Abstract
The Genomes OnLine Database (GOLD) (https://gold.jgi.doe.gov/) is a manually curated, daily updated collection of genome projects and their metadata accumulated from around the world. The current version of the database includes over 1.17 million entries organized broadly into Studies (45 770), Organisms (387 382) or Biosamples (101 207), Sequencing Projects (355 364) and Analysis Projects (283 481). These four levels contain over 600 metadata fields, which includes 76 controlled vocabulary (CV) tables containing 3873 terms. GOLD provides an interactive web user interface for browsing and searching by a wide range of project and metadata fields. Users can enter details about their own projects in GOLD, which acts as a gatekeeper to ensure that metadata is accurately documented before submitting sequence information to the Integrated Microbial Genomes (IMG) system for analysis. In order to maintain a reference dataset for use by members of the scientific community, GOLD also imports projects from public repositories such as GenBank and SRA. The current status of the database, along with recent updates and improvements are described in this manuscript.
INTRODUCTION
Genomes OnLine Database (GOLD) is an open-access repository of genome and metagenome sequencing projects with their associated metadata. Login-free access is provided to a growing catalogue of manually curated public projects from all over the world. Starting from whole genome sequencing of cultured bacteria, uncultured single cells, complex eukaryotes or metagenome sequencing of environmental samples, the world of sequencing is advancing at a rapid pace. In overcoming the limitations of culturing microbial isolates, improved sequencing and analysis methods have broadened our understanding of the microbial world. Metagenome-assembled genomes (MAGs) and single-amplified genomes (SAGs) (1,2) are two such examples of genomes of uncultivated organisms that have recently expanded our knowledge about microorganisms.
In any sequencing project, metadata or accessory information about the sample being sequenced plays a key role. In order to interpret raw sequence data and make accurate scientific predictions, the presence of a wide variety of metadata is extremely important. For example, comparative analyses of the microbial community of soil samples from multiple locations would be very difficult if the geographic location information or soil type is not recorded. On the other hand, the scientific value of the same sequence will likely increase several-fold if detailed environmental and physicochemical properties of the soil are collected. Once the bacterial and archaeal members of the soil community are identified it is important to classify them taxonomically (3) and assign proper nomenclature (4). Finally, environmental gradient information can be combined with genomic and phenotypic traits of the microbial community to get a more complete understanding of the soil ecosystem (5).
Now in its eighth release since its first publication (6) followed by regular updates (7–9), GOLD has been instrumental in providing a rich and reliable resource of metadata that is widely used by the genomics community. This is evident from the over 2500 citations that different publications of GOLD have received over the years. For example, GOLD metadata has been applied to study resistance, biogeochemistry and metabolism of arsenic-related genes in a global survey of soil microbiomes (10). In a recent article, Guittar et al. (11) used trait-based metadata from GOLD Organisms such as Gram stain, motility, oxygen tolerance, pH and sporulation to infer patterns of infant gut microbiome succession. Ever since genome sequencing has become a routine technique, efforts to develop bioinformatics tools to store and analyze sequence data are also growing. GOLD, through its established metadata standards and wealth of metadata fields, plays a key role in this effort to make sense of sequence information. GOLD follows the Minimum Information about any (x) Sequence (MIxS) standards (12) established by The Genomic Standards Consortium (GSC) (13) and also works in close collaboration with The Environment Ontology (ENVO) community (14). There are three different ways in which projects and their associated metadata are entered into GOLD: (i) projects that are added by individual GOLD users; (ii) projects for samples sequenced at the Joint Genome Institute (JGI) as part of the JGI User Programs and (iii) projects sourced from public databases such as the NCBI GenBank (15) and Sequence Read Archive (SRA) (16). Well-defined genome and metagenome projects in GOLD are a prerequisite for submitting sequence data to the Integrated Microbial Genomes with Microbiome (IMG/M) (17) system for annotation and analysis (18,19). GOLD and IMG work closely to provide a seamless experience to our broad user base of scientists, students, and policy makers as well as novice science enthusiasts. Users can browse through several metadata fields in GOLD and take advantage of the multiple filters and advanced search tools to identify a subset of genomes or metagenomes of interest. They can then use the IMG identifiers from GOLD and directly go to the IMG user interface and conduct further research and comparative analyses. In this paper, we give an overview of the database for first-time users of the database and provide detailed descriptions of our new features that were added since its last publication.
OVERVIEW OF GOLD
GOLD Organization and Current Status
GOLD is organized in a four-level hierarchical system to describe the overall proposal, samples or organisms studied, sequencing projects undertaken and their analysis process. These levels include Study, Organism or Biosample, Sequencing Project (SP), and Analysis Project (AP). All four levels, along with their complex list of metadata fields and controlled vocabulary (CV) terms, are connected to each other in a lucid framework to enhance scientific discovery. A brief description of each of these four levels is provided below.
Study
The overall research objectives and goals are captured in a Study, which lies at the helm of the four-level organization structure. A Study is similar to the concept of NCBI’s umbrella BioProject and comprises one or many Organism(s) or Biosample(s) as well as their respective Sequencing and Analysis Projects. Studies can vary in the type of samples collected. For instance, they may include a group of cultured bacteria or soil sample(s) from a rainforest or a mixture of both, provided they answer a common research question. Subsequently, a single Study may have several Sequencing and Analysis Projects that differ in their methodology and application such as Whole Genome Sequencing (WGS) and analysis, metagenome analysis or a combination of both. The number of new Studies has been steadily increasing over the years. As of August 2020, there are 45 767 studies in GOLD, representing a 37% increase from the 33 400 Studies in its previous release.
Organism/Biosample
A GOLD Organism or Biosample contains descriptive information about the biological or physical material that is being sequenced. Any living entity such as bacteria, archaea, fungi, virus, plant or animal may constitute an Organism in GOLD. An Organism may be cultured, uncultured (such as single cells) or even bioinformatically predicted (such as Metagenome-Assembled Genomes or MAGs). The most important metadata feature of a GOLD Organism is its Taxonomic assignment including phylum, genus, species, strain etc. To promote interconnectedness with other databases, all GOLD Organisms have the NCBI Taxonomy ID, which is a numerical unique identifier of the NCBI taxonomy database (20). With a sudden surge in the number of bioinformatically predicted organisms, it has become essential to define taxonomic standards that include these organisms (21,22). Until such a standard is established, GOLD will continue to follow its own naming standards for MAGs that is described in detail here https://gold.jgi.doe.gov/resources/Metagenome-Assembled-Genomes-Naming-Standards.pdf. As of August 2020, there were 387 480 Organisms in GOLD representing a 23% increase compared to the previous release. Of these Organisms, 22,946 are uncultured, including 57% MAGs and 43% single-cells.
The majority (88%) of these organisms are bacteria, followed by eukaryotes (8.5%), viruses (2.5%) and archaea (1%) spread across 305 different phyla and candidate phyla. The recent COVID-19 pandemic has led to a renewed interest in studying viruses and hundreds of viral genomes and metagenomes are being sequenced all over the world. GOLD currently has 77 Sequencing Projects for viruses belonging to the Coronaviridae family. We expect this number to increase significantly in the coming months. While it is necessary to study a large number of coronaviruses to follow the progress of this outbreak, it is equally important to accurately record their metadata such as host-specificity, collection date, isolation site and more (23).
A physical sample from the environment consisting of a community of microorganisms is referred to as a Biosample in GOLD. This is slightly different from NCBI’s concept of BioSample, which may include individual organisms, environmental samples, cell lines etc. A GOLD Biosample is defined specifically in the context of metagenome and metatranscriptome projects. Contrary to most organisms which have a well-defined name, there are no strict rules governing the nomenclature of environmental samples. GOLD, however, follows a standardized nomenclature in which the name of a Biosample is constructed using a combination of habitat, microbial community, geographic location and a unique identifier. The rules for naming a Biosample, along with examples, are described here: https://gold.jgi.doe.gov/resources/Standardized_Metagenome_Naming.pdf. To correctly define a Biosample in GOLD, a user must provide its habitat, collection site, geographic location, and isolation country along with some additional mandatory metadata, in accordance to the Minimum Information standards as defined by the Genomic Standards Consortium. Over the last few years, GOLD has seen a phenomenal increase in the number of new Biosamples. As compared to 49 821 in the v.7 release of the database (September 2018), GOLD now has 101,561 Biosamples, an increase of over 100%. These Biosamples are spread across Host-associated (55%), Environmental (38%) and Engineered (7%) Ecosystems (24).
Sequencing Project
The process of generating sequencing data from a Biosample or Organism is described in a Sequencing Project (SP). GOLD currently has 15 different types of SPs, from which whole-genome sequencing (WGS) and metagenome are most commonly used. The input material for an SP can either be DNA or RNA corresponding to a genome or transcriptome project, respectively. This material can come from either an organism, in the case of WGS and transcriptomes, or from a Biosample, in the case of metagenomes and metatranscriptomes. A cultured organism can sometimes be sequenced by more than one institution at different times, resulting in multiple SPs for the same organism. The same Organism entity will be the basis for these SPs. In the case of environmental samples, the same Biosample may be used for both metagenome (DNA) and metatranscriptome (RNA) SPs. Some of the critical metadata present in Sequencing Projects include the type of nucleic acid, sequencing instrument, library method, sequencing institution and funding agency as well as NCBI identifiers such as BioProject/BioSample Accession and SRA Experiment IDs. In its current version, GOLD has 354 270 Sequencing Projects compared to 215 881 SPs in 2018, representing a 64% increase.
Analysis Project
The information about the processing of sequence data and its analysis is captured in a GOLD Analysis Project (AP). A GOLD AP is required in order to submit the corresponding sequence data to IMG for annotation. There are 13 different types of Analysis Projects that can be defined in the current version, namely: Genome, Metagenome, Metagenome—Cell Enrichment, Metagenome—Single Particle Sort, Metatranscriptome, Single Cell (Unscreened), Single Cell (Screened), Metagenome-Assembled Genome, as well as five different types of Combined Assembly. GOLD APs follow the standards set by the Genomic Standards Consortium (GSC) including Minimum Information about a Single Amplified Genome (MISAG) and Minimum Information about a Metagenome-Assembled Genome (MIMAG) (25), facilitating their community wide adoption. Some of the key metadata fields of a GOLD AP are assembly method, sequencing depth, estimated size, binning method (for MAG AP), contamination screening method [for Single Cell (Screened) AP] etc. NCBI Identifiers such as GenBank ID, Assembly Accession as well as SRA Run IDs can also be found in a GOLD AP. As of August 2020, there were 283 764 Analysis Projects, an increase of 62% compared to the previous version of the database (September 2018). Genome analysis APs constitute the major chunk (58%) of the current GOLD APs, followed by metagenome analysis AP (26%), metatranscriptome AP (7%) and MAG AP (4.5%).
Database architecture and processes
The GOLD production database is a highly normalized relational database hosted on an Oracle server behind a firewall. The production database is optimized for warehousing public data from NCBI, loading data, automatic data updates from internal JGI systems as well indexing for optimal search performance. The internal database schema is not publicly accessible. The public web interface is powered by a normalized database and Apache Lucene indexes to support fast, complex searches and data downloads. Standard HTML and JavaScript technologies are used on our web pages, and Java-based technologies are used to drive our back-end business logic and web servers.
GOLD’s production database is updated in real time, from user-entered data as well as from automated processes. Because of numerous simultaneous data input streams, Apache Lucene indexes are updated in real-time, as well as re-generated nightly from the production database, to ensure all updates are caught. Precomputed statistics are automatically generated daily, to populate different sections of GOLD site, including our home page tables, NCBI import tracker, distribution graphs, and statistics pages.
BROWSING AND SEARCHING GOLD
Public data in GOLD are freely accessible through the website (https://gold.jgi.doe.gov). The homepage provides key statistics on different entities, along with links to related metadata. For example, the number of Biosamples associated with Environment, Host-Associated or Engineered ecosystems are displayed. A user can click on each of these classifications and look at the list of Biosamples from that specific ecosystem. The total count of public Studies, Biosample, Organisms, Sequencing Projects and Analysis Projects is also displayed on the top left corner. These numbers are updated daily and are presented as clickable links. GOLD imports genome and metagenome projects from NCBI periodically. The home page has a dedicated NCBI Import Tracker that displays the number of projects that are in NCBI, GOLD and IMG at any given time. The projects in the tracker are broken down by prokaryotes, eukaryotes, viruses, metagenomes and metatranscriptomes. It is to be noted that the number of projects in GOLD and IMG on a given day is usually less than that of NCBI. This is due to our focus on prioritizing data relevant to diversity, energy and the environment. That being said, we regularly receive and prioritize user requests to add projects of interest into GOLD/IMG systems. Another important metadata tracker available on the home page is the number of bacterial and archaeal type strains and their WGS projects in GOLD and IMG. A Type strain is a strain that represents an organism and is the strain used when a species is first described. They are important reference points in taxonomy and comparative genomics analyses and usually have a significant amount of metadata associated with them. Below we describe some of the menu tabs through which a user can browse the database.
Search
The Search tab has two options: Advanced Search and Metadata Search. While there are different ways to search the database, one of the most popular and commonly used features is the ‘Advanced Search.’ It provides a quick and easy way to search across all the different levels using several metadata fields. Users can add multiple search filters to their query or selectively remove them after a search is completed, giving them an opportunity to refine searches. The Metadata Search option uses a combination of tables and graphs to present search results using specific metadata fields.
Distribution Graphs
The Distribution Graphs section gives a top-down view of different fields in which the user receives a snapshot of the metadata scale and their diversity. A brief summary of the different types of Sequencing Projects for genome and metagenome projects and their sequencing statuses are displayed as pie-charts in the Distribution Graphs section. Additionally, a user can look at the phylogenetic distribution of projects both as pie charts and in hyperlinked expandable table formats. Finally, it also shows the breakdown of Biosamples by each of the five different Ecosystem levels.
Biogeographical Metadata
The Biogeographical Metadata section shows the isolation location of Biosamples and Organisms as placeholders on interactive maps. This part of the user interface received a fresh makeover in this latest release where ecosystem classification is used to filter select Biosample or Organisms that are displayed on a dynamic map. More description of this Biogeographical Metadata section and details about the update is provided later in the feature update section.
SRA Explorer
NCBI Sequence Read Archive (SRA) stores a collection of sequence data from all forms of life including metagenomic samples. The NCBI SRA Explorer provides a way to search SRA data. Using a combination of free text fields (such as organism name or SRA Experiment Title) and fields with predefined or controlled vocabulary based values (such as sequencing technologies or library strategies), the different types of data available in SRA can be reviewed. The SRA Explorer also has a filter where users can see which SRA data are currently present in GOLD. While we do import genomes and metagenomes from NCBI on a regular basis, our import process is often based on specific filters which change over time. For example, only metagenomes that are sequenced on the Illumina platform are currently imported. Through the SRA Explorer, Illumina metagenomes can be selected, as well as those sequenced by other platforms, such as 454 or PacBio etc.
Statistics
The Statistics tab contains graphical descriptions of several Sequencing Project metadata fields. This ranges from the phylogenetic distribution and relevance of bacterial genome projects, breakdown of projects by sequencing centers or the yearly growth of sequencing projects by organism domain. The charts and pre-computed graphs on this page are updated every week to ensure that users have access to the latest data in GOLD.
Downloads
The Downloads page gives users the ability to access different types of data, ranging from a list of several controlled vocabulary terms, pubic metagenome and metatranscriptome projects from SRA that were annotated in IMG or a data dump of GOLD-containing select fields from various database entities. These files can be downloaded in Excel format and the underlying data is updated daily. We recently added this section and more details on this Downloads page is provided in the ‘Feature Updates’ section of the manuscript.
CREATING SEQUENCING PROJECTS IN GOLD
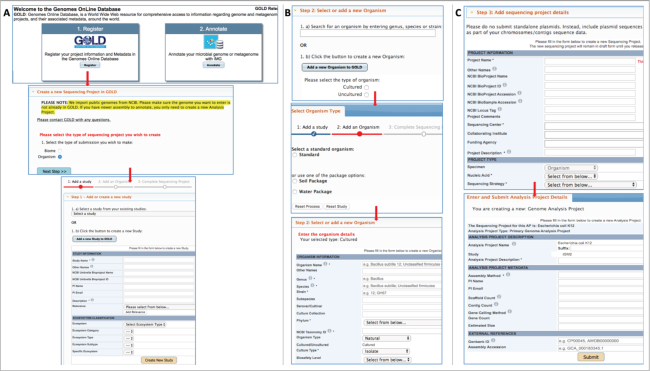
There are two broad aspects of accessing the database: a login-free public access and a secure login option for users who want to enter private individual projects. A user account is required to access this feature. Figure 1 summarizes the steps to create an isolate genome Sequencing and Analysis Project. To initiate the project entry process and go to the main project creation page, a user should begin by clicking the ‘Register’ button on the homepage and then select ‘Create a new Sequencing Project in GOLD’ (Figure 1). After clicking on the ‘Organism’ radio button, which selects Organism-specific forms behind the scenes, users need to define their Study. A Study describes the overall objective of a research proposal or a set of sequencing projects and is placed at the top of the GOLD’s four-level classification system. Once a Study is created, the next step is to define an Organism, which can be done by selecting from >386 000 public Organisms currently available in the database or by creating a new Organism using the intuitive Organism entry form. At this step, a user has the option to choose from a list of metadata packages such as Soil, Water, Hydrocarbon etc. to include additional environment-specific metadata or select the standard set of metadata fields. While some of the metadata fields are optional for an Organism, there are certain fields such as Genus, Species, Strain, phylogeny, NCBI Taxonomy ID etc. that are mandatory in the Organism form. After an Organism is defined, a user can proceed to create a Sequencing Project. The form to define a SP is broadly divided into three sections, namely: Project Information, Project Type and Sequencing Information under which the fields for Project Name, Nucleic Acid, Sequencing Strategy, Sequencing Center, Project Description and Sequencing Status are mandatory. Finally, the user needs to define an Analysis Project, which is required for submitting sequence data to IMG. Detailed, step-by-step instructions on how to enter different types of Sequencing and Analysis Projects are available in the Project Entry Help Document, which can be accessed through the Help menu bar or directly by using the URL: https://gold.jgi.doe.gov/resources/project_help_doc.pdf.
Figure 1.
Steps to create isolate WGS projects. (A) Study creation form is accessed by clicking on ‘Register’ button on homepage followed by selecting ‘Organism’ radio button. (B) Step 2 involves creating an Organism (C) Details about Sequencing and Analysis Projects are entered in steps 3 and 4 respectively.
UPDATES SINCE LAST RELEASE
Expanded downloads
One of the main goals of the database is to provide easy access to the metadata of genomes and metagenomes for the scientific community. This is aligned with the Findable, Accessible, Interoperable and Reusable (FAIR) guidelines (26). As the number of projects and metadata fields has increased over the years, so has our commitment to sharing this information with users. We have added a new Downloads section to the homepage where users have access to four separate files that are updated daily. One file contains an export of the public data, along with a pre-selected list of key metadata fields. Each tab in this file represents the four levels of the database: Study, Biosample/Organism, Sequencing Project and Analysis Project. The second file provides a list of public SRA-based metagenome and metatranscriptome projects that are in GOLD and IMG with some key identifiers. The third Excel file contains a list of all the Controlled Vocabulary (CV) tables and their respective CV terms. There are currently 3873 CV terms from 76 CV tables that are spread across all of the four different levels. For example, ‘Relevance’ of a Study or Sequencing Project is an important metadata descriptor which summarizes the broad goal or application of a particular research project. For a single Study/Project, a user can select one or more ‘Relevance’ terms from a list of 175 CV terms. The fourth downloadable file contains a unique list of GOLD’s five-level ecosystem classification paths. These downloadable files are not aimed at creating a database dump, but their purpose is to provide easy access to the bulk of normalized data to our users.
Search results download
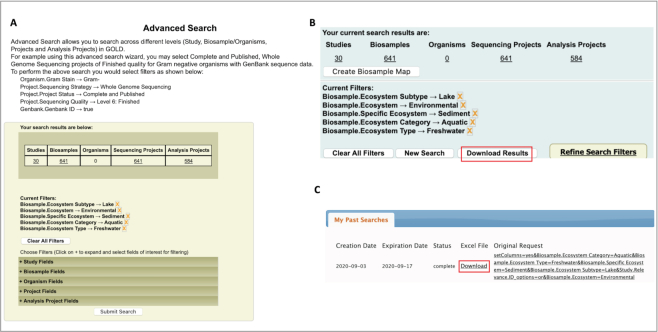
The Advanced Search feature was developed to help users search and apply filters to query a wide range of project features and metadata fields. Since its inception, this has been one of the most widely accessed features in GOLD. While this was an excellent first step for a user to seamlessly search across all of the four levels at the same time, it did not allow users to download and save their search results. The current version addressed this by adding a new feature in which users can download the results of their advanced search with the click of a button (Figure 2). The search results are provided as an Excel file that is saved in a new section called ‘My Past Searches’ which can be accessed on the menu bar after a user is logged in. These downloadable search result files are stored for 2 weeks before the link expires. While the downloadable file is purged, the user searches are available in the ‘My Past Searches’ section as a clickable link (Figure 2C); thus, if a user decides to execute the same search at a later time, one can simply click on the link and they will be taken to the Advanced Search page with preselected filters from the saved search. It should be noted that the maximum allowable row count for this downloadable file is 20 000; in order to avail this feature, a user needs to refine his search filters so that the results do not exceed 20 000 rows.
Figure 2.
Advanced Search results download. (A) Performing an Advanced Search query by selecting filters for freshwater lake sediment metagenomes. (B) Search results are shown with the number of Studies, Biosamples, Sequencing Projects and Analysis Projects appearing as clickable numbers. The option to download the search results is highlighted. (C) ‘My Past Searches’ tab displaying the search history with creation date, result expiration date, status of search, search query and a link to download the results as an Excel file.
Environmental packages
Minimum Information about any (x) Sequence (MIxS) environmental packages, released by the Genomic Standards Consortium (GSC), provide the list of fields and parameters to describe a biological sample and other related information about sequencing (27). Incorporation of the specific MIxS environmental packages in GOLD along with a standard set of metadata fields gives the opportunity to capture environment-specific details. In the current version, we have updated the previously incorporated MIxS Soil and Water packages from version 4 to the current version 5. We also added five more MIxS packages; namely, Plant-associated, Hydrocarbon resources-cores and Hydrocarbon resources-fluids/swabs, Sediment and Microbial mat/biofilm.
The MIxS Plant-associated package has 166 fields to describe samples that are from plants. Some of the fields such as plant product (substance produced by the plant, where the sample was obtained from) and various treatment regimens including fertilizer regimen (information about treatment involving the use of fertilizers), growth hormone regimen (information about treatment involving the use of growth hormones), antibiotic regimen (information about treatment involving antibiotic administration) are specific to the Plant-associated package and are useful to contextualize plant samples. Samples from hydrocarbon-rich environments such as oil reservoir, gas reservoir, oil sand, and coal bed are described using two similar, but distinct MIxS packages: Hydrocarbon resources-cores package (HCR-C) and Hydrocarbon resources-fluids/swabs package (HCR-FS). The HCR-C package contains 173 fields and the HCR-FS package contains 177 fields; they share 163 fields between them. Fields such as permeability (the measure of the ability of a hydrocarbon resource to allow fluids to pass through it) and source rock lithology are unique in HCR-C, while water cut [the current amount of water (%) in a produced fluid stream or the average of the combined streams] and water production rate (water production rates per well) are unique to HCR-FS. Thus, unique fields among these two MIxS HCR packages are useful in describing specific biological samples from their respective environments. The MIxS Sediment package consists of 161 fields to describe sediment samples from various environments including marine, lake, estuary, river, etc. The Sediment package shares 153 fields with the MIxS water package. However, the remaining eight unique fields help to describe and distinguish sediment samples from water, particularly: sediment type, porosity, particle classification and water content. The MIxS Microbial mat/biofilm package contains a list of 155 fields to describe microbial mat and biofilm samples studied from various environments including hot springs, deep subsurface biofilm, stromatolite mat and ice sheets. A total of 150 out of 155 fields of the MMB package are also provided in the Water package. Five fields namely water content, methane, total carbon, total organic carbon and total nitrogen content provided in the MMB package differentiate it from the Water package and are useful fields to describe the Organisms and Biosample isolated from microbial mat and biofilm environments.
Ecosystem classification explorer
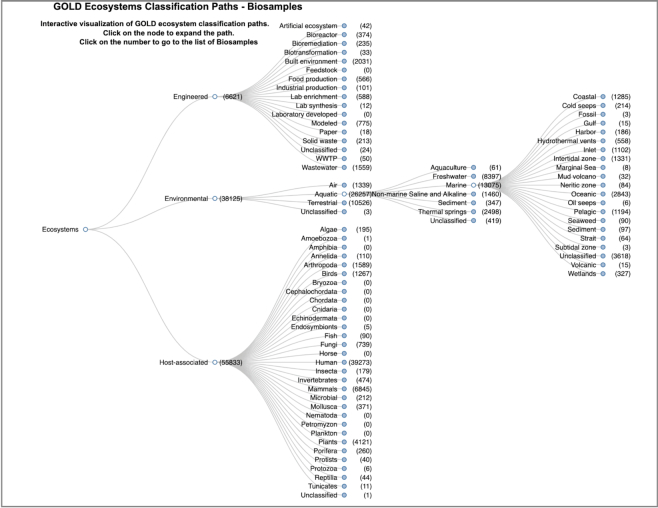
The five-level ecosystem classification system describes the environment from which a particular Biosample or Organism is sourced. At the top of this classification chain is Ecosystem, which consists of Environmental, Host-Associated and Engineered, which are further classified into four different levels, namely Ecosystem Category, Ecosystem Type, Ecosystem Subtype and Specific Ecosystem. All Biosamples and most Organisms have a defined ecosystem classification path. For example, a Biosample from a lake sediment environment will have the following Ecosystem Classification: Ecosystem: Environment, Ecosystem Category: Aquatic, Ecosystem Type: Freshwater, Ecosystem Subtype: Lake and Specific Ecosystem: Sediment. In order to provide an interactive visualization of all the different combinations of nearly 800 distinct ecosystem classification paths, we developed a new Ecosystem Classification Explorer (Figure 3). This new feature can be accessed from the main landing page of the website. A user can click on Biosample Ecosystem paths or Organism Ecosystem paths to view the ecosystem classifications of the respective entities. Each node of the interactive Ecosystem Tree can be expanded further to view its connected leaves. The number of public Biosamples or Organisms connected to each node is displayed as a hyperlinked number within parenthesis. Clicking on these numbers takes the user to a page from where they can access the linked entities as well as their associated Studies, Sequencing and Analysis Projects.
Figure 3.
Ecosystem Classification Explorer. Interactive visualization of GOLD Ecosystem classification path for Biosample with few representative nodes expanded for reference. Number of Biosamples with a particular ecosystem classification is shown within parenthesis.
Interactive Biosample/Organism map
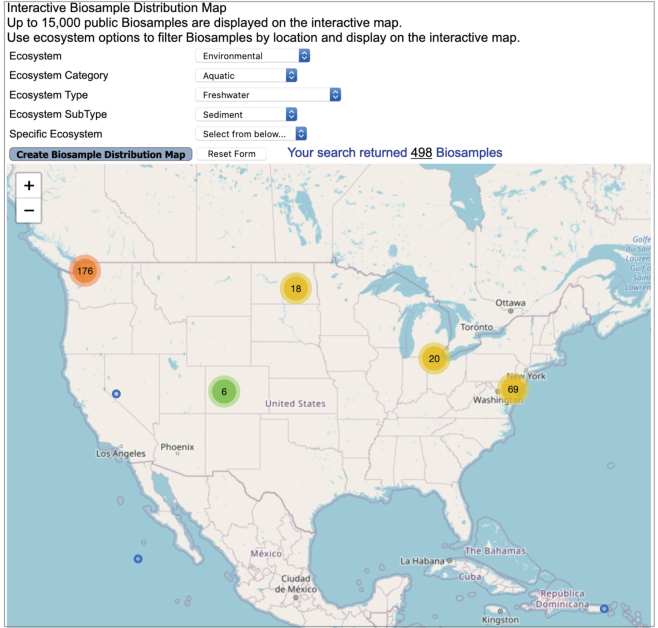
In the latest release, we revamped the Biogeographical Metadata feature to include interactive, user-friendly Biosample and Organism Distribution maps. Each of these maps now allows a user to select different ecosystem classification levels as a filter to display Biosample and Organisms on an interactive biogeographical map. A user can zoom in or out to focus on a specific location and then click on the numeric placeholders in the map to reveal the particular Biosample or Organism that was isolated from that region. Figure 4 shows the distribution of freshwater lake Biosamples spread across the USA and Canada.
Figure 4.
Interactive Biosample distribution map. Distribution of freshwater lake Biosamples over parts of the USA and Canada. Number of Biosamples from a specific location is displayed in a heatmap format where green represents a small number of Biosamples, orange depicts a high number whereas yellow indicates an intermediate number.
Updated help pages
We have updated the Help Pages with more useful information to further improve user experience. For example, our FAQ section was updated to include detailed answers to the most common questions that we receive from users. Additionally, the GOLD Terminology page was updated with descriptions of several terms that a new user may have trouble understanding or not know about. We believe that the above updates will hasten the project entry process significantly, since a user may avoid needing to contact curators with common questions. If a user still has a question or prefers to contact GOLD with his feedback or concern, we have added a direct link within the Help Page where they can send a detailed message.
Mapping Environment Ontology triad to GOLD metadata
In recent years we have seen a large increase in the volume of microbiome sequence data produced by different institutions on multiple sequencing platforms and analyzed using their favorite bioinformatics tools and databases. While this has led to several key individual discoveries, the lack of interoperability and crosstalk among these databases continues to be a key bottleneck for meta-analysis or cross-study comparisons in microbiome research (28). To address this drawback, we have embarked on an effort to curate Biosamples with Environment Ontology (EnvO) terms. EnvO is a resource that provides a semantically controlled description of environmental entities (14).
EnvO describes environmental samples using three primary axes: the biome, environmental feature and environmental material that are mandated by the GSC MIxS (version 5) as a triad, namely broad-scale environmental context, local environmental context and environmental medium respectively (https://gensc.org/mixs/). In GOLD, the environmental features of a Biosample or Organism are described by a five-level ecosystem classification path that includes Ecosystem, Ecosystem Category, Ecosystem Type, Ecosystem Subtype and Specific Ecosystem. For example, for sediment Biosample from a lake, the five-level classification would be Environmental/Aquatic/Freshwater/Lake/Sediment while the MIxS-EnvO triad can be freshwater lake biome (broad-scale environmental context), Mesotrophic Lake (local environmental context) and lake sediment (environmental medium). This example shows that the GOLD five-level classification path provides an option to mine all aquatic Biosamples at the Ecosystem Category level and Lake Biosamples at the Ecosystem Subtype level. Whereas EnvO provides an additional option to choose a specific type of lake from its local environmental context. Thus, using GOLD metadata terms in conjunction with MIxS-EnvO triads gives users more options for data mining.
Mapping of the MIxS-EnvO triad for each of the Biosamples was carried out by taking various GOLD metadata fields into account. Fields like sample collection site, habitat, identifier, Biosample name, Biosample description and the GOLD ecosystem classification path terms were used in MIxS-EnvO triad curation. In the current version, 40 619 Biosamples were mapped to EnvO triad and our initial focus was on the environmental samples. A total of 38 broad-scale environmental contexts, 227 local environmental contexts and 115 environmental medium EnvO terms were used to curate 40 619 Biosamples. All of the Biosamples that are classified under the ecosystem category Aquatic are mapped to 24 different EnvO broad-scale environmental contexts including freshwater biome, freshwater lake biome, freshwater river biome and marine biome. Similarly, all the terrestrial Biosamples are described by 17 different EnvO broad-scale environmental contexts including desert biome, forest biome, cropland biome, grassland biome and urban biome. Three biomes namely, polar biome, polar desert biome and urban biome have been used to describe both aquatic and terrestrial Biosamples. Aquatic and Terrestrial are two different broad GOLD Ecosystem categories that are described by 38 different MIxS/EnvO broad-scale environmental context terms for mapped Biosamples, demonstrating metadata enrichment by the combined use of both GOLD and EnvO.
Search engine update
The growing amount of data and increased complexity of the metadata fields were causing the search queries to run slowly. To address this problem, we have updated our search engine to provide an improved search experience by leveraging the capabilities of the Apache Lucene search engine software library (https://lucene.apache.org/). This has significantly improved overall search speeds across the website, leading to less waiting time between multiple queries and improved user experience. For example, searches on SRA Explorer that took ∼50 s are now returning results in a few seconds.
UPCOMING FEATURES
Exponentially growing genome and metagenome projects around the world and the appreciation for metadata and metadata standards by the research community, necessitates both to curate/import a large number of projects as well as expand and organize the metadata as per evolving community standards. Our future plans include the continuation of importing genome and metagenome projects with pertinent metadata as well as the following extensions.
NCBI virus genomes
The current release includes close to 8000 virus genomes. There are over 30 000 viral genomes at NCBI, including 12 000 RefSeq viral genomes. Our semi-automated isolate genome imports from NCBI rely on the NCBI BioProject/BioSample accessions, Assembly Accessions and GenBank IDs for a given genome. However, virus genomes at NCBI do not necessarily exist with these well-defined accessions that are standard for isolate genomes. As a result, our existing import pipeline is not suitable for handling virus genomes from NCBI. To address this challenge, we plan to work on a separate virus import process to facilitate the import of NCBI viral genomes.
Improving semi-automated metadata curation
One key bottleneck to including thousands of new sequencing projects and their metadata in GOLD is the time and effort required for manual curation. While some level of manual intervention is necessary to maintain the quality and accuracy of data, its volume cannot be increased significantly without automating some of the redundant curation steps. Over the last few years, we have automated several different parts of the project-addition process including parsing of NCBI BioSample attributes, automatically mapping them to GOLD metadata fields, semi-automatically mapping institutes and sequencing instruments to curated sequencing centers and sequencing technologies etc. We hope to incorporate text mining and machine learning algorithms to further automate additional curation steps. Additionally, we are working on mapping latitude/longitude values obtained from public samples to their respective geographic locations and/or countries and displaying them on a map.
Expanding downloadable search results
In the current implementation, a downloadable search results file is limited to a maximum of 20 000 records. This has been a very useful feature for our users, and we understand the need to increase this number. Along these lines we are investigating options to expand this limit so that users can download more of their search results. We also plan to increase our storage capacity so that search results are saved longer than the current limit of 2 weeks, after which they expire.
Additional environmental packages
As described above, we expanded the number of environmental packages from 2 to 7, in the current release. This has significantly increased the number and diversity of metadata fields, specifically for Biosample and Organisms. We plan to implement additional MIxS environmental packages in the future to support our users who study samples from specific environments such as air or wastewater sludge.
MIxS-ENVO triad assignment to existing GOLD Biosamples
As outlined above nearly 41,000 environmental Biosamples have been curated with MIxS-ENVO triads. We plan to continue curating all of the remaining Biosamples.
ACKNOWLEDGEMENTS
The authors would like to thank our broad user base and members of the research community for submitting projects and metadata to GOLD. We are also thankful to members of the JGI project management team, microbial genomics and metagenomics group and our leadership team for their constant support, feedback and encouragement. We thank members of the microbial genomics and metagenomics research and standards communities for their constant support, feedback and helpful discussions. Visualizations displayed in this manuscript have been created using MS-Office Suite, GNU Image Manipulation Program (GIMP) v 2.10 and Adobe Acrobat Professional.
Contributor Information
Supratim Mukherjee, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Dimitri Stamatis, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Jon Bertsch, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Galina Ovchinnikova, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Jagadish Chandrabose Sundaramurthi, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Janey Lee, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Mahathi Kandimalla, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
I-Min A Chen, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Nikos C Kyrpides, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
T B K Reddy, DOE Joint Genome Institute, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
FUNDING
U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility [DE-AC02-05CH11231]. Funding for open access charge: Office of Science of the U.S. Department of Energy [DE-AC02-05CH11231]; J.C.S. is supported by National Microbiome Data Collaborative (NMDC); Genomic Science Program in the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (BER) [DE-AC02-05CH11231 to LBNL, 89233218CNA000001 to LANL, DE-AC05-00OR22725 to ORNL, DE-AC05-76RL01830 to PNNL].
Conflict of interest statement. None declared.
REFERENCES
- 1. Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W.. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017; 2:1533–1542. [DOI] [PubMed] [Google Scholar]
- 2. Alneberg J., Karlsson C.M.G., Divne A.-M., Bergin C., Homa F., Lindh M.V., Hugerth L.W., Ettema T.J.G., Bertilsson S., Andersson A.F. et al.. Genomes from uncultivated prokaryotes: a comparison of metagenome-assembled and single-amplified genomes. Microbiome. 2018; 6:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parks D.H., Chuvochina M., Chaumeil P.-A., Rinke C., Mussig A.J., Hugenholtz P.. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020; 38:1079–1086. [DOI] [PubMed] [Google Scholar]
- 4. Murray A.E., Freudenstein J., Gribaldo S., Hatzenpichler R., Hugenholtz P., Kämpfer P., Konstantinidis K.T., Lane C.E., Papke R.T., Parks D.H. et al.. Roadmap for naming uncultivated Archaea and Bacteria. Nat. Microbiol. 2020; 5:987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Madin J.S., Nielsen D.A., Brbic M., Corkrey R., Danko D., Edwards K., Engqvist M.K.M., Fierer N., Geoghegan J.L., Gillings M. et al.. A synthesis of bacterial and archaeal phenotypic trait data. Sci. Data. 2020; 7:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kyrpides N.C. Genomes OnLine Database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics. 1999; 15:773–774. [DOI] [PubMed] [Google Scholar]
- 7. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Verezemska O., Isbandi M., Thomas A.D., Ali R., Sharma K., Kyrpides N.C. et al.. Genomes OnLine Database (GOLD) v.6: data updates and feature enhancements. Nucleic Acids Res. 2017; 45:D446–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mukherjee S., Stamatis D., Bertsch J., Ovchinnikova G., Katta H.Y., Mojica A., Chen I.-M.A., Kyrpides N.C., Reddy T.. Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic Acids Res. 2019; 47:D649–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reddy T.B.K., Thomas A.D., Stamatis D., Bertsch J., Isbandi M., Jansson J., Mallajosyula J., Pagani I., Lobos E.A., Kyrpides N.C.. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015; 43:D1099–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunivin T.K., Yeh S.Y., Shade A.. A global survey of arsenic-related genes in soil microbiomes. BMC Biol. 2019; 17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guittar J., Shade A., Litchman E.. Trait-based community assembly and succession of the infant gut microbiome. Nat. Commun. 2019; 10:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Field D., Garrity G., Gray T., Morrison N., Selengut J., Sterk P., Tatusova T., Thomson N., Allen M.J., Angiuoli S.V. et al.. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 2008; 26:541–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Field D., Sterk P., Kottmann R., De Smet J.W., Amaral-Zettler L., Cochrane G., Cole J.R., Davies N., Dawyndt P., Garrity G.M. et al.. Genomic standards consortium projects. Stand. Genomic Sci. 2014; 9:599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buttigieg P.L., Morrison N., Smith B., Mungall C.J., Lewis S.E. the ENVO Consortium . The environment ontology: contextualising biological and biomedical entities. J. Biomed. Semant. 2013; 4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., Sayers E.W.. GenBank. Nucleic Acids Res. 2016; 44:D67–D72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen I.-M.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J.R., Seshadri R. et al.. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019; 47:D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen I.-M.A., Markowitz V.M., Chu K., Palaniappan K., Szeto E., Pillay M., Ratner A., Huang J., Andersen E., Huntemann M. et al.. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017; 45:D507–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huntemann M., Ivanova N.N., Mavromatis K., Tripp H.J., Paez-Espino D., Palaniappan K., Szeto E., Pillay M., Chen I.-M.A., Pati A. et al.. The standard operating procedure of the DOE-JGI Microbial Genome Annotation Pipeline (MGAP v.4). Stand. Genomic Sci. 2015; 10:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chuvochina M., Rinke C., Parks D.H., Rappé M.S., Tyson G.W., Yilmaz P., Whitman W.B., Hugenholtz P.. The importance of designating type material for uncultured taxa. Syst. Appl. Microbiol. 2019; 42:15–21. [DOI] [PubMed] [Google Scholar]
- 22. Konstantinidis K.T., Rosselló-Móra R., Amann R.. Uncultivated microbes in need of their own taxonomy. ISME J. 2017; 11:2399–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schriml L.M., Chuvochina M., Davies N., Eloe-Fadrosh E.A., Finn R.D., Hugenholtz P., Hunter C.I., Hurwitz B.L., Kyrpides N.C., Meyer F. et al.. COVID-19 pandemic reveals the peril of ignoring metadata standards. Sci. Data. 2020; 7:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ivanova N., Tringe S.G., Liolios K., Liu W.-T., Morrison N., Hugenholtz P., Kyrpides N.C.. A call for standardized classification of metagenome projects. Environ. Microbiol. 2010; 12:1803–1805. [DOI] [PubMed] [Google Scholar]
- 25. Bowers R.M., Kyrpides N.C., Stepanauskas R., Harmon-Smith M., Doud D., Reddy T.B.K., Schulz F., Jarett J., Rivers A.R., Eloe-Fadrosh E.A. et al.. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017; 35:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood-Charlson E.M., Anubhav Auberry D., Blanco H., Borkum M.I., Corilo Y.E., Davenport K.W., Deshpande S., Devarakonda R., Drake M. et al.. The National Microbiome Data Collaborative: enabling microbiome science. Nat. Rev. Microbiol. 2020; 18:313–314. [DOI] [PubMed] [Google Scholar]
- 27. Yilmaz P., Kottmann R., Field D., Knight R., Cole J.R., Amaral-Zettler L., Gilbert J.A., Karsch-Mizrachi I., Johnston A., Cochrane G. et al.. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any sequence (MIxS) specifications. Nat. Biotechnol. 2011; 29:415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Su X., Jing G., Zhang Y., Wu S.. Method development for cross-study microbiome data mining: challenges and opportunities. Comput. Struct. Biotechnol. J. 2020; 18:2075–2080. [DOI] [PMC free article] [PubMed] [Google Scholar]