Abstract
PULs (polysaccharide utilization loci) are discrete gene clusters of CAZymes (Carbohydrate Active EnZymes) and other genes that work together to digest and utilize carbohydrate substrates. While PULs have been extensively characterized in Bacteroidetes, there exist PULs from other bacterial phyla, as well as archaea and metagenomes, that remain to be catalogued in a database for efficient retrieval. We have developed an online database dbCAN-PUL (http://bcb.unl.edu/dbCAN_PUL/) to display experimentally verified CAZyme-containing PULs from literature with pertinent metadata, sequences, and annotation. Compared to other online CAZyme and PUL resources, dbCAN-PUL has the following new features: (i) Batch download of PUL data by target substrate, species/genome, genus, or experimental characterization method; (ii) Annotation for each PUL that displays associated metadata such as substrate(s), experimental characterization method(s) and protein sequence information, (iii) Links to external annotation pages for CAZymes (CAZy), transporters (UniProt) and other genes, (iv) Display of homologous gene clusters in GenBank sequences via integrated MultiGeneBlast tool and (v) An integrated BLASTX service available for users to query their sequences against PUL proteins in dbCAN-PUL. With these features, dbCAN-PUL will be an important repository for CAZyme and PUL research, complementing our other web servers and databases (dbCAN2, dbCAN-seq).
INTRODUCTION
CAZymes (Carbohydrate Active EnZymes) are enzymes that act upon specific glycosidic linkages to degrade, synthesize or modify polysaccharides (1). They are found in essentially all organisms on Earth, and particularly abundant in plant-associated microbes. The reason is that these microbes feed on plant cell-wall carbohydrates, which requires a significant proportions of their genomes encoding CAZymes (2).
In the past 15 years, experimental studies have shown that CAZymes often work in conjunction with other CAZymes and proteins to fully digest complex carbohydrates, and that genes encoding these proteins tend to form physically linked gene clusters (3). In Bacteroidetes genomes, these gene clusters have been termed ‘Polysaccharide Utilization Loci’ (4), or PULs, and contain pairs of homologs of signature genes as their hallmark: an outer membrane Ton-B dependent transporter (TBDT, susC) and a cell surface glycan-binding protein (SGBP, susD) (5). As Bacteroidetes is one of the most important bacterial phyla in the human gut, susC-susD PULs are key players in modulating host-diet-microbiome interactions.
However, PULs have been increasingly discovered in other bacterial phyla (6–8), such as Proteobacteria (9), Firmicutes (10), Actinobacteria (11), metagenomes (12), and even Archaea (13) from different ecological environments including ocean, soil, rhizosphere, termite gut, cow rumen, etc. Notably, another term ‘CUT’, short for CarbohydrateUtilization containingTBDT loci (14), also appeared in the literature for PULs containing no susD. However, all these experimentally verified PULs/CUTs except those susC-susD PULs from Bacteroidetes (see below) are buried in the literature.
We have previously developed dbCAN (15) to predict CAZymes, and CGC-Finder (16) to identify CAZyme gene clusters (CGCs). CGC is a term that we coined in 2018 for physically linked gene clusters that encode at least one CAZyme, one transporter, one transcriptional regulator, and one signaling transduction protein (16). There are also other bioinformatics tools recently developed to predict PULs from genomic data (2,3,17). Unlike experimentally characterized PULs, CGCs are computationally predicted without known carbohydrate substrate information. A comprehensive repository of functionally characterized PULs will be extremely useful to help predict a substrate for CGCs, and thus facilitate the experimental characterization of new PULs in nature. Extending our previous work on CAZymes and CGCs (2,15,16), we have developed dbCAN-PUL (http://bcb.unl.edu/dbCAN_PUL/) to collect all experimentally verified PULs, by manually curating 1,100+ PubMed papers (see below). We have further obtained the corresponding GenBank sequences of these PULs and presented a variety of metadata and sequence-derived data on the website.
The only similar web resource as dbCAN-PUL is the Polysaccharide Utilization Database, or PULDB (http://www.cazy.org/PULDB/) (18). It is a repository of PULs that must contain susC-susD gene pairs from Bacteroidetes. PULDB largely consists of predicted PULs (98.5% of PULs are predicted and not curated from literature), and is taxonomically biased towards Bacteroidetes (>99% of PULs are from the Bacteroidetes phylum). Of the 1.5% literature curated PULs, 100% are from Bacteroidetes, and only 28.9% have substrate information. Overall, PULDB is centered on susC-susD gene pairs from Bacteroidetes and not all PULs contain CAZymes.
dbCAN-PUL aims to provide a comprehensive online database of experimentally characterized CAZyme-containing PULs with known target substrates for efficient retrieval of annotated sequences and metadata. We were motivated by the fact that there exist no repositories that: (i) only experimentally characterized PULs are collected; (ii) all PULs must have a target substrate elucidated; (iii) all PULs must contain CAZyme genes; (iv) PULs are not restricted to Bacteroidetes but broadly cover characterized PULs from all taxa in the literature.
DATABASE CONTENT
Literature curation and data collection
We performed two rounds of queries against PubMed database using (i) a general query and (ii) a query that included specific substrates:
General query: (oligosaccharide [Title/Abstract] OR polysaccharide [Title/Abstract] OR carbohydrate [Title/Abstract]) AND (utilization [Title/Abstract] OR degrad* [Title/Abstract] OR catabolism [Title/Abstract]) AND (cluster [Title/Abstract] OR locus [Title/Abstract] OR loci [Title/Abstract] OR operon [Title/Abstract])
Specific query: (SPECIFIC SUBSTRATE)AND (utilization [Title/Abstract] OR degrad* [Title/Abstract] OR catabolism [Title/Abstract]) AND (cluster [Title/Abstract] OR locus [Title/Abstract] OR loci [Title/Abstract] OR operon [Title/Abstract])
The list of substrate keywords (Table 1) in the Specific query do not mean to be the most complete. Instead, it was primarily used to complement the General query, categorize PULs based on substrates, and can be expanded in our future updates.
Table 1.
Substrate keywords for specific query against PubMed
| β-glucans & hemicelluloses | α-glucans | Algal Glycans | Animal & Fungal Glycans | Other Glycans |
|---|---|---|---|---|
| Arabinogalactan | Amylopectin | Alginate | Alpha-mannan | Fructan |
| Arabinoxylan | Amylose | Carrageenan | Chitin | Inulin |
| Beta-glucan | Arabinan | Galactan | Chondroitin sulfate | Levan |
| Beta-mannan | Dextran | Laminarin | Heparin | Homogalacturonan |
| Cellobiose | Glycogen | Porphyran | Hyaluronan | Pectic galactan |
| Galactomannan | Pullulan | Ulvan | Mucin | Rhamnogalacturonan |
| Glucomannan | Starch | N-glycans | Melibiose | |
| Xylan | O-glycans | Raffinose | ||
| Xyloglucan | Sialoglycoconjugates |
Papers that were hits from the queries were retrieved and manually curated using the following criteria: (i) organism(s) of study were prokaryotic (Bacteria or Archaea), (ii) the gene cluster in question contained at least one CAZyme and (iii) investigators experimentally verified that at least one gene in the PUL that synthesizes or degrades a carbohydrate substrate with experimental methods, such as enzymatic assays, sugar utilization assay, RNA-seq or expression microarrays. Regarding the last criterion, we also added ‘sequence homology analysis’ as one of the characterization methods. PULs characterized by ‘sequence homology analysis’ were found with a high sequence homology to previously characterized PULs by investigators in their papers. These PULs differ from the large number of computationally predicted PULs in PULDB, as the homology search was carried out by the original investigators followed by their manual inspection and peer-reviewed publication. It should be noted that dbCAN-PUL focuses on CAZyme-containing PULs, in which PULs must contain at least one CAZyme but do not have to contain susC-susD gene pairs. Additionally, we note that the final dbCAN-PUL database contains more target substrates than listed in Table 1 due to the search terms matching papers across a wide spectrum of possible target substrates from the General query.
A total of 1,113 papers were yielded from both rounds of the PubMed searches (as of January 2020), and after manual curation, 294 papers remained meeting our curation criteria (Table 2). These papers contained a total of 602 PULs from prokaryotic genomes or metagenomic sequences across 87 genera, 10 phyla (including 1 archaeal phylum), 74 characterization methods and 126 unique substrates. Of the 602 PULs, 104 were biosynthetic PULs and 498 were degradative PULs. Additionally, there is an overlap between PULs of dbCAN-PUL and those of PULDB (Figure 1). Approximately 469 PULs from PULDB were literature curated but not necessarily experimentally verified or with a substrate characterized. For example, we excluded Zobelia galactivorans DsijT PULs curated from (19) which are included in PULDB, because substrates were not characterized experimentally; the authors of (19) had inferred putative substrates from CAZyme family activities. We also consolidated PULs from between two assemblies of Bacteroides ovatus ATCC 8483, rather than kept PULs of both like in PULDB, to reduce redundancy. Thus, 138 PULs overlapped between PULDB and dbCAN-PUL (Table 2 and Figure 1).
Table 2.
Comparison of entries in PULDB vs. dbCAN-PUL
| Counts of data entries | PULDB | dbCAN-PUL |
|---|---|---|
| # of Literature curated PULs | 691 | 602 |
| # of Literature curated PULs with CAZymes | 469 | 602 |
| # of Literature curated PULs with CAZymes + experimentally verified substrate | 138 | 602 |
| # of Papers containing literature curated PULs | 28 | 294 |
| # of Phyla of experimentally verified + substrate identified CAZyme containing PULs | 1 | 10 |
| # of Genera of experimentally verified + substrate identified CAZyme containing PULs | 5 | 87 |
| # of Species/metagenomes of experimentally verified + substrate identified CAZyme containing PULs | 10 | 173 |
| # of General substrate categories | 37 | 126 |
| # of Unique substrate categories (not found in the other database) | 12 | 101 |
Figure 1.
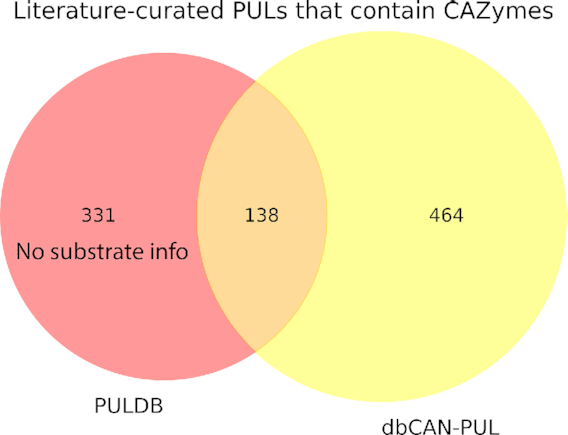
Venn diagram comparing the number of literature-curated PULs in PULDB and dbCAN-PUL. PULDB contains 469 literature-curated PULs encoding CAZymes. However, 331 of them do not have curated substrates. The remaining 138 PULs overlap with dbCAN-PUL, which contains additional 464 PULs with CAZymes and curated substrates.
CAZyme annotation and CAZyme Gene Cluster (CGC) prediction
For each of these 602 curated PULs, we created a GenBank format flat file that contains various data about the PUL (Supplementary Table S1). The PUL genomic location was extracted from the corresponding literature during curation. From the GenBank flat file, we parsed the nucleotide sequence, protein sequences of coding genes (if available) and GFF3 formatted file of coding genes (if available).
It should be noted that the 602 PULs have already been characterized to encode CAZymes and meet the PUL definition according to their original experimental papers. However, the nomenclature and annotation of PUL signature genes vary among these papers. In order to standardize the signature gene annotation in PULs including CAZymes, transporters, transcription factors, and signal transduction proteins, all PULs were subsequently ran through the ‘run_dbcan’ standalone tool of dbCAN2 (https://github.com/linnabrown/run_dbcan, version 2.0.11) for signature gene annotation and CGC prediction (through CGC-Finder that is coded within run_dbcan) (16). Default parameters were used for run_dbcan to annotate signature genes and predict CGCs in most PULs. In other cases, more relaxed parameters were employed as noted in Supplementary Table S1.
Regarding CGC prediction, in contrast to PULs, CGCs are a broader term that is intended for the study of CAZyme gene clustering in genomes without any implications on their functions or target substrate. CGC prediction requires that at least one CAZyme and one other specific signature genes, namely either transporters, transcription factors and/or signal transduction proteins, be present within a certain number of intergenic distances to predict a potential gene cluster (16). Predicted CGCs (from run_dbcan) are therefore not necessarily identical to PULs (from literature) in terms of genomic boundaries.
As illustrated in Figure 2, there are different scenarios of CGC predictions in PULs. Among the 602 PULs, 524 (87.0%) had at least 1 CGC predicted: (i) in 115 PULs, the predicted CGCs have the same genomic boundaries; (ii) in 403 PULs, the predicted CGCs are shorter; (iii) in 6 PULs, more than one CGCs are predicted within the PUL range. Additionally, there are also 78 PULs with CAZymes but without any CGCs predicted due to CGC-Finder not finding other signatures genes (namely transporters), possibly because the original experimental papers did not require it or due to the limitation of CGC-Finder. These 78 PULs are included in dbCAN-PUL and annotated just like other PULs except no CGC predictions available for them (Supplementary Table S1).
Figure 2.
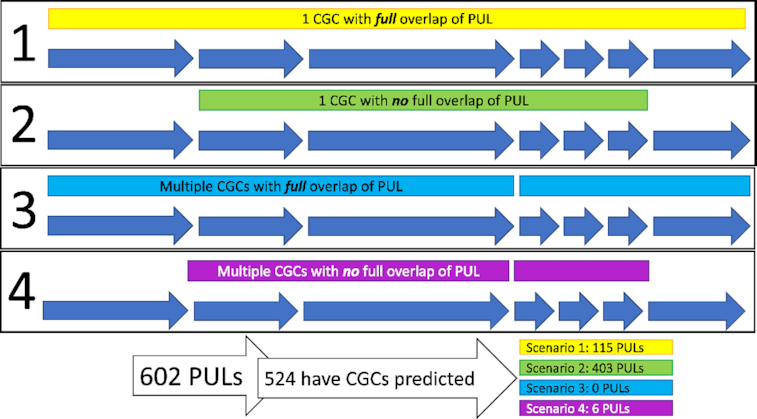
Different scenarios of CGC prediction overlap with PUL genes. Scenario 1: One CGC is predicted for the PUL, and the predicted CGC encompasses all genes in the PUL; 115 PULs had Scenario 1-type CGC predictions. Scenario 2: One CGC is predicted for the PUL, and it does not encompass all genes of the PUL; 403 PULs had Scenario 2-type CGC predictions. Scenario 3: Multiple CGCs are predicted that encompass all of the genes in the PUL; there were no PULs with Scenario 3-type CGC predictions. Scenario 4: Multiple CGCs are predicted that do not encompass all of the genes in the PUL; 6 PULs had Scenario 4-type CGC predictions.
WEB DESIGN
The dbCAN-PUL website is powered by SQLite + Django + JavaScript + Apache + HTML. The following web features (Figure 3) are unique to dbCAN-PUL compared to PULDB:
Figure 3.
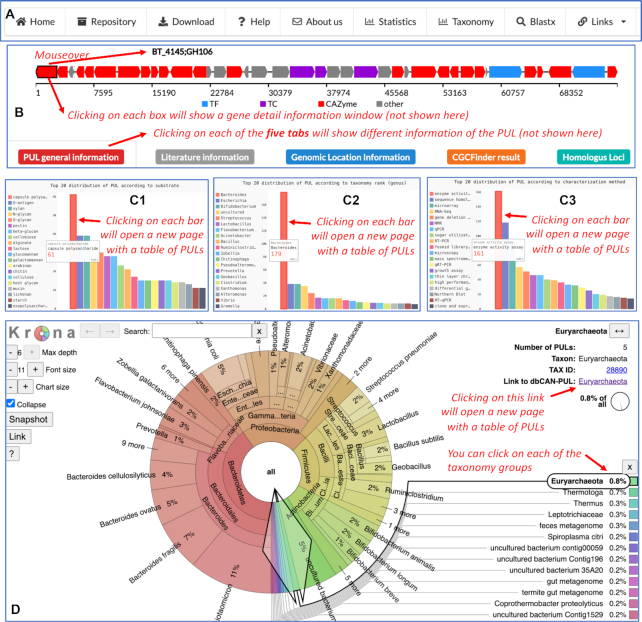
Screenshots of dbCAN-PUL website. (A) The navigation menu provides links to different webpages. (B) The top part of an example PUL page (http://bcb.unl.edu/dbCAN_PUL/CGC?clusterid=PUL0564) includes the gene cluster graphic and the five tabs that are clickable to show different information about the PUL. (C) In Home page, there are three bar graphs that allow to quickly navigate to PULs categorized according to substrates (C1), taxonomic genera (C2), and methods of characterization (C3). More complete bar graphs are provided in the Statistics page. (D) The Taxonomy page is made by Krona (20), which allows a quick navigation of different taxonomic groups across different levels. Each taxonomic group can be clicked and conveniently link to the corresponding group of PULs by clicking on a link in the top-right corner.
Browse PULs by metadata
Whereas PULDB only allows for filtering PULs by knowing beforehand the protein, CAZyme family or substrate of interest, dbCAN-PUL allows for browsing and filtering of PULs by all available substrates, species/genomes and characterization methods (Figure 3A). Browsing can be facilitated by either the summary bar plots (Figure 3C) on the Home page or Statistics page, or by NCBI Taxonomy based browsing via the Krona chart (20) on the Taxonomy page (Figure 3D). We also provide a Repository page where users can click on metadata links for PUL browsing (Figure 3A). This will help facilitate user navigation to PULs of interest by shared substrates, organisms or characterization method.
Download page
The download page (Figure 3A) features a selection of tarballs containing data for each PUL grouped by specific characterization methods, substrates or taxonomic groupings. Users can also download spreadsheets detailing the metadata for each PUL (‘dbCAN-PUL.xlsx’) and the PUL-CGC congruence for each PUL (‘dbCAN-PUL_CGC_vs_PUL_coverage.xlsx’), as well as the FASTA format database (‘PUL.faa’) of protein sequences in PULs of dbCAN-PUL that can be used in BLASTX search (see below).
Help page
The help page (Figure 3A) features explanations of database functionality and data offerings, with labeled figures aiding users in navigating the database website as well as explanations of terms and the methodology of data curation.
PUL annotation page
For each PUL, the PUL annotation page features information about the PUL, as well as an interactive graphic of the PUL (Figure 3B). If one or more CGCs were predicted in the PUL, the genes predicted in the CGCs will be displayed in the gene cluster graphic; but if no CGCs were predicted, all genes of the PUL will be displayed in the gene cluster graphic. The PUL annotation page has five information tabs for the PUL:
PUL General Information illustrates metadata about the PUL such as organism, target substrate, GenBank/JGI accession, nucleotide position range. Links are provided to direct users to external databases such as NCBI GenBank or Taxonomy databases.
Literature Information displays the PubMed ID which links to the paper(s) on PubMed, as well as the literature information and abstract for the paper. Keywords that were hits from the two PubMed literature queries are highlighted in colors.
Genomic Location Information tabulates the proteins encoded in the PUL, with links to NCBI for their protein sequence, as well as displays nucleotide positions in the GenBank accession and Enzyme Commission (EC) Numbers with links to ExPASy, if available.
CGCFinder Result charts which proteins in the PUL were predicted to be part of a PUL as well as the CGC signature gene annotations with links to external databases of protein annotations.
Homologous Loci presents the output of a homologous loci search of PUL proteins against GenBank nucleotide database using MultiGeneBlast (version 1.1.13) (21). Parameters specified for the search for each PUL were the following: allowable hits per gene: 500, maximum kilobase distance between two blast hits to be considered as belonging to the same locus: 20 (kb), and maximum number of output pages = 10, with all other parameters set to default.
BLASTX page
Users can query nucleotide sequences against proteins in PULs in dbCAN-PUL either by pasting or uploading FASTA format nucleotide sequences. The results page illustrates which proteins and PULs that sequences have hits against, as well as summary data detailing which PUL in dbCAN-PUL has the most hits to the user's queried sequences and how many CAZymes, signal transduction proteins, transcription factors and transporters were hits to the user's queried sequences. This is designed to help researchers to quickly identify the best experimentally verified PUL matches to their query nucleotide sequences.
CONCLUSIONS
In conclusion, dbCAN-PUL offers a comprehensive repository of functionally characterized PULs with rich metadata. Compared with other available resources, dbCAN-PUL focuses on CAZyme-containing PULs curated from literature: all PULs must contain at least one CAZyme but do not have to contain susC-susD gene pairs. In addition, dbCAN-PUL has several unique features that aim to enhance the study of CAZymes and PULs in prokaryotic organisms. This includes available batch download of all PUL sequences and metadata, the ability to view homologous loci across nucleotide sequences contained in GenBank or allow users to query their own DNA sequences against proteins in PULs within dbCAN-PUL using a BLASTX search. Furthermore, dbCAN-PUL currently contains the greatest number of functionally characterized PULs with substrates determined for all PULs within the repository. dbCAN-PUL aims to propel CAZyme and PUL research by providing a reference point of characterized PULs to facilitate the discovery of PULs in prokaryotic isolates and metagenomic sequences.
FUTURE WORK
We plan to update dbCAN-PUL annually with newly characterized PULs from literature. dbCAN-PUL will complement our popular dbCAN2 web server (15,16) and dbCAN-seq database (2). It provides a highly accurate and literature-supported reference database for future development of new tools to infer substrates for CGCs predicted by CGC-Finder.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the help of our previous lab members: Yiyi Cheng and Fei Xing for their help with the literature curation process and helpful discussions. This work was partially completed utilizing the Holland Computing Center of the University of Nebraska, which receives support from the Nebraska Research Initiative.
Contributor Information
Catherine Ausland, Department of Biological Sciences, Northern Illinois University, DeKalb, IL 60115, USA.
Jinfang Zheng, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
Haidong Yi, Department of Computer Science, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Bowen Yang, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
Tang Li, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
Xuehuan Feng, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
Bo Zheng, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
Yanbin Yin, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska, Lincoln, NE 68588, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) CAREER award [DBI-1933521]; United States Department of Agriculture (USDA) award [58-8042-9-089]; start-up grant of UNL [2019-YIN to Y.Y.]. Funding for open access charge: NSF CAREER award [DBI-1933521].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B.. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014; 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang L., Zhang H., Wu P., Entwistle E., Li X., Yohe T., Yi H., Yang Z., Yin Y.. dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 2018; 46:D516–D521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terrapon N., Lombard V., Gilbert H.J., Henrissat B.. Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics. 2015; 31:647–655. [DOI] [PubMed] [Google Scholar]
- 4. Bjursell M.K., Martens E.C., Gordon J.I.. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J. Biol. Chem. 2006; 281:36269–36279. [DOI] [PubMed] [Google Scholar]
- 5. Lapebie P., Lombard V., Drula E., Terrapon N., Henrissat B.. Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 2019; 10:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cockburn D.W., Koropatkin N.M.. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 2016; 428:3230–3252. [DOI] [PubMed] [Google Scholar]
- 7. Grondin J.M., Tamura K., Dejean G., Abbott D.W., Brumer H.. Polysaccharide utilization loci: fueling microbial communities. J. Bacteriol. 2017; 199:e00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hemsworth G.R., Dejean G., Davies G.J., Brumer H.. Learning from microbial strategies for polysaccharide degradation. Biochem. Soc. Trans. 2016; 44:94–108. [DOI] [PubMed] [Google Scholar]
- 9. Koch H., Durwald A., Schweder T., Noriega-Ortega B., Vidal-Melgosa S., Hehemann J.H., Dittmar T., Freese H.M., Becher D., Simon M. et al.. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 2019; 13:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. La Rosa S.L., Leth M.L., Michalak L., Hansen M.E., Pudlo N.A., Glowacki R., Pereira G., Workman C.T., Arntzen M.O., Pope P.B. et al.. The human gut Firmicute Roseburia intestinalis is a primary degrader of dietary beta-mannans. Nat. Commun. 2019; 10:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sela D.A., Chapman J., Adeuya A., Kim J.H., Chen F., Whitehead T.R., Lapidus A., Rokhsar D.S., Lebrilla C.B., German J.B. et al.. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:18964–18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu N., Li H., Chevrette M.G., Zhang L., Cao L., Zhou H., Zhou X., Zhou Z., Pope P.B., Currie C.R. et al.. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J. 2019; 13:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou J., Han J., Cai L., Zhou J., Lu Y., Jin C., Liu J., Xiang H.. Characterization of genes for chitin catabolism in Haloferax mediterranei. Appl. Microbiol. Biotechnol. 2014; 98:1185–1194. [DOI] [PubMed] [Google Scholar]
- 14. Blanvillain S., Meyer D., Boulanger A., Lautier M., Guynet C., Denance N., Vasse J., Lauber E., Arlat M.. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One. 2007; 2:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin Y., Mao X., Yang J., Chen X., Mao F., Xu Y.. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012; 40:W445–W451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z., Busk P.K., Xu Y., Yin Y.. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018; 46:W95–W101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart R.D., Auffret M.D., Roehe R., Watson M.. Open prediction of polysaccharide utilisation loci (PUL) in 5414 public Bacteroidetes genomes using PULpy. 2018; bioRxiv doi:18 September 2018, preprint: not peer reviewed 10.1101/421024. [DOI]
- 18. Terrapon N., Lombard V., Drula E., Lapebie P., Al-Masaudi S., Gilbert H.J., Henrissat B.. PULDB: the expanded database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018; 46:D677–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barbeyron T., Thomas F., Barbe V., Teeling H., Schenowitz C., Dossat C., Goesmann A., Leblanc C., Oliver Glockner F., Czjzek M. et al.. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacterium Zobellia galactanivorans Dsij(T). Environ. Microbiol. 2016; 18:4610–4627. [DOI] [PubMed] [Google Scholar]
- 20. Ondov B.D., Bergman N.H., Phillippy A.M.. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011; 12:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medema M.H., Takano E., Breitling R.. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 2013; 30:1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.