Abstract
Viruses are integral components of all ecosystems and microbiomes on Earth. Through pervasive infections of their cellular hosts, viruses can reshape microbial community structure and drive global nutrient cycling. Over the past decade, viral sequences identified from genomes and metagenomes have provided an unprecedented view of viral genome diversity in nature. Since 2016, the IMG/VR database has provided access to the largest collection of viral sequences obtained from (meta)genomes. Here, we present the third version of IMG/VR, composed of 18 373 cultivated and 2 314 329 uncultivated viral genomes (UViGs), nearly tripling the total number of sequences compared to the previous version. These clustered into 935 362 viral Operational Taxonomic Units (vOTUs), including 188 930 with two or more members. UViGs in IMG/VR are now reported as single viral contigs, integrated proviruses or genome bins, and are annotated with a new standardized pipeline including genome quality estimation using CheckV, taxonomic classification reflecting the latest ICTV update, and expanded host taxonomy prediction. The new IMG/VR interface enables users to efficiently browse, search, and select UViGs based on genome features and/or sequence similarity. IMG/VR v3 is available at https://img.jgi.doe.gov/vr, and the underlying data are available to download at https://genome.jgi.doe.gov/portal/IMG_VR.
INTRODUCTION
Viruses occupy all of Earth's biomes, and are known to infect all organisms across the tree of life, including animals, plants, protists, fungi, bacteria and archaea (1–4). While the full extent of viral diversity and the influence they exert on their environment is poorly understood, viruses are broadly recognized as critical agents of health and disease, as well as key regulators of microbiomes. Viruses have now been described, and their impacts evaluated, in a broad range of ecosystems from the world's oceans to acidic hot springs, human gut, and thawing permafrost (2,5–7). Collectively, these studies have highlighted a substantial influence of viral lysis in reshaping microbial communities and nutrient cycling, a large potential for viruses to act as lateral gene transfer agents influencing long-term evolution of cellular organisms, and fundamental alterations of cellular pathways during viral infections (8–10).
Viruses lack a conserved single-copy universal marker gene such as the 16S ribosomal RNA gene which is frequently used to detect, identify and classify bacteria and archaea, or the mitochondrial cytochrome c oxidase gene often used to survey eukaryotes. Consequently, uncultivated viruses cannot be readily and comprehensively identified through amplicon sequencing approaches (11). Instead, uncultivated viral diversity is primarily explored through metagenomics, i.e. shotgun sequencing of DNA or RNA extracted directly from a sample (12,13). In particular, recent advances in sequencing technologies and bioinformatics analyses now enable the recovery of large fragments and even complete viral genomes from metagenomes (14,15). Viral sequences can be assembled from metagenomes specifically targeting the viral fraction of environmental samples (i.e. ‘viromes’), but also from untargeted samples, even if the latter are often dominated by the cellular components of the community (14). Complementarily, viral genomes residing in the host cell, either integrated in the host chromosome or extrachromosomal, can also be successfully recovered from whole genome shotgun sequencing data (16–19).
The critical importance of viral sequences obtained without cultivation and isolation of the virus, i.e. Uncultivated Virus Genomes or UViGs, has now become undeniable (14,20). UViGs are routinely used for (i) taxonomic classification, and exploration of the viral sequence space, (ii) estimation of viral taxa distribution across samples and biomes, (iii) evaluation of virus:host networks via in silico host prediction and (iv) study of virus functional potential via analysis of gene content. In turn, comprehensive databases and analysis frameworks of UViGs, such as IMG/VR, are foundational resources enabling these studies.
The first installment of IMG/VR, a database gathering viral sequences identified from genomes and metagenomes hosted in the IMG/M database (21), was released in 2016 and included 264 413 sequences (22). A second version followed three years later, which included 735 112 sequences, along with improved taxonomic classification, an automatic identification of high-quality genomes, and new search capabilities to query the database based on sequence similarity (23). Here we present the third version of the database, which is currently the largest collection of viral genomes publicly available, totaling 2 332 702 genomes. This genome collection is now compiled from multiple sources including published studies which described UViGs from specific virus groups (24–26) or types (17) that were under-represented in databases thus far. IMG/VR UViGs are functionally annotated using the IMG pipeline (21), and their quality assessed using the new CheckV tool (27). Viral taxonomic classification of IMG/VR UViGs now reflects the updated ICTV framework (28), and in silico host predictions are provided based on sequence similarity to isolate and metagenome-assembled genomes (29) as well as matches to predicted CRISPR spacers (30). Finally, a redesigned interface allows for fast and intuitive browsing and search of the IMG/VR database using sequence or annotation queries.
MATERIALS AND METHODS
Sequence origin and selection criteria
Sequences in IMG/VR v3 (release #5) were gathered from 6 primary sources (Supplementary Table S1):
2 028 310 sequences were obtained by mining public IMG/M (21) metagenomes (up to 15 December 2019) using the Earth's Virome protocol (31), which includes a minimum contig size of 5 kb (hereafter ‘EVP’).
30 759 sequences were previously obtained by mining public IMG/M metagenomes (downloaded on 1 April 2019) for circular sequences (≥1 kb) based on direct terminal repeats (DTR), and viral prediction with VirFinder v1.1 (32) and custom marker genes (27) (hereafter ‘CheckV DTR’).
9445 sequences originated from a search of public genomes and metagenomes for inovirus sequences (≥500 bp), both extrachromosomal genomes and integrated proviruses, based on specific inovirus marker genes and gene content features analyzed using a custom random forest classifier (24) (hereafter ‘Inovirus’). This search was conducted in 2018, and included 141 sequences not available in IMG.
12 498 sequences originated from a search of public NCBI RefSeq/WGS genomes for integrated and extrachromosomal proviruses (≥800 bp) using VirSorter v1.0 (33), conducted in 2014 (17) (hereafter ‘Prophage’). This study targeted the active prophages and lytic viruses, so all predictions lacking a viral hallmark gene or a viral gene enrichment, and all prophage detections displaying viral gene enrichment only and lacking viral hallmark genes, were discarded. Among these, 5474 were not obtained from IMG/M.
1475 sequences (≥5 kb) were obtained from a custom search of public IMG/M metagenomes for virophages, conducted in 2018, and based on virophage marker genes (26) (hereafter ‘Virophage’).
47 356 sequences originated from a custom search of public IMG/M metagenomes for nucleocytoplasmic large DNA viruses (NCLDVs), i.e. giant viruses, conducted in 2018 (25) (hereafter ‘Giant Virus’). Because NCLDV genomes are much larger than other viruses, the identification and recovery of these sequences used a different approach including a genome binning step to identify contig(s) belonging to the same genome. Overall, these 47 356 sequences corresponded to a total of 2059 NCLDV genomes.
In addition, a set of 293 759 reference viral sequences, not currently available in IMG, were included in the IMG/VR database to enhance taxonomic classification and host prediction at the viral Operational Taxonomic Units (vOTU) level (see below). These included 12 182 and 6880 sequences from NCBI Viral RefSeq and GenBank respectively (34,35), 20 234 reference sequences from the CheckV database (27), and 254 463 medium-quality, high-quality and complete genomes from the GOV2, GVD and MGV (https://github.com/snayfach/MGV_catalog) datasets (36,37). GOV2 includes UViGs obtained from ocean viromes, while GVD and MGV sequences were derived from human gut viromes (GVD) and metagenomes (MGV).
The EVP, Inovirus and Prophages datasets were processed through CheckV (v0.4.0, May 2020), to identify and remove any host-derived regions from the viral contigs. For EVP and prophages, CheckV-cleaned viral sequences shorter than 5kb and 1kb (respectively) were discarded. This clean-up step was not used for the Giant Viruses or Virophages sequences because these were already curated as part of their original analysis. Sequences detected across multiple datasets (e.g. proviruses included in both Inovirus and Prophages datasets) were identified and only a single copy was retained in the final IMG/VR database.
Once cleaned, compiled, and reconciled, the final IMG/VR database (hereafter ‘IMG/VR-db’) consisted of 2 332 702 distinct UViGs, including 2 033 220 sequences available through the IMG/VR web interface (as of 25 August 2020, hereafter ‘IMG/VR-online’, Supplementary Table S1). The entire IMG/VR database (i.e. IMG/VR-db) is available for download at https://genome.jgi.doe.gov/portal/IMG_VR (release #5).
vOTU clustering
The entire dataset (2 332 702 sequences) was clustered into vOTUs following the MIUViG guidelines (14) (95% ANI - Average Nucleotide Identity and 85% AF – Aligned Fraction). Briefly, an all-vs-all blastn (v2.5.0+) was computed with the following options: -task megablast, -max_target_seqs 25000, and -perc_identity 90 (38). Then, custom python scripts were used to calculate ANI and AF between all pairs of sequences based on the cumulated blast hits, and generate viral OTUs (‘vOTUs’) using a greedy clustering approach with sequences sorted by decreasing length. This led to the identification of 933,352 vOTUs.
A separate approach was required to cluster the giant virus genome bins because they are composed of multiple contigs which may be split across different clusters in an ANI-based contig clustering. Instead, dRep v2.6.1 (39) was used to group the 2059 giant virus bins (module dereplicate, option –ignoreGenomeQuality) into 2010 vOTUs. Then, these vOTUs defined based on genome bins were reconciled with vOTUs previously defined based on individual contigs ANI as follows: for all contigs-based vOTUs containing one or more sequences from one of the the giant virus bin, the taxonomic classification of the vOTU members was observed. If a majority of sequences were taxonomically classified as NCLDV (i.e. Nucleocytoviricota), all sequences within this contig-based vOTU were included in the genome bin-based vOTU. Otherwise, only the sequence(s) from the giant virus genome bin was moved to the genome bin-based vOTU, while the other formed a separate contig-based vOTU.
Finally, some of the datasets mined to build the IMG/VR database include replicates, either biological or technical (e.g. multiple assemblies of the same original metagenome), which could lead to duplicated sequences. To avoid artificially inflating the size of vOTUs, pairs of sequences with ≥99.8% ANI and ≥99.9% AF, or ≥99.9% ANI and assembled from the same original sample, were considered as duplicates, and only counted once.
Genome quality assessment
CheckV v0.4.0 (27) was used to assess genome quality through estimation of genome completeness, except for Giant Virus, Inovirus and Virophage sequences (see below). For each UViG, CheckV AAI-based estimation of completeness was used if this estimation was qualified as medium or high confidence, or the HMM-based estimate was used otherwise if available. For the Giant Virus dataset, we instead obtained completeness estimates from the original publication, where it was calculated based on the detection of single-copy marker genes (25). For the Inovirus and Virophage datasets, an alternative completeness estimation was calculated based on a maximum length known for these types of viruses, i.e. 30 kb for inoviruses and 35 kb for virophages. This alternative estimation was used instead of the CheckV value when (i) there was no CheckV estimated completeness, or (ii) CheckV estimated the completeness to be <50%, as we found it to be more reliable for partial genomes.
The genomes were furthered quality-checked by searching for three types of artifacts. First, all contigs with ≥50 ambiguous bases (i.e. ‘N’) were flagged as ‘low quality’ (n = 21 668 sequences). These could derive from suboptimal assemblies or scaffolding of multiple contigs with gaps of unknown length. The completeness of these assemblies typically cannot be accurately estimated, and was set to ‘unknown’. Similarly, all contigs with terminal repeats representing ≥20% of the sequence length or that were identified as exact palindromes were flagged as ‘concatemers’, which represent low-quality assemblies (n = 1959 sequences). Finally, several sequences were further identified as originating from cellular rather than viral genomes. This is expected, as many viral sequence detection approaches can misidentify some eukaryotic genome sequences as viral (40). These sequences were automatically detected from the EVP based on the CheckV contamination information as contigs with ≥5 host markers and more than twice the number of host markers than viral markers (n = 6102), as well as contigs encoding 16S and/or 23S rRNA genes, often linked to neighboring decayed prophage regions (n = 232). Further, for all vOTUs whose seed sequence was identified as a putative contaminant based on this non-viral gene content signal, all the vOTU members were also removed from the IMG/VR database (n = 2930 vOTUs).
Genome quality was assigned following the MIUViG standards (14). Genomes with direct terminal repeats, i.e. ‘circular’ genomes predicted as complete, and genomes estimated to be ≥90% and ≤120% complete based on CheckV, and not flagged as low-quality or concatemers, were considered as high-quality genomes. Genomes <90% complete were considered as ‘Genome fragment’, while the quality of genomes estimated to be >120% complete was set as ‘Unsure (completeness > 120%)’.
Taxonomic classification
Two complementary approaches were used for taxonomic classification of IMG/VR sequences. First, predicted proteins from IMG/VR sequences were compared to NCBI Viral RefSeq proteins v200 (34) using diamond v0.9.25 with options ‘blastp –evalue 1e–5 –query-cover 50 –subject-cover 50 -k 10000’. For IMG/VR sequences with ≥30% of proteins having a significant hit to Viral RefSeq, a consensus affiliation was obtained based on the best hits of individual proteins (≥50% majority rule).
Second, a taxonomic classification was determined based on the detection of 588 marker genes identified in the VOG database v97 (http://vogdb.org, Supplementary Table S2). Predicted proteins from IMG/VR sequences were compared to the 588 selected VOG HMM profiles using hmmsearch v3.2.1 (41) with option ‘-E 1.0e–02’, and a minimum score of 40 and maximum E-value of 1e–05 for individual hits. If multiple conflicting markers were detected, a consensus taxonomy was obtained based on the individual markers detected (simple plurality rule).
For both RefSeq- and VOG-based classification, the reference taxonomy used was the 2019 ICTV Release, i.e. including ranks from domain to genus. In addition, the lowest common ancestor (LCA) of non-singleton vOTUs was obtained and used as taxonomic classification for any member of that vOTU not already classified.
In silico host prediction
All UViGs identified as proviruses in IMG/M bacteria and archaea genomes were associated with the corresponding IMG/M host taxonomy. For other UViGs, two main approaches were used to link them to a putative host: sequence similarity to a microbial genome, and matches to IMG/M CRISPR spacers. For matches to microbial genomes, all IMG/VR sequences were compared to 95 012 bacterial and archaeal genomes in IMG/M (release from 4 June 2020) as well as 52 515 bacterial and archaeal genome bins from the Genome from Earth's Microbiomes (GEM) dataset (29), using blastn (options ‘-task megablast -evalue 0.001 -max_target_seqs 25000 -perc_identity 90’) (38). For genome bins (GEM), contigs which were mainly viral (i.e. hit to an IMG/VR sequence at ≥90% identity over ≥ 50% of the host contig) were excluded as these can be incorrectly binned (29). Similarly, all contigs from IMG/M genomes matching at ≥90% identity and ≥50% of their length to a viral genome from RefSeq, a giant virus, or a virophage were also excluded as these are typically contaminants of whole genome sequence datasets (e.g. PhiX174 genomes). Host predictions were then based on matches of ≥90% nucleotide identity covering ≥2 kb of the virus and (putative) host sequences. When multiple matches to different hosts were obtained, an 80% consensus rule at each rank was used to predict host taxonomy. Applied to the NCBI Viral RefSeq (v200) genomes, this approach yielded 96.2%, 95.3% and 91.2% of correct host prediction at the order, family, and genus ranks respectively, consistent with previous benchmarks (29).
For CRISPR matches, SpacePharer 2.fc5e668 (42) was used to compare all IMG/VR sequences to the database of CRISPR spacers derived from IMG/M genomes (21). Individual viral genomes and CRISPR spacers for which a hit was obtained with SpacePharer were further realigned using blastn (v2.5.0+) with options ‘-task blastn-short -evalue 1 -dust no -word_size 7’, as the alignment provided by SpacePharer did not always extend over the full length of the spacer (30). For each pair of viral sequence and putative host genome (i.e. set of CRISPR spacers), a host prediction was made when (i) at least one hit had 0 or 1 mismatch over the entire spacer length (‘CRISPR (near)identical’) or (ii) two hits or more had ≥80% identity over the entire spacer length (‘CRISPR multiple partial’). For each viral sequence, a CRISPR-based prediction was then derived based on an 80% consensus of all ‘CRISPR (near)identical’ predictions, or if no such prediction was available, an 80% consensus of ‘CRISPR multiple partial’ predictions. Applying this pipeline to NCBI Viral RefSeq (v200) genomes suggested that 97.5%, 94.9% and 88.2% of the predictions were correct at the order, family, and genus ranks respectively, for the ‘CRISPR (near)identical’ prediction, and 88.5%, 84.1% and 70.9% were correct at the order, family, and genus ranks respectively for the ‘CRISPR multiple partial’ prediction, consistent with previously published benchmarks (30).
Next, a predicted host taxonomy was obtained for each vOTU as the LCA of provirus-based predictions (if available), or CRISPR-based predictions. This LCA was used as putative host taxonomy for any sequence in these vOTUs not already associated with a specific host prediction. Finally, virophage sequences were associated with putative ‘host’ as follows: for virophage UViGs which were also identified in a Giant Virus genome bin, the corresponding genome bin and taxonomy was used as ‘host prediction’ for the virophage UViG. On the IMG/VR interface, a summarized host taxonomy is also displayed based on the following priority: provirus in a known genome or detection in a Giant Virus genome bin for virophage, match to host genome(s) if available at the genus rank, match to CRISPR spacer(s) if available at the genus rank, match to host genome(s) if available above the genus rank, match to CRISPR spacer(s) if available the genus rank, and host taxonomy of the corresponding vOTU otherwise and if available.
Identification of similar UViGs based on gene content and/or sequence similarity (IMG/VR-online)
Two methods of sequence comparison are available on the IMG/VR web interface. First, users can query the IMG/VR database using nucleotide or protein sequence(s) as input. The sequence comparison is computed using blast+ 2.6.0 (38) with default parameters, and users can select a specific E-value cutoff (from 1e–50–10). A second method of comparison between IMG/VR UViG is available through the IMG/VR web interface (tab ‘Similar UViGs’ in UViG detail page). This comparison is based on the affiliation of UViG genes to PFAM (43), marker VOGs (http://vogdb.org, see above) and VPF (Viral Protein Families, (23)) HMM profiles. PFAM affiliation were obtained from the IMG/M database, marker VOGs were obtained from the taxonomic classification pipeline (see above), while for VPFs, a comparison of all IMG/VR UViGs predicted proteins to the VPF database was performed using hmmsearch v3.2.1 (41) with option ‘-E 1.0e–02’, and minimum score of 40 and maximum E-value of 1e–05 for individual hits. For each pair of UViGs, a gene content similarity score (as in (44)) is computed by counting the number of predicted cds from genome A with a corresponding PFAM, VOG, or VPF hit in genome B, and dividing it by the total number of predicted cds with a hit to a PFAM, VOG or VPF domain in genome A. The same score is computed from genome B to genome A, and the final pairwise similarity is calculated as the average of the two scores. The similarity score is set at 0 if there are no genes affiliated to PFAM, VOG or VPF in any UViGs. Users can select a minimum cutoff (applied to the similarity) from 0.2 to 0.9, and the pairwise similarities can be visualized as a table or an interactive network.
RESULTS
Multiple sources of uncultivated virus genomes are used to build the IMG/VR database
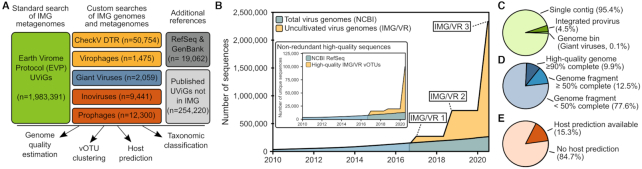
The IMG/VR database contains a large and broad collection of 2 332 702 isolates and uncultivated viral genomes (UViGs) compiled from 21 075 public genomes, metagenomes, and published datasets (Figure 1A and B, Supplementary Table S1, and see Methods). The vast majority of these UViGs (85%) were identified through a systematic search of public IMG/M metagenomes (45) using a standard approach to detect bacteriophages and archaeovirus sequences (31). This dataset was then complemented with the results of custom searches focused on specific virus or contig types including inoviruses (24), giant viruses (25), virophages (26), prophages (17), and predicted complete viral genomes (27) as well as additional viral reference sequences including 19,062 references from NCBI Viral RefSeq and GenBank (34,35) and 254 220 previously published UViGs (36,37). A small subset of these sequences are not currently available in the IMG/M database, and are thus only included in the downloadable IMG/VR files but not available on the web interface (see below ‘Data Availability’). Overall, 2 033 220 UViGs are available to browse, search, and analyze through the IMG/VR web interface, and will be designated hereafter as the ‘IMG/VR-online’ subset (Supplementary Table S1).
Figure 1.
Overview of the origin, number, and annotation of IMG/VR v3 sequences. (A) Origin and annotation of sequences for IMG/VR v3. The three columns represent three types of data, and individual boxes represent individual searches or reference databases (see Materials and Methods). Datasets are colored according to their primary type of sequences, i.e. individual viral contigs in green (standard search), yellow (custom search), and gray (additional references), genome bins in blue, and proviruses in red. The total number of sequences from each search or reference database is indicated in each box. Published UViGs not in IMG were obtained from the Global Ocean Virome 2 dataset (36), the Gut Virome Database (37), and the Metagenomic Gut Viruses dataset. (B) Number of sequences in the IMG/VR database compared to the number of viral genomes in NCBI over time. The main plot compares the total number of sequences in both databases. The total number of viral genomes in NCBI was obtained as in (14), i.e. querying GenBank and excluding human viruses for which an exceedingly large number of near-identical genomes are available (Influenza A/B and HIV). The inset shows the number of non-redundant high-quality sequences (i.e. HQ vOTUs) in IMG/VR compared to the number of viral genomes in NCBI Viral RefSeq. (C) Proportion of each genome type in IMG/VR v3. (D) Fraction of high-quality genomes (≥90% estimated completeness), and genome fragments of ≥50% and < 50% estimated completeness, as based on CheckV (27), in IMG/VR v3. (E) Proportion of sequences with and without a host prediction in IMG/VR v3.
IMG/VR includes tens of thousands of high-quality genomes
The IMG/VR v3 database is mostly composed of metagenome contigs predicted as entirely viral (∼95%), while ∼5% of the sequences were identified as integrated proviruses, i.e. on a contig including both viral and host region(s) (Figure 1C). Genome quality was estimated for all UViGs based on completeness estimation calculated with CheckV (27) as well as the detection of technical artifacts such as concatemers (see Methods). Overall, ∼22% of IMG/VR v3 UViGs were predicted as being ≥50% complete, and ∼10% were predicted as ≥90% complete, i.e. are considered as ‘High-quality’ according to the MIUViG standards (14) (Figure 1D). Since the last version of IMG/VR (released on 1 July 2018), the total number of UViGs has more than doubled, and the number of non-redundant high-quality UViGs, i.e. number of viral OTUs (vOTUs) including at least one high-quality genome (see below), increased by a factor of 5 (Figure 1B). This growth rate reflects both the increase in number and size of public metagenomes as well as the improvement in metagenome assembly and virus sequence detection tools (14).
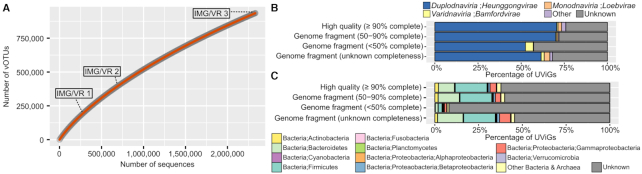
Global clustering of UViGs in vOTUs reveals ubiquitous and prevalent groups of uncultivated viruses
All IMG/VR UViGs, i.e. IMG/M sequences and non-IMG references, were clustered into 935,362 viral OTUs (vOTUs) using established standard cutoffs (95% ANI & 85% AF, (14), Figure 1B). Overall, 188,930 vOTUs included two or more members (excluding exact duplicates, see Materials and Methods), while 746 432 were singletons. This ratio of 32% of singletons is lower than observed for previous versions of IMG/VR (45.2% and 36.4% for IMG/VR v1 and v2, respectively), but suggests there is still a large portion of viral diversity to be explored (Figure 2A).
Figure 2.
Clustering and annotation results of IMG/VR v3 sequences. (A) Accumulation curve of IMG/VR sequences showing the number of vOTUs (y-axis) observed for different number of sequences (x-axis). A total of 50 random shuffling of the sequences was performed (gray dots), and the average value is plotted as the orange curve. The number of sequences corresponding to the three versions of IMG/VR are indicated on the plot. (B) Taxonomic distribution of IMG/VR v3 sequences, displayed separately for each quality tier, at the Kingdom rank. The Heunggongvirae kingdom includes notably the Caudovirales family, the Bamforvirae includes the Megaviricetes class encompassing most of the NCLDV, and the Loebvirae includes the Inoviridae family of filamentous phages (see Supplementary Table S3). (C) Host taxonomy prediction of IMG/VR v3 sequences, displayed separated for each quality tier. For panels B and C, the groups noted as ‘unknown completeness’ include UViGs for which no reliable completeness could be estimated with CheckV, e.g. because of assembly artifacts (see Materials and Methods).
This vOTU clustering also suggested the existence of some ubiquitous and highly prevalent viruses. Notably, the 13 largest vOTUs each included >1000 UViGs (range: 1043–3606) and are all associated with human gut and wastewater samples. Only two of these vOTUs included a reference genome: vOTU_043225 includes Faecalibacterium phage FP_Mushu initially identified as a prophage in Faecalibacterium prausnitzii genomes and putatively associated with inflammatory bowel disease patients (46), while vOTU_002247 includes the ‘Uncultured crAssphage’ reference sequence which was originally obtained from viral metagenome assembly and identified as a highly-prevalent member of the human gut virome (47). Meanwhile, all 13 largest vOTUs included >100 high-quality genomes. This highlights how the most prevalent viruses in a highly-sampled environment (human gut microbiome) still lack isolate representatives, and can only be identified via metagenomics.
Members of the Caudovirales order dominate the IMG/VR database
Taxonomic classification for individual UViGs is based on an affiliation of individual proteins to Viral RefSeq references, or on selected marker genes if no closely related reference exists in Viral RefSeq (see Materials and Methods). Such direct taxonomic classification was possible for 1 081 998 UViGs. For UViGs that remained unaffiliated, we reasoned some of these sequences were likely too short to display enough (marker) genes for direct classification, but may be clustered in vOTUs from which a taxonomic classification could be derived. We confirmed that the taxonomic classification of individual UViGs was consistent for 83.6% of the qualified vOTU at the family level (123 955 of 148 155 vOTUs with > 1 classified member, excluding giant virus MAGs), and thus affiliated unclassified sequences to the lowest common ancestor (LCA) of the vOTU classified members, when available. This approach enabled us to taxonomically classify 337,383 additional UViGs.
The vast majority of classified IMG/VR UViGs (91.8%) were affiliated with the Caudovirales order, followed by Megaviricetes (4.9%), Microviridae (0.7%) and Tubulavirales (0.7%; Figure 2B). This is consistent with previous reports highlighting Caudovirales as the most prevalent and frequently detected viral taxon across biomes (1,23). As could be expected, genomes with high (≥50%) completeness tend to be more classified (73.6% with a taxonomic classification) compared with small genome fragments (<50% complete, 57.2% assigned, Figure 2B). However, 24.1% of high-quality UViGs with ≥90% predicted completeness remain unclassified, and likely include representatives of entirely novel viral taxa.
Multi-feature approaches link IMG/VR sequences to a broad diversity of putative hosts
Prediction of host taxonomy for IMG/VR UViGs is based on (i) long (≥2 kb) sequence similarity between a UViG and a putative host genome, and (ii) sequence similarity between UViGs and predicted CRISPR spacers (see Materials and Methods). Similar to viral taxonomic classification, host predictions were consistent at the genus level for 96.4% of qualified vOTUs (32 791 of 34 016 vOTUs with >1 member with a host prediction). Thus, host prediction at the genus level was propagated within vOTUs using an LCA approach. Host prediction was obtained for 15.3% of all UViGs, however this was strongly driven by UViG completeness: host taxonomy could be predicted for 38.9% of UViGs estimated to be 50% complete or more, compared to 8.1% of UViGs estimated to be <50% complete (Figure 2C). The majority (85.8%) of these host taxonomy predictions could be achieved down to the genus rank.
Overall, IMG/VR UViGs were associated with a broad diversity of hosts spanning across 1481 putative host genera and 92 putative host phyla, according to the current IMG/M taxonomy. These were not evenly distributed however, and 64.1% of the host predictions were restricted to only five families: Clostridiales, Bacteroidales, Enterobacterales, Lactobacillales and Pseudomonadales (Figure 2C). This likely reflects the combined biases in the type of samples from which viruses are extracted, the host reference genome database, and the methods used to link viruses to hosts. Additional technological developments may be needed to more comprehensively identify virus:host pairs across microbial diversity.
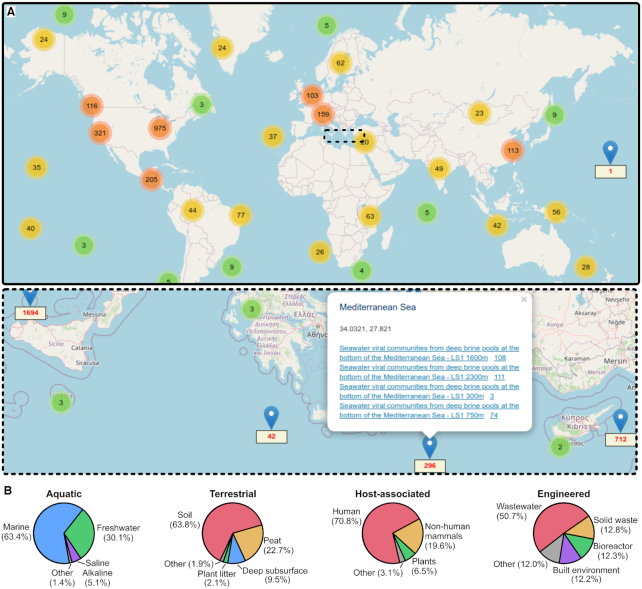
IMG/VR UViGs are derived from diverse biomes and geographic locations
Sequences in the IMG/VR v3 database were obtained from 5582 genomes and 15 493 metagenomes. The latter originated from a broad range of geographic locations spanning across all continents and oceans (Figure 3A), and represented a large diversity of biomes classified in IMG/VR using the GOLD five-level ecosystem framework (48,49) (Figure 3B).
Figure 3.
Geographical and environmental distribution of IMG/VR-online sequences. (A) Overview map of the IMG/VR v3 sequences based on the location of their original sample in IMG/M (reference sequences not currently in IMG/M are not included on the map). The two insets (bottom) display zoomed-in views of two regions with a high sample count. Colored samples indicate groups of geographic locations too close to be represented separately in the current zoom level, colored according to the number of locations, which is also indicated in the circle. Once zoomed in, individual locations are displayed using blue pins, with the number of UViGs indicated in red. Clicking on the pin brings in a new window including links to the datasets and UViGs at this location. (B) Proportion of IMG/VR v3 sequences per sample type for four major biomes. The pie charts are based on all UViGs regardless of quality, i.e. n = 1 592 032 aquatic, n = 122 133 terrestrial, n = 487 433 host-associated and n = 75 228 engineered sequences.
Overall, IMG/VR UViGs are primarily derived from three sample types: marine (44.0%), freshwater (20.8%) and human-associated (15.0%). The same trend was observed when excluding small genome fragments and redundant UViGs: counting vOTUs with at least one member estimated to be ≥50% complete, the three main sample types were marine (29.4%), human-associated (28.8%) and freshwater (12.7%). This suggests that the large number of UViGs in these samples is not an artifact due to fragmented assemblies and redundant assembly of identical genomes, but that these environments are the primary source of viral genome diversity available in the IMG/VR v3 database.
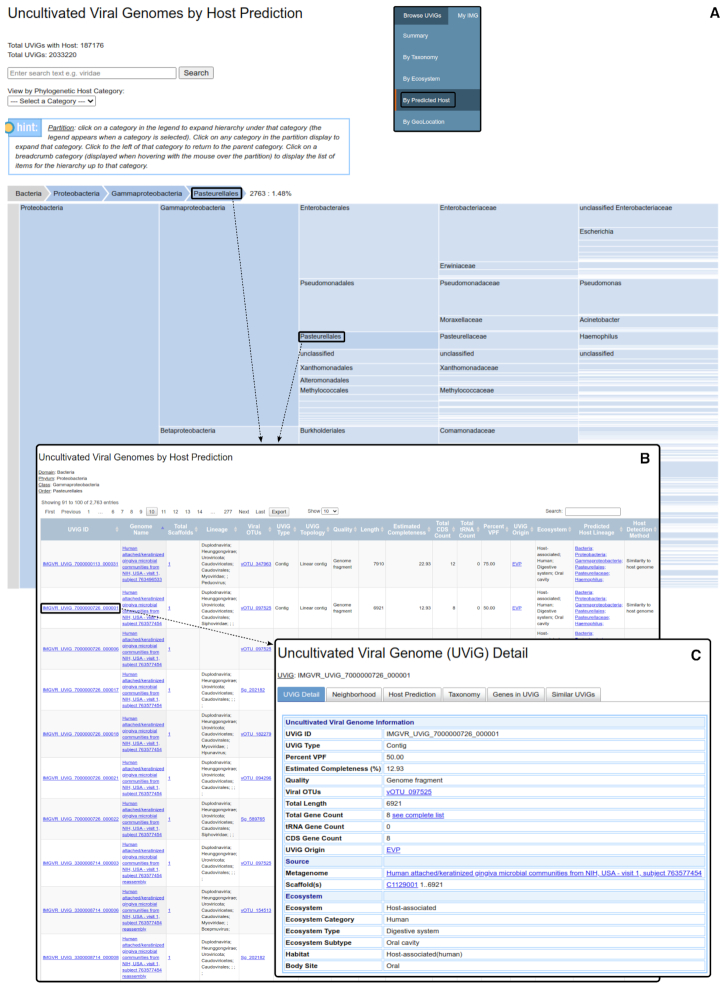
IMG/VR provides intuitive data browsing of millions of UViGs
The 2 033 220 UViGs available online can be browsed through the redesigned interface (https://img.jgi.doe.gov/vr/, Supplementary Figure S1). From the home page, the ‘Browse UViGs’ menu lists the different parameters that can be used to explore and select subsets of IMG/VR data (Supplementary Figure S2). Users can browse UViGs based on ecosystem, taxonomy, or predicted host taxonomy classifications using new interactive treemaps (Figure 4A). A summary of the group currently selected including number and percentage of UViGs in the group is provided above the treemap, along with a search bar allowing the user to search the treemap for specific (partial) terms. Alternatively, the same data can be browsed through a table by selecting a specific rank within the hierarchical classification (Supplementary Figure S3). Clicking on a selected group will provide a table of the corresponding UViGs including basic characteristics which can be exported as an excel spreadsheet (Figure 4B). UViG identifiers in the table link to individual UViG pages which include detailed information and annotation (Figure 4C).
Figuer 4.
Browsing and selection of a subset of IMG/VR-online sequences. (A) Example of a treemap representation of IMG/VR-online UViGs based on predicted host taxonomy. Similar treemaps are available for taxonomic and ecosystem classification. The inset shows how to access this treemap using the main IMG/VR menu. (B) UViG table result corresponding to the search in panel A. (C) Example of a UViG Detail page, obtained by clicking on a UViG identifier in the result table.
Users can also browse UViGs based on geographical location or human body site using an interactive world map and human body diagram, respectively, as in IMG/VR v2 (Figure 3A). UViGs selected through these maps are then presented using the same table as for the treemap selection (Figure 4B). Finally, UViGs can now be browsed based on features such as the detection of a specific pfam domain (Supplementary Figure S4), or UViG length (Supplementary Figure S5). For the latter, groups are defined on fixed intervals, which are progressively narrowed as the user selects individual groups until the number of UViGs selected is reasonable to be displayed in a table (≤5000, Supplementary Figure S5).
IMG/VR interface includes novel search capabilities
In addition to the new UViG browsing, new search capabilities have also been implemented in the IMG/VR v3 interface (Supplementary Figure S6). UViGs can first be searched through individual UViG or scaffold identifier(s) (Supplementary Figure S7). As with browsing, the search result is a table listing the basic characteristics of each UViG, and allowing the user to navigate to each UViG detail page. Searches by identifiers also accept comma-separated lists, allowing users to retrieve a specific set of UViGs.
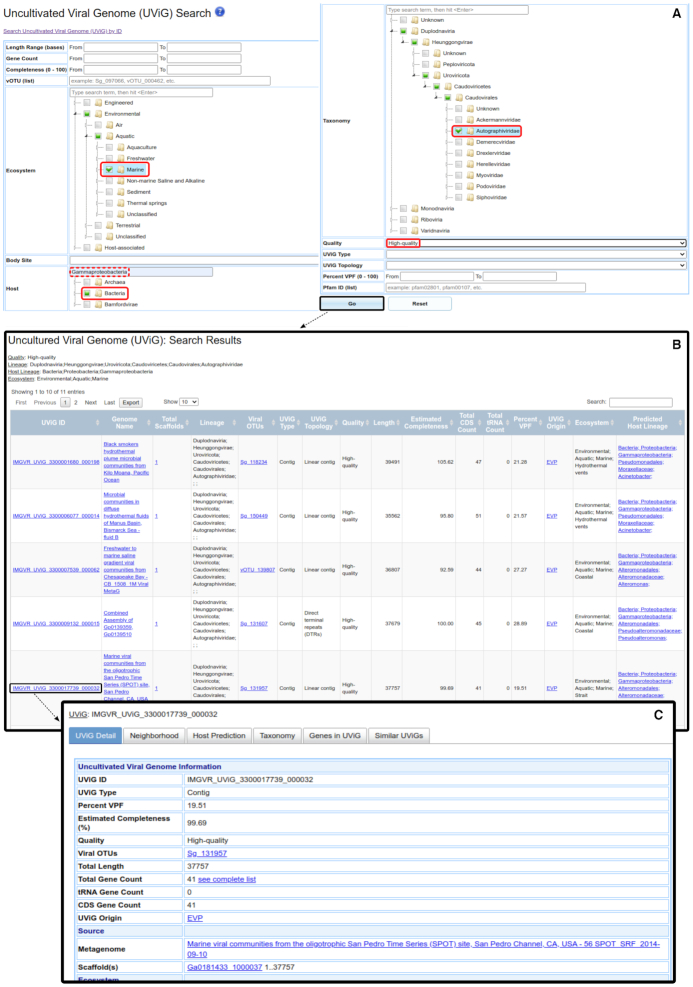
UViGs can also be searched based on a combination of attributes (Figure 5). Searchable attributes include any combination of length, number of genes, estimated completeness, vOTU identifier(s), ecosystem, taxonomy, predicted host taxonomy, percentage of VPFs, and detection of pfam domain(s). Search terms are combined using an ‘AND’ logic operator, i.e. UViGs will be selected only if they fulfill all conditions specified in the search form. Search results are displayed through a downloadable UViG table (Figure 5B), from which a user can navigate to individual UViG detail pages (Figure 5C).
Figure 5.
Searching IMG/VR-online sequences based on a combination of UViG attributes. (A) Example of a search of IMG/VR-online UViGs based on multiple attributes including ecosystem, host taxonomy prediction, taxonomic classification and quality. The inset shows how to access this search page using the main IMG/VR menu. The selected search parameters are outlined in red, and the host taxon was selected using the built-in search field (dashed red rectangle). (B) UViG table result corresponding to the search in panel A. (C) Example of a UViG Detail page, obtained by clicking on the corresponding UViG identifier in the result table.
Sequence comparison to and within IMG/VR
As with previous versions of IMG/VR, two types of sequence-based searches are available in IMG/VR v3 (Supplementary Figure S8). First, a user can compare their own sequence(s) against IMG/VR-online UViGs based on blastn (for nucleotide sequences) or blastp (for protein sequences). Summarized blast results are provided through an interactive table (Supplementary Figure S8) while complete result files are provided for download. User sequences can also be compared to the IMG/M CRISPR spacer databases using the same search form.
UViGs can also be compared based on the detection of common functional domains (Supplementary Figure S9). This tool is available through the ‘Compare UViGs’ section on individual UViG pages, and is meant to provide a quick way to identify UViGs with similar gene content as the one currently selected (Supplementary Figure S9). Similarity between UViGs is based on gene affiliation to pfam, VOG and VPF databases, and is measured for each pair of UViG using a gene content similarity score (see Methods). The results are displayed through a table or an interactive network with edge length proportional to similarity value and node colored by UViG attribute, enabling a user to explore the viral sequence space in the vicinity of the UViG initially selected (Supplementary Figure S9).
Bulk download
The entire IMG/VR v3 database is freely available to download at https://genome.jgi.doe.gov/portal/pages/dynamicOrganismDownload.jsf?organism=IMG_VR (accessible through the ‘Download IMG/VR Database’ link on the main page, Supplementary Figure S10). Downloadable files include fasta files for contigs and predicted proteins, a master table listing all UViGs with their main attributes such as ecosystem and taxonomic affiliation, and a host prediction table listing all connections between UViG and putative hosts. The fasta files and tables include both UViGs available from IMG/M, and external UViGs used as reference in IMG/VR (see Methods).
The IMG/VR download portal includes present and past versions of the IMG/VR database, identified using the database release date. Files corresponding to the IMG/VR v3 (release 5) described in this manuscript are available under the ‘IMG_VR_2020-09-10_5’ folder.
DISCUSSION
Since its initial release in 2016, IMG/VR continues to expand as the largest database of viral genomes assembled from metagenomes, and has been extensively leveraged by the research community (50–55). Compared to its initial release, the new IMG/VR v3 database includes ∼16 times as many sequences overall, and ∼20 times as many sequences meeting the MIUViG criteria to be categorized as ‘high quality’ (i.e. ≥90% complete and non-redundant). Notably, IMG/VR v3 includes sequences identified from the extensive set of genomes and metagenomes available in IMG, but also from other previously published collections of UViGs. Given the accelerating pace at which new uncultivated viral genomes are collected, we anticipate the exponential growth of IMG/VR to continue for the foreseeable future. This will be enabled particularly by technological improvements such as long-read sequencing (56) and new bioinformatics approaches (57), which are expected to yield longer and more complete viral genomes from metagenomes.
This new version of IMG/VR includes an updated database, new analyses using recently developed tools such as CheckV to estimate genome completeness (27), and a redesigned user interface to accommodate the scale and diversity of UViG data. In addition to viral contigs, the IMG/VR database now includes proviruses (i.e. viral region within a host contig) and genome bins, which are both critical capabilities for exploring specific areas of the virosphere such as filamentous phages and giant viruses (24,25). New browsing and search capabilities were introduced, providing users with a set of tools to identify sequences of interest within the large IMG/VR database. These new tools are paired with the functionalities of the broader IMG/M platform including ‘Scaffold carts’ and ‘Scaffold sets’ enabling further analyses and easy export of the corresponding data.
While IMG/VR represents a unique and unprecedented resource for exploration and characterization of the global virosphere, improvements in key features are expected to increase its usability and usefulness. First, while a majority of UViGs are taxonomically classified, most of them are assigned to a large ‘unknown Caudovirales’ group. As ICTV progressively refines the taxonomy within this order (58) and as classification tools improve, we expect taxonomy in future releases of IMG/VR to be more informative. Second, host prediction remains available only for a minority of UViGs, limiting the range of analyses possible and preventing a comprehensive integration of viral diversity in microbiome models. We anticipate that improvements to host reference databases, especially through the recovery of genomes and CRISPR arrays from metagenomes (29), paired with new methods to robustly integrate multiple signals in a single host prediction (e.g. (42,59)) will eventually enable a broader linkage between UViGs and microbial hosts. Finally, as more tools and infrastructure are developed for viral ecogenomics (60,61), we plan to further integrate the IMG/VR database across different platforms including IMG, iVirus(61) and KBase (62), to broaden the scope of analyses that can be performed on these data and to make it easily accessible to the community at large.
DATA AVAILABILITY
The complete IMG/VR database is available for download at https://genome.jgi.doe.gov/portal/pages/dynamicOrganismDownload.jsf?organism=IMG_VR. IMG/VR-online sequences can also be browsed, searched, and analyzed at https://img.jgi.doe.gov/vr/.
Supplementary Material
Contributor Information
Simon Roux, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
David Páez-Espino, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
I-Min A Chen, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Krishna Palaniappan, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Anna Ratner, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Ken Chu, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
T B K Reddy, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Stephen Nayfach, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Frederik Schulz, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Lee Call, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Russell Y Neches, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Tanja Woyke, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Natalia N Ivanova, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Emiley A Eloe-Fadrosh, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Nikos C Kyrpides, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]. Funding for open access charge: U.S. Department of Energy Joint Genome Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Páez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., Rubin E., Ivanova N.N., Kyrpides N.C.. Uncovering Earth's virome. Nature. 2016; 536:425–430. [DOI] [PubMed] [Google Scholar]
- 2. Williamson K.E., Fuhrmann J.J., Wommack K.E., Radosevich M.. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017; 4:201–219. [DOI] [PubMed] [Google Scholar]
- 3. Brum J.R., Sullivan M.B.. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 2015; 13:doi:10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 4. Hatfull G.F. Dark matter of the biosphere: the amazing world of bacteriophage diversity. J. Virol. 2015; 89:8107–8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Prangishvili D. The wonderful world of archaeal viruses. Annu. Rev. Microbiol. 2013; 67:565–585. [DOI] [PubMed] [Google Scholar]
- 6. Shkoporov A.N., Hill C.. Bacteriophages of the human gut: the “Known Unknown” of the microbiome. Cell Host Microbe. 2019; 25:195–209. [DOI] [PubMed] [Google Scholar]
- 7. Suttle C.A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 2007; 5:801–812. [DOI] [PubMed] [Google Scholar]
- 8. Brüssow H., Canchaya C., Hardt W.. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004; 68:560–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Breitbart M., Bonnain C., Malki K., Sawaya N.A.. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018; 3:754–766. [DOI] [PubMed] [Google Scholar]
- 10. Sullivan M.B., Weitz J.S., Wilhelm S.W.. Viral ecology comes of age. Environ. Microbiol. Rep. 2017; 9:33–35. [DOI] [PubMed] [Google Scholar]
- 11. Sullivan M.B. Viromes, not gene markers for studying dsDNA viral communities. J. Virol. 2015; 89:2459–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edwards R.A., Rohwer F.. Viral metagenomics. Nat. Rev. Microbiol. 2005; 3:504–510. [DOI] [PubMed] [Google Scholar]
- 13. Dutilh B.E., Reyes A., Hall R.J., Whiteson K.L.. Virus discovery by metagenomics: the (im)possibilities. Front. Microbiol. 2017; 8:5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roux S., Adriaenssens E.M., Dutilh B.E., Koonin E. V., Kropinski A.M., Krupovič M., Kuhn J.H., Lavigne R., Brister J.R., Varsani A. et al.. Minimum information about an Uncultivated Virus Genome (MIUViG). Nat. Biotechnol. 2019; 37:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y.Z., Chen Y.M., Wang W., Qin X.C., Holmes E.C.. Expanding the RNA virosphere by unbiased metagenomics. Annu. Rev. Virol. 2019; 6:119–139. [DOI] [PubMed] [Google Scholar]
- 16. Roux S., Hawley A.K., Torres Beltran M., Scofield M., Schwientek P., Stepanauskas R., Woyke T., Hallam S.J., Sullivan M.B.. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell- and meta- genomics. Elife. 2014; 3:e03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roux S., Hallam S.J., Woyke T., Sullivan M.B.. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. 2015; 4:e08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Labonté J.M., Swan B.K., Poulos B.T., Luo H., Koren S., Hallam S.J., Sullivan M.B., Woyke T., Wommack E.K., Stepanauskas R.. Single cell genomics-based analysis of virus-host interactions in marine surface bacterioplankton. ISME J. 2015; 9:2386–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jarett J.K., Džunková M., Schulz F., Roux S., Paez-Espino D., Eloe-Fadrosh E., Jungbluth S.P., Ivanova N., Spear J.R., Carr S.A. et al.. Insights into the dynamics between viruses and their hosts in a hot spring microbial mat. ISME J. 2020; 14:2527–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simmonds P., Adams M.J., Benkő M., Breitbart M., Brister J.R., Carstens E.B., Davison A.J., Delwart E., Gorbalenya A.E., Harrach B. et al.. Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 2017; 15:161–168. [DOI] [PubMed] [Google Scholar]
- 21. Chen I.M.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J.R., Seshadri R. et al.. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019; 47:D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Páez-Espino D., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Szeto E., Pillay M., Huang J., Markowitz V.M., Nielsen T. et al.. IMG/VR: a database of cultured and uncultured DNA viruses and retroviruses. Nucleic Acids Res. 2016; 45:D457–D465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paez-Espino D., Roux S., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Huntemann M., Reddy T.B.K., Pons J.C., Llabrés M. et al.. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 2018; 47:D678–D686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roux S., Krupovic M., Daly R.A., Borges A.L., Nayfach S., Schulz F., Sharrar A., Matheus Carnevali P.B., Cheng J.-F.F., Ivanova N.N. et al.. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth's biomes. Nat. Microbiol. 2019; 4:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schulz F., Roux S., Paez-Espino D., Jungbluth S., Walsh D.A., Denef V.J., McMahon K.D., Konstantinidis K.T., Eloe-Fadrosh E.A., Kyrpides N.C. et al.. Giant virus diversity and host interactions through global metagenomics. Nature. 2020; 578:432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paez-Espino D., Zhou J., Roux S., Nayfach S., Pavlopoulos G.A., Schulz F., McMahon K.D., Walsh D., Woyke T., Ivanova N.N. et al.. Diversity, evolution, and classification of virophages uncovered through global metagenomics. Microbiome. 2019; 7:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nayfach S., Camargo A.P., Schulz F., Eloe-fadrosh E., Roux S., Kyrpides N.. CheckV: assessing the quality of metagenome-assembled viral genomes. Nat. Biotechnol. 2020; doi:10.1101/2020.05.06.081778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koonin E. V., Dolja V.V., Krupovič M., Varsani A., Wolf Y.I., Yutin N., Zerbini F.M., Kuhn J.H.. Global organization and proposed megataxonomy of the viru world. Microbiol. Mol. Biol. Rev. 2020; 84:e00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., Wu D., Paez-Espino D., Chen I.-M.A., Huntemann M. et al.. A genomic catalogue of Earth's microbiomes. Nat. Biotechnol. 2020; in press. [Google Scholar]
- 30. Edwards R.A., McNair K., Faust K., Raes J., Dutilh B.E.. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol. Rev. 2016; 40:258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Páez-Espino D., Pavlopoulos G.A., Ivanova N.N., Kyrpides N.C.. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat. Protoc. 2017; 12:1673–1682. [DOI] [PubMed] [Google Scholar]
- 32. Ren J., Ahlgren N.A., Lu Y.Y., Fuhrman J.A., Sun F.. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017; 5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roux S., Enault F., Hurwitz B.L., Sullivan M.B.. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015; 3:e985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2020; 48:D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gregory A.C., Zayed A.A., Conceição-Neto N., Temperton B., Bolduc B., Alberti A., Ardyna M., Arkhipova K., Carmichael M., Cruaud C. et al.. Marine DNA viral Macro- and microdiversity from pole to pole. Cell. 2019; 177:1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gregory A.C., Zablocki O., Zayed A.A., Howell A., Bolduc B., Sullivan M.B., Gregory A.C., Zablocki O., Zayed A.A., Howell A. et al.. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. 2020; 28:doi:10.1016/j.chom.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olm M.R., Brown C.T., Brooks B., Banfield J.F.. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017; 11:2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ponsero A.J., Hurwitz B.L.. The promises and pitfalls of machine learning for detecting viruses in aquatic metagenomes. Front. Microbiol. 2019; 10:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011; 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang R., Mirdita M., Karin E.L., Norroy C., Galiez C., Soeding J.. SpacePHARER: sensitive identification of phages from CRISPR spacers in prokaryotic hosts. 2020; bioRxiv doi:15 May 2020, preprint: not peer reviewed 10.1101/2020.05.15.090266. [DOI] [PMC free article] [PubMed]
- 43. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jacobs-Sera D., Abad L.A., Alvey R.M., Anders K.R., Aull H.G., Bhalla S.S., Blumer L.S., Bollivar D.W., Alfred Bonilla J., Butela K.A. et al.. Genomic diversity of bacteriophages infecting Microbacterium spp. PLoS One. 2020; 15:e0234636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou K., Zhang R., Sun J., Zhang W., Tian R., Chen C., Kawagucci S., Xu Y., Kong H., Kong H. et al.. Potential SUP05-Phage interactions in hydrothermal vent sponges. Appl. Environ. Microbiol. 2019; 85:e00992-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cornuault J.K., Petit M.A., Mariadassou M., Benevides L., Moncaut E., Langella P., Sokol H., De Paepe M.. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018; 6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G.Z., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K. et al.. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014; 5:4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.M.A., Kyrpides N.C., Reddy T.B.K.. Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic Acids Res. 2019; 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ivanova N., Tringe S.G., Liolios K., Liu W.-T.T., Morrison N., Hugenholtz P., Kyrpides N.C.. A call for standardized classification of metagenome projects. Environ. Microbiol. 2010; 12:1803–1805. [DOI] [PubMed] [Google Scholar]
- 50. Rosario K., Fierer N., Miller S.L., Luongo J., Breitbart M.. Diversity of DNA and RNA viruses in indoor air as assessed via metagenomic sequencing. Environ. Sci. Technol. 2018; 52:1014–1027. [DOI] [PubMed] [Google Scholar]
- 51. Dyall-Smith M., Palm P., Wanner G., Witte A., Oesterhelt D., Pfeiffer F.. Halobacterium salinarum virus ChaoS9, a novel halovirus related to PhiH1 and PhiCh1. Genes (Basel). 2019; 10:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Szafrański S.P., Kilian M., Yang I., Bei der Wieden G., Winkel A., Hegermann J., Stiesch M.. Diversity patterns of bacteriophages infecting Aggregatibacter and Haemophilus species across clades and niches. ISME J. 2019; 13:2500–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang Y., Wang L., Wang Z., Zhao J., Yang Q., Wang M., Yang K., Zhang L., Jiao N., Zhang Y.. Metagenomic analysis of the diversity of DNA viruses in the surface and deep sea of the south China sea. Front. Microbiol. 2019; 10:1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jahn M.T., Arkhipova K., Markert S.M., Stigloher C., Lachnit T., Pita L., Kupczok A., Ribes M., Stengel S.T., Rosenstiel P. et al.. A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host Microbe. 2019; 26:542–550. [DOI] [PubMed] [Google Scholar]
- 55. Malone L.M., Warring S.L., Jackson S.A., Warnecke C., Gardner P.P., Gumy L.F., Fineran P.C.. A jumbo phage that forms a nucleus-like structure evades CRISPR–Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat. Microbiol. 2020; 5:48–55. [DOI] [PubMed] [Google Scholar]
- 56. Warwick-Dugdale J., Solonenko N., Moore K., Chittick L., Gregory A.C., Allen M.J., Sullivan M.B., Temperton B.. Long-read viral metagenomics captures abundant and microdiverse viral populations and their niche-defining genomic islands. PeerJ. 2019; 2019:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Antipov D., Raiko M., Lapidus A., Pevzner P.A.. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics. 2020; 36:4126–4129. [DOI] [PubMed] [Google Scholar]
- 58. Adriaenssens E.M., Sullivan M.B., Knezevic P., van Zyl L.J., Sarkar B.L., Dutilh B.E., Alfenas-Zerbini P., Łobocka M., Tong Y., Brister J.R. et al.. Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2020; 165:1253–1260. [DOI] [PubMed] [Google Scholar]
- 59. Wang W., Ren J., Tang K., Dart E., Ignacio-Espinoza J.C., Fuhrman J.A., Braun J., Sun F., Ahlgren N.A.. A network-based integrated framework for predicting virus-host interactions. NAR Genomics Bioinforma. 2019; 2:lqaa044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bolduc B., Youens-Clark K., Roux S., Hurwitz B.L., Sullivan M.B.. iVirus: facilitating new insights in viral ecology with software and community data sets imbedded in a cyberinfrastructure. ISME J. 2017; 11:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao N.L., Zhang C., Zhang Z., Hu S., Lercher M.J., Zhao X., Bork P., Liu Z., Chen W.. MVP: a microbe – phage interaction database. Nucleic Acids Res. 2018; 4:D700–D707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., Dehal P., Ware D., Perez F., Canon S. et al.. KBase: the United States Department of Energy systems biology knowledgebase. Nat. Biotechnol. 2018; 36:566–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete IMG/VR database is available for download at https://genome.jgi.doe.gov/portal/pages/dynamicOrganismDownload.jsf?organism=IMG_VR. IMG/VR-online sequences can also be browsed, searched, and analyzed at https://img.jgi.doe.gov/vr/.