via https://www.ncbi.nlm.nih.gov/research/coronavirus/.
ACKNOWLEDGEMENTS
The authors would like to thank Chih-Hsuan Wei, Robert Leaman, W. John Wilbur and David Landsman for their help on this project.
Contributor Information
Qingyu Chen, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20892, USA.
Alexis Allot, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20892, USA.
Zhiyong Lu, National Center for Biotechnology Information (NCBI), National Library of Medicine (NLM), National Institutes of Health (NIH), Bethesda, MD 20892, USA.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Chen Q., Allot A., Lu Z.. Keep up with the latest coronavirus research. Nature. 2020; 579:193. [DOI] [PubMed] [Google Scholar]
- 2. Palayew A., Norgaard O., Safreed-Harmon K., Andersen T.H., Rasmussen L.N., Lazarus J.V.. Pandemic publishing poses a new COVID-19 challenge. Nat. Hum. Behav. 2020; 4:666–669. [DOI] [PubMed] [Google Scholar]
- 3. Fiorini N., Canese K., Starchenko G., Kireev E., Kim W., Miller V., Osipov M., Kholodov M., Ismagilov R., Mohan S. et al.. Best match: new relevance search for PubMed. PLoS Biol. 2018; 16:e2005343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fiorini N., Leaman R., Lipman D.J., Lu Z.. How user intelligence is improving PubMed. Nat. Biotechnol. 2018; 36:937–945. [DOI] [PubMed] [Google Scholar]
- 5. Leaman R., Wei C.H., Allot A., Lu Z.. Ten tips for a text-mining-ready article: how to improve automated discoverability and interpretability. PLoS Biol. 2020; 18:e3000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vergoulis T., Kanellos I., Chatzopoulos S., Karidi D.P., Dalamagas T.. BIP4COVID19: Releasing impact measures for articles relevant to COVID-19. 2020; bioRxiv doi:12 April 2020, preprint: not peer reviewed 10.1101/2020.04.11.037093. [DOI]
- 7. Wang L.L., Lo K., Chandrasekhar Y., Reas R., Yang J., Eide D., Funk K., Kinney R., Liu Z., Merrill W.. CORD-19: the Covid-19 open research dataset. ACL NLP-COVID Workshop. 2020; 2020: [Google Scholar]
- 8. International Society for Biocuration Biocuration: distilling data into knowledge. PLoS Biol. 2018; 16:e2002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Q., Britto R., Erill I., Jeffery C.J., Liberzon A., Magrane M., Onami J.I., Robinson-Rechavi M., Sponarova J., Zobel J. et al.. Quality matters: biocuration experts on the impact of duplication and other data quality issues in biological databases. Genomics Proteomics Bioinform. 2020; doi:10.1101/788034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chakraborti S., Srinivasan N.. Drug repurposing approach targeted against main protease of SARS-CoV-2 exploiting ‘neighbourhood behaviour’in 3D protein structural space and 2D chemical space of small molecules. 2020; chemRxiv doi:01 April 2020, preprint: not peer reviewed 10.26434/chemrxiv.12057846.v1. [DOI]
- 11. Galmés S., Serra F., Palou A.. Current state of evidence: influence of nutritional and nutrigenetic factors on immunity in the COVID-19 pandemic framework. Nutrients. 2020; 12:2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee K., Famiglietti M.L., McMahon A., Wei C.H., MacArthur J.A.L., Poux S., Breuza L., Bridge A., Cunningham F., Xenarios I. et al.. Scaling up data curation using deep learning: an application to literature triage in genomic variation resources. PLoS Comput. Biol. 2018; 14:e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Chen Q., Yang Z., Lin H., Lu Z.. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci. Data. 2019; 6:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen Q., Peng Y., Lu Z.. BioSentVec: creating sentence embeddings for biomedical texts. 2019 IEEE International Conference on Healthcare Informatics (ICHI). 2019; 1–5. [Google Scholar]
- 15. Lee J., Yoon W., Kim S., Kim D., Kim S., So C.H., Kang J.. BioBERT: pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2019; 36:1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei C.H., Allot A., Leaman R., Lu Z.. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 2019; 47:W587–W593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei C.H., Kao H.Y., Lu Z.. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013; 41:W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pérez-Iglesias J., Pérez-Agüera J.R., Fresno V., Feinstein Y.Z.. Integrating the probabilistic models BM25/BM25F into Lucene. 2009; arXiv doi:01 December 2009, preprint: not peer reviewedhttps://arxiv.org/abs/0911.5046.
- 19. Hanson K.E., Caliendo A.M., Arias C.A., Englund J.A., Lee M.J., Loeb M., Patel R., El Alayli A., Kalot M.A., Falck-Ytter Y.. Infectious diseases society of america guidelines on the diagnosis of COVID-19. Clin. Infect. Dis. 2020; doi:10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thorlund K., Dron L., Park J., Hsu G., Forrest J.I., Mills E.J.. A real-time dashboard of clinical trials for COVID-19. Lancet Digit Health. 2020; 2:e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Janiaud P., Axfors C., van't Hooft J., Saccilotto R., Agarwal A., Appenzeller-Herzog C., Contopoulos-Ioannidis D.G., Danchev V., Dirnagl U. et al.. The worldwide clinical trial research response to the COVID-19 pandemic-the first 100 days. F1000Research. 2020; 1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yeganova L., Islamaj R., Chen Q., Leaman R., Allot A., Wei C.-H., Comeau D.C., Kim W., Peng Y., Wilbur W.J. et al.. Navigating the landscape of COVID-19 research through literature analysis: a bird's eye view. 2020; arXiv doi:11 September 2020, preprint: not peer reviewedhttps://arxiv.org/abs/2008.03397.
- 23. Chen Q., Lee K., Yan S., Kim S., Wei C.H., Lu Z.. BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale. PLoS Comput. Biol. 2020; 16:e1007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poux S., Arighi C.N., Magrane M., Bateman A., Wei C.-H., Lu Z., Boutet E., Bye-A-Jee H., Famiglietti M.L., Roechert B.. On expert curation and sustainability: UniProtKB/Swiss-Prot as a case study. Bioinformatics. 2017; 33:3454–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su D., Xu Y., Yu T., Siddique F.B., Barezi E.J., Fung P.. CAiRE-COVID: a question answering and multi-document summarization system for COVID-19 research. 2020; arXiv doi:16 October 2020, preprint: not peer reviewedhttps://arxiv.org/abs/2005.03975.
- 26. Allot A., Chen Q., Kim S., Vera Alvarez R., Comeau D.C., Wilbur W.J., Lu Z.. LitSense: making sense of biomedical literature at sentence level. Nucleic Acids Res. 2019; 47:W594–W599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X., Guan Y., Liu W., Chauhan A., Jiang E., Li Q., Liem D., Sigdel D., Caufield J., Ping P.. Evidenceminer: Textual evidence discovery for life sciences. Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics: System Demonstrations. 2020; 56–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
LitCovid is free and open to all users and there is no login requirement. LitCovid can be accessed via https://www.ncbi.nlm.nih.gov/research/coronavirus/.