Abstract
tRNA-derived small RNAs (tsRNAs) are a class of novel small RNAs, ubiquitously present in prokaryotes and eukaryotes. It has been reported that tsRNAs exhibit spatiotemporal expression patterns and can function as regulatory molecules in many biological processes. Current tsRNA databases only cover limited organisms and ignore tsRNA functional characteristics. Thus, integrating more relevant tsRNA information is helpful for further exploration. Here, we present a tsRNA database, named tsRBase, which integrates the expression pattern and functional information of tsRNAs in multiple species. In tsRBase, we identified 121 942 tsRNAs by analyzing more than 14 000 publicly available small RNA-seq data covering 20 species. This database collects samples from different tissues/cell-lines, or under different treatments and genetic backgrounds, thus helps depict specific expression patterns of tsRNAs under different conditions. Importantly, to enrich our understanding of biological significance, we collected tsRNAs experimentally validated from published literatures, obtained protein-binding tsRNAs from CLIP/RIP-seq data, and identified targets of tsRNAs from CLASH and CLEAR-CLIP data. Taken together, tsRBase is the most comprehensive and systematic tsRNA repository, exhibiting all-inclusive information of tsRNAs from diverse data sources of multiple species. tsRBase is freely available at http://www.tsrbase.org.
INTRODUCTION
tRNA-derived small RNAs (tsRNAs) are a group of novel small non-coding RNAs that arise from either tRNA precursors or mature tRNAs. In the late 1970s, tsRNAs were originally discovered and considered as random degradation byproducts of tRNAs, so they were ignored for a long time (1). Until 2005, Lee et al. reported that certain tsRNAs were induced by starvation in Tetrahymena thermophile, indicating that tsRNA expression is regulated under specific conditions (2). In 2008, Dr Qu's group identified the stress-induced tsRNAs in the primitive eukaryote Giardia lamblia and first proposed these tRNA fragments as a novel class of small RNAs (3). With the rapid development and application of high-throughput sequencing technology, tsRNAs have been found to universally exist in all kingdoms of life (4–6).
tsRNAs are usually 16–40 nt in length, and are divided into three distinct categories according to the cleavage sites within the source tRNAs: (i) tRNA-derived fragments (tRFs), which are generated through the endo-nucleolytic cleavage of mature tRNAs (7,8); (ii) 3′U tRF, which are cleaved from 3′ end of pre-tRNAs by RNase Z during tRNA maturation (9–11) and (iii) tRNA halves (tRHs or tiRNAs), which are produced from ribonucleolytic cleavage in the anti-codon loop of mature tRNAs by angiogenin (ANG, an RNase A family member) in mammals (12,13) and by an RNase T2 family member, Rny1, in yeast (7).
Numerous studies have demonstrated that tsRNA expression is tightly regulated during development or under stress conditions (3,14,15). For example, the expression of 5′ tRHs of tRNA-Gly increased during post-testicular sperm maturation (16). In addition, ANG-mediated tsRNAs have been reported to accumulate during different stress treatments, such as heat shock, UV irradiation and nutrition deficiency (12,13). Intriguingly, tsRNAs are dysregulated in multiple cancers and could be potential biomarkers for cancer diagnosis (17–20).
Increasing evidence indicates that tsRNAs play important roles in many biological processes, such as cell proliferation, translation inhibition, genome stability maintenance and intergenerational epigenetic inheritance (4,6,9,11,16,21–27). Similar to miRNA or piRNA, tsRNA was found to bind AGO or PIWI proteins to suppress the expression of target genes at transcriptional and/or post-transcriptional levels (5,28,29). However, the detailed molecular mechanism of tsRNAs in vivo requires further exploration.
Currently, seven tsRNA databases are available, including tRFdb (30), PtRFdb (31), tRex (32), MINTbase 2.0 (33), tRF2Cancer (34), tRFexplorer (35) and OncotRF (36). Among them, tRFdb was the first tsRNA database, collecting tsRNAs from approximately 200 small RNA-seq data of eight different species, but it has not been updated for several years. The PtRFdb database only provides basic information, such as tsRNA type and sequence, from 10 different plant species. The tRex database reports tsRNA profiling of the model plant Arabidopsis thaliana from 300 samples of different tissues, ecotypes, genotypes or stress conditions. The following four databases focus on human cells or cancers. MINTbase 2.0 and tRF2Cancer provide tsRNAs’ diversity and expression patterns in human cancer samples from The Cancer Genome Atlas (TCGA) database, tRFexplorer mainly exhibits differential expression and correlation analyses of tsRNAs from NCI-60 cancer cell lines and TCGA samples, while OncotRF contains tsRNAs’ expression, tsRNA-gene correlations and survival analyses in cancer. Altogether, these databases have increased our knowledge of tsRNAs in different species, but do not provide any information about tsRNA function. Hence, it is essential to construct a comprehensive database covering tsRNAs’ profiling in different biological processes and in different organisms, especially combining functional information of tsRNAs with their targets.
Herein, we developed ‘tsRBase’, a comprehensive tsRNA database that integrates over 14 000 public small RNA-seq data across 20 organisms. With a friendly user interface, people can easily search for tsRNAs of interest, and compare tsRNA expression levels under different conditions. Furthermore, we not only collected functional tsRNA from the published literatures, but also identified protein-binding tsRNAs and their targets from publicly available datasets for the first time (37,38). We believe that tsRBase will be a valuable and comprehensive resource for scientists who study tsRNAs from different fields.
MATERIALS AND METHODS
Pre-processing of the small RNA-seq
The small RNA sequencing data were downloaded from the Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/sra) (39). The adapter sequences from the raw data were removed using Cutadapt (40). Clean reads with length of 16–40 bases were kept for subsequent analyses.
Building tRNA index
The sequences of the tRNA genes from 20 species were downloaded from the ‘Genomic tRNA database’ (GtRNAdb) (http://gtrnadb.ucsc.edu) (41). Only high confident tRNAs with conserved secondary cloverleaf structure were used. Reference genomes with the same genome assembly as GtRNAdb were downloaded (Supplementary Table S1). Mature tRNA sequences were obtained by removing intron sequences and adding ‘CCA’ tail at the end of the original sequences of tRNAs. 100 nucleotides downstream sequences of tRNAs were extracted from reference genome based on their genomic coordinates.
Mapping small RNA
Species-specific tRNA BLAST databases were built for small RNA sequences query. We then used BLAST-2.2.16 to find the tRNA-related RNA sequences in each library (42). We only considered those small RNAs that were perfectly mapped to the tRNA sequence as tsRNAs. Then the expression value of tsRNA was quantified using our custom script. Next, we filtered tsRNAs with expression value lower than one count-per-million (CPM) and/or expressed in less than five samples to eliminate random degradation sequences.
Annotation of tsRNAs
The types of tsRNAs were determined according to where tsRNAs were generated from their corrsesponding tRNAs. In brief, sequences that map to the 5′ or 3′ extreme end of mature tRNAs and do not overlap with anticodon-loop were classified as 5′ tRFs or 3′ tRFs, respectively. Sequences that map to the 5′ or 3′ extreme end of mature tRNAs and also overlap with anticodon-loop were defined as 5′ tRHs and 3′ tRHs, respectively. The sequences exclusively from internal regions of mature tRNAs were annotated as inter tRFs. Sequences mapping the downstream of mature tRNAs and ending with or without UUU were annotated as 3′U tRFs.
tsRNA target identification through CLASH/CLEAR data analysis
Crosslinking, ligation, and sequencing of hybrids (CLASH) and Covalent ligation of endogenous Argonaute-bound RNAs (CLEAR)-CLIP are two new techniques that designed to directly observe miRNA-target interaction in vivo through AGO binding (37,38). In addition, some studies have demonstrated that tsRNAs can recognize specific RNA targets through AGO protein in a miRNA-like way (5,43,44). Therefore, we used a custom pipeline to explore tsRNA-target interaction in CLASH/CLEAR-CLIP data. Three datasets were used: (i) CLASH data of human HEK293 cells (GSE50452); (ii) CLEAR-CLIP data of human Huh7.5 cells (GSE73057); (iii) CLEAR-CLIP data of mouse cortex (GSE73058). We first used Cutadapt to remove adapters and filter reads less than 16 nt. Then, we collapsed reads with the same sequence and counted the number of each unique read. The reads were then mapped to the tRNA reference. Reads that partially match (16–40 nt of the reads mapped to tRNAs perfectly and the unmapped part was >8 nt) to tRNAs were kept as candidate tsRNA-target chimeras. Next, we mapped the candidates to the genome using bowtie-1.2.2 and blat v.35, to mark and remove fake chimeras that can mapped to other sites. At the same time, we mapped the candidates to the NT database to remove pollutants. Then, the candidate tsRNA–target chimeras were split into tsRNAs and target sequences. The target sequences were mapped to the genome to obtain their corresponding genome positions, and then the 100 nt downstream of the ligation sites were assigned as target sequences. The duplex structure predictions for tsRNAs and target regions were made using RNAhybrid (45). Target genes were annotated using bedtools (46).
Protein binding tsRNAs exploration from CLIP/RIP data
The small RNA-seq data of protein CLIP/RIP samples were first pre-processed as previously mentioned. Only reads with a length between 16 nt and 40 nt were kept. Each unique read was then collapsed and counted. Next, we used BLAST-2.2.16 to map these reads to their corresponding tsRNA reference to get the protein binding tsRNAs and their expression value. At last, we normalized each tsRNA’s expression value to CPM.
Mining tsRNA literatures
Since articles that describe tsRNAs’ specific expression patterns or biological functions are limited, it is feasible to use a text-mining approach to search for the information of tsRNAs in the full-text of open access articles. We first collected all articles that mentioned the tsRNAs by searching keywords such as ‘tsRNA’, ‘tRF’ or ‘tRNA’ in the NCBI PubMed database. Then we identified tsRNAs with validated specific expression or function by full-text mining. All the information of these tsRNAs was recorded and corresponded to tsRBase ID.
Implementation
The tsRBase was built under the XAMPP environment on the Linux system, which is comprised of Apache HTTP server version 2.4.33 with MariaDB 10.1.33 at the back end, and the PHP 7.2.6, HTML and JavaScript at the front end. The BLAST function was set up based on ViroBLAST (47).
RESULTS
Data content of tsRBase
tsRBase characterized and integrated tsRNAs from more than 14 000 small RNA-seq data. In total, 121 942 tsRNAs were identified, belonging to 20 species, including human (Homo sapiens), Arabidopsis thaliana, cow (Bos taurus), nematode (Caenorhabditis elegans), Clostridium acetobutylicum, zebra fish (Danio rerio), fruit fly (Drosophila melanogaster), Escherichia coli, chicken (Gallus gallus), soybean (Glycine max), mouse (Mus musculus), rice (Oryza sativa), sheep (Ovis aries), Physcomitrella patens, rat (Rattus norvegicus), yeast (Schizosaccharomyces pombe), pig (Sus scrofa), grape (Vitis vinifera), frog (Xenopus tropicalis) and corn (Zea mays) (Table 1).
Table 1.
Samples and number of tsRNAs of 20 species in tsRBase
| Kingdom | Species | Samples | Number of tsRNAs |
|---|---|---|---|
| Animal | Homo sapiens | 7619 | 24 773 |
| Mus musculus | 2304 | 13 310 | |
| Rattus norvegicus | 1034 | 9035 | |
| Bos taurus | 569 | 5091 | |
| Ovis aries | 73 | 1946 | |
| Sus scrofa | 190 | 3995 | |
| Gallus gallus | 142 | 5262 | |
| Xenopus tropicalis | 13 | 84 | |
| Danio rerio | 122 | 3538 | |
| Drosophila melanogaster | 402 | 5914 | |
| Caenorhabditis elegans | 292 | 7026 | |
| Plant | Arabidopsis thaliana | 592 | 7747 |
| Glycine max | 279 | 5836 | |
| Oryza sativa | 204 | 5739 | |
| Vitis vinifera | 148 | 5063 | |
| Zea mays | 217 | 4382 | |
| Physcomitrella patens | 33 | 2027 | |
| Fungi | Schizosaccharomyces pombe | 244 | 10 981 |
| Bacteria | Escherichia coli | 39 | 173 |
| Clostridium acetobutylicum | 84 | 20 | |
| Total | 14 600 | 121 942 |
Importantly, tsRBase attaches the important information about biological functions of tsRNAs. For this purpose, we collected and analyzed the following different sources that contributed to functional research, and these results were embedded in the database: (i) the published literatures about tsRNAs with experimentally validated expression patterns or biological functions; (ii) protein-binding tsRNAs identified from 501 CLIP/RIP small RNA-seq data crossing nine species (human, Arabidopsis thaliana, nematode, zebra fish, fruit fly, mouse, rice, rat and frog); (iii) tsRNA-target pairs based on the CLASH data (human) and CLEAR-CLIP data (human and mouse) analysis (Table 2).
Table 2.
Number of data used to reveal functional tsRNAs in tsRBase
| Species | CLIP/RIP | CLASH/CLEAR-CLIP |
|---|---|---|
| Arabidopsis thaliana | 103 | - |
| Caenorhabditis elegans | 6 | - |
| Danio rerio | 2 | - |
| Drosophila melanogaster | 137 | - |
| Homo sapiens | 153 | 22 |
| Mus musculus | 96 | 26 |
| Oryza sativa | 3 | - |
| Rattus norvegicus | 1 | - |
| Xenopus tropicalis | 4 | - |
Web interface and usage
Search
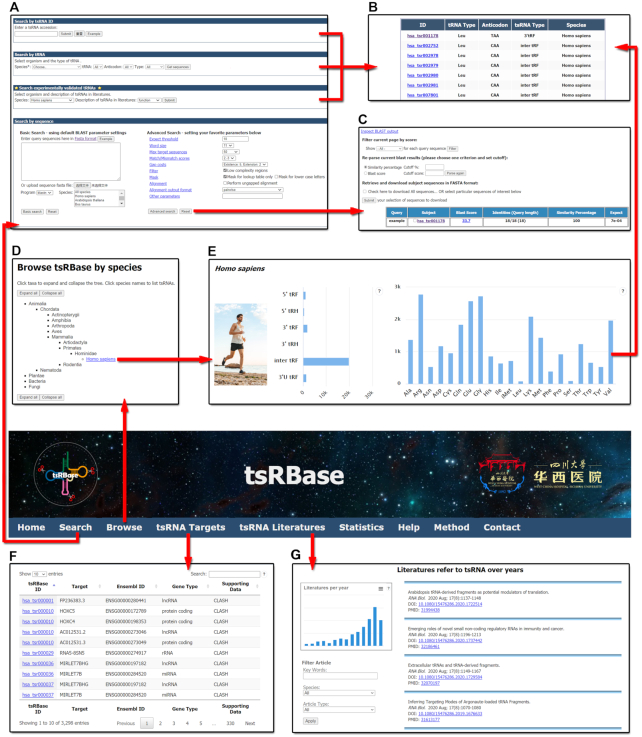
In order to fulfill different users’ requirements, tsRBase provides users with three basic ways to query information of tsRNAs in the module ‘Search’ (Figure 1A). First, users can search for tsRNAs by their unique tsRBase IDs from the interface. Second, users can search for tsRNAs based on biological traits such as the corresponding amino acid, anticodon type, and co-ordinates of the tRNA from which they are derived. Third, tsRBase also allows the performance of BLAST tool to find out whether the sequences are tsRNAs and obtain relevant information. In this function, users can input the sequences through either loading a local file or directly pasting the sequences in the input box. In addition to performing BLAST with default parameters, tsRBase also provides additional options for fine-tuning parameters. For example, users can set the mismatch scores, the mode of alignment (with or without indels), the gap costs of matches and the statistical significance expect threshold to get the final output. Subsequently, users can filter the result using the BLAST score or similarity percentage of sequences.
Figure 1.
Overview of tsRBase database. (A) The page with searching tools in tsRBase. (B) The result table of tsRNAs after searching by tsRNA ID, tRNA type or experimentally validated tsRNAs. (C) The summary page of tsRNAs resulted from the input sequence and BLAST searching. (D) The page for browsing tsRBase by species. (E) Summary of tsRNAs in Homo sapiens. (F) The page displaying the tsRNA targets. (G) The page showing the published tsRNA literatures.
Since some tsRNAs have been reported to exhibit certain biological functions or expression patterns, we excavated all the scientific literatures and collected tsRNAs with experimentally validated biological functions or expression patterns. Moreover, we provided users with a tool to obtain these validated tsRNAs by searching both species and known biological traits (biological function or expression pattern) (Figure 1A).
All the tsRNA search outputs are displayed in tables. For direct searches (ID based, tRNA based, and experimentally validated tsRNAs searching), a summary table will be provided on the result page displaying five items: tsRBase ID (hyperlinks to the page of detailed tsRNA information), tRNA type, anticodon, tsRNA type and species (Figure 1B). For the BLAST search, the result tables include query sequence accession, subjected tsRNA ID, BLAST score, identities of matching, similarity percentage and expect value (Figure 1C). For each item, the hyperlinks of ‘BLAST score’ and ‘Subject’ are further hyperlinked to the detailed information of alignment and a certain tsRNA, respectively.
Browse
The browse page presents users a species phylogenetic list (Figure 1D). Users can click taxa to expand and collapse the tree. When clicking the Latin name of selected species, the link will direct users to a summary page of the species, which shows the histogram of different types of tsRNAs in this species (Figure 1E). Furthermore, each bar of the histogram hyperlinks to the list of corresponding types of tsRNAs.
tsRNA Targets
Understanding targets of tsRNA is crucial for uncovering tsRNA’s role in cellular biological implication. Thus, we explored tsRNA-target interactions through AGO protein in the CLASH and CLEAR-CLIP datasets, and found 3298 tsRNA-target pairs. The results are shown in the ‘tsRNA Targets’ module (Figure 1F). This table shows the tsRBase ID of tsRNA, gene name, Ensembl ID and biological type of target gene, and the dataset from which this result was derived.
tsRNA Literatures
Currently, tsRNAs are attracting the attention of scientists more than ever, and fruitful research achievements have been made. In order to sort out the developmental skeleton of tsRNA research and help follow research progress, we collected as many tsRNA-related literatures as we can and displayed them in the ‘tsRNA Literatures’ module (Figure 1G). At present, we have collected 364 literatures on our website.
Detailed information page of tsRBase
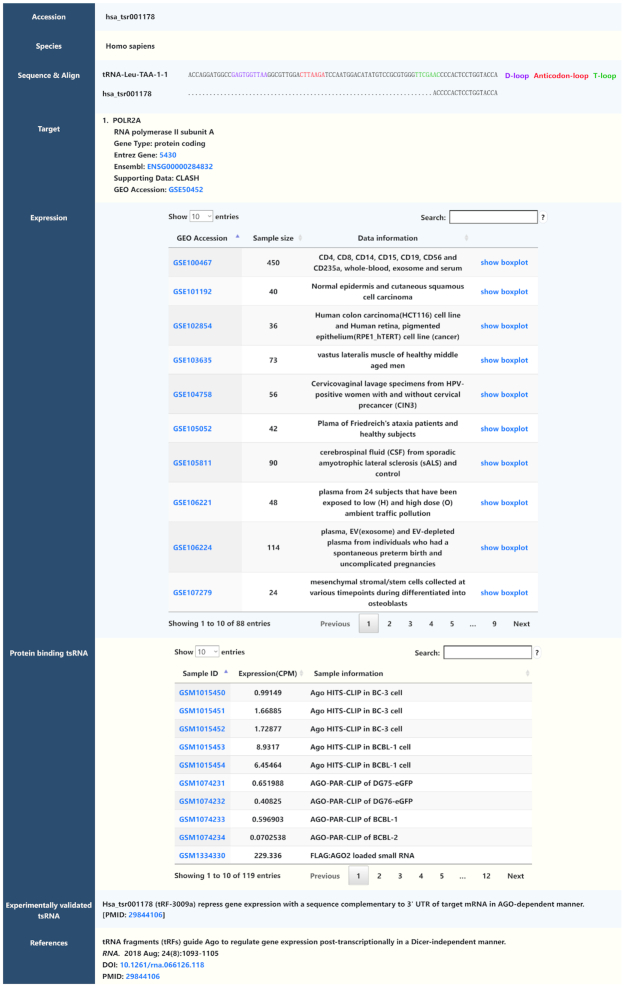
Detailed information about each tsRNA is integrated and displayed on the result page. Here, we take hsa_tsr001178 as an example to describe each section of the result page (Figure 2). First, the page displays the unique tsRBase ID (hsa_tsr001178) of this tsRNA and the Latin name of the species (Homo sapiens). The ‘Sequence & Align’ section shows the sequence of hsa_tsr001178 and demonstrates that it comes from the 3′ end of tRNA-Leu-TAA-1-1. The ‘Target’ provides the predicted targets of tsRNAs identified from CLASH or CLEAR-CLIP data. Here we know that hsa_tsr001178 might target RNA polymerase II subunit A (POLR2A) gene from the CLASH data. Moreover, users can acquire more information about POLR2A gene through the hyperlinks to NCBI, Ensemble and Uniprot databases.
Figure 2.
An example of the webpage showing the detailed information of hsa_tsr001178 in tsRBase.
The ‘Expression’ part provides the expression value of hsa_tsr001178 in different GEO datasets. The GEO accession hyperlinks to the GEO database and can display the detailed information of each dataset. Furthermore, users can click the ‘show boxplot’ button to see the boxplot of this tsRNA between different groups (the grouping method is according to the specific GEO dataset, which can be different cell/tissue or different treatment). In addition, users can download the expression data of the tsRNA embedded in each boxplot.
The ‘Protein binding tsRNA’ section has a table showing the abundance of the tsRNA in different CLIP/RIP-seq data. For hsa_tsr001178, the table displays its expression value in a total of 119 data, such as Ago HITS-CLIP in BC-3 cells and BCBL-1 cells.
The ‘Experimentally validated tsRNA’ part describes the biological function or specific expression pattern of tsRNA. Here, we can see that hsa_tsr001178 repress gene expression with a sequence complementary to the 3′ UTR of the target mRNA in an AGO-dependent manner (26). Lastly, the ‘References’ section exhibits published articles related to this tsRNA.
DISCUSSION
In the past few years, studies on tsRNAs have increased rapidly. It has been demonstrated that tsRNAs have specific expression patterns and important biological functions. However, current tsRNA databases merely focused on certain basic biological traits of tsRNAs, such as sequences or expression levels in limited organisms, thus this is far from meeting the needs of actual tsRNA research. Hence, we built a new tsRNA database, named tsRBase, which not only collects basic traits of tsRNAs from wide range of species, but also focuses on the targets and biological functions of tsRNAs. Besides, full-featured searching tools are provided for users to conduct diverse tsRNAs analyses.
Compared with existing tsRNA databases, tsRBase has made the following important adjustments. First, tsRBase provides genome-wide view of tsRNAs’ expression in various tissues/cell-lines or under specific conditions covering as many as 20 organisms, including animals, plants, fungi and bacteria. This makes tsRBase hitherto a database of the highest species diversity, thus it will assist tsRNA researchers from different fields. tsRBase also integrates functional information of tsRNAs from three different sources: (i) tsRBase provides tsRNAs with experimentally validated functions and expression patterns by full-text mining on scientific papers; (ii) tsRBase displays credible tsRNA-target pairs from analysis of CLASH and CLEAR-CLIP data; (iii) tsRBase exhibits the abundance of protein-binding tsRNAs in different CLIP/RIP data. Moreover, tsRBase collects the latest literatures about tsRNAs, and this will assist researchers stay up to date with the progress of this field.
Overall, we aim to make tsRBase a comprehensive and systematic tsRNA database, covering full-ranged information of tsRNA for various species, which is helpful for understanding tsRNA’s functions and advancing the progress of tsRNA study.
DATA AVAILABILITY
tsRBase is freely available at http://www.tsrbase.org.
Supplementary Material
Contributor Information
Yuanli Zuo, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China; Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of life Sciences, Sichuan University, Chengdu 610041, China.
Lei Zhu, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Zhixin Guo, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Wenrong Liu, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Jiting Zhang, Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of life Sciences, Sichuan University, Chengdu 610041, China.
Zhen Zeng, Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of life Sciences, Sichuan University, Chengdu 610041, China.
Qingbin Wu, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Jian Cheng, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Xin Fu, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Yang Jin, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
Yun Zhao, Key Laboratory of Bio-Resource and Eco-Environment of Ministry of Education, College of life Sciences, Sichuan University, Chengdu 610041, China.
Yong Peng, Laboratory of Molecular Oncology, Frontiers Science Center for Disease-related Molecular Network, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu 610064, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2016YFA0502204 and 2017YFA0504304]; National Natural Science Foundation of China [81772960, 81902819]; Science and Technology Program of Sichuan Province [2019JDTD0013, 2020YFS0222]; 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University [ZYJC18030, ZYGD20008]. Funding for open access charge: 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University.
Conflict of interest statement. None declared.
REFERENCES
- 1. John Speer M.D., Gehrke C.W., Ms K.C.K., Waalkes T.P., Borek E.. tRNA breakdown products as markers for cancer. Cancer. 1979; 44:2120–2123. [DOI] [PubMed] [Google Scholar]
- 2. Lee S.R., Collins K.. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005; 280:42744–427449. [DOI] [PubMed] [Google Scholar]
- 3. Li Y., Luo J., Zhou H., Liao J.Y., Ma L.M., Chen Y.Q., Qu L.H.. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008; 36:6048–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kumar P., Kuscu C., Dutta A.. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem. Sci. 2016; 41:679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar P., Anaya J., Mudunuri S.B., Dutta A.. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014; 12:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu L., Ow D.W., Dong Z.. Transfer RNA-derived small RNAs in plants. Sci. China Life Sci. 2018; 61:155–161. [DOI] [PubMed] [Google Scholar]
- 7. Thompson D.M., Parker R.. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009; 185:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gebetsberger J., Polacek N.. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013; 10:1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu L., Liu X., Pu W., Peng Y.. tRNA-derived small non-coding RNAs in human disease. Cancer Lett. 2018; 419:1–7. [DOI] [PubMed] [Google Scholar]
- 10. Balatti V., Nigita G., Veneziano D., Drusco A., Stein G.S., Messier T.L., Farina N.H., Lian J.B., Tomasello L., Liu C.G. et al.. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:8071–8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee Y.S., Shibata Y., Malhotra A., Dutta A.. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009; 23:2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X.. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009; 583:437–442. [DOI] [PubMed] [Google Scholar]
- 13. Yamasaki S., Ivanov P., Hu G.F., Anderson P.. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009; 185:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao J.Y., Guo Y.H., Zheng L.L., Li Y., Xu W.L., Zhang Y.C., Zhou H., Lun Z.R., Ayala F.J., Qu L.H.. Both endo-siRNAs and tRNA-derived small RNAs are involved in the differentiation of primitive eukaryote Giardia lamblia. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fricker R., Brogli R., Luidalepp H., Leander W., Michel F., Oliver J., Marek Z., Mark H., André S., Marina C. et al.. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 2019; 10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. et al.. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G., Rassenti L.Z., Pass H.I., Kipps T.J., Liu C.G. et al.. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5071–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.Q. et al.. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019; 18:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Veneziano D., Tomasello L., Balatti V., Palamarchuk A., Rassenti L.Z., Kipps T.J., Pekarsky Y., Croce C.M.. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:24252–24258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun C., Fu Z., Wang S., Li J., Li Y., Zhang Y., Yang F., Chu J., Wu H., Huang X. et al.. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018; 414:16–25. [DOI] [PubMed] [Google Scholar]
- 21. Raina M., Ibba M.. tRNAs as regulators of biological processes. Front. Genet. 2014; 5:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim H.K., Fuchs G., Wang S., Wei W., Zhang Y., Park H., Roy-Chaudhuri B., Li P., Xu J., Chu K. et al.. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017; 552:57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schorn A.J., Gutbrod M.J., LeBlanc C., Martienssen R.. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017; 170:61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. et al.. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016; 351:397–400. [DOI] [PubMed] [Google Scholar]
- 25. Mo D., Jiang P., Yang Y., Mao X., Tan X., Tang X., Wei D., Li B., Wang X., Tang L. et al.. A tRNA fragment, 5′-tiRNA, suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019; 457:60–73. [DOI] [PubMed] [Google Scholar]
- 26. German M., Choudury S.G., Keith S.R.. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017; 45:5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu L., Ge J., Li T., Shen Y., Guo J.. tRNA-derived fragments and tRNA halves: the new players in cancers. Cancer Lett. 2019; 452:31–37. [DOI] [PubMed] [Google Scholar]
- 28. Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A.. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018; 24:1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Keam S.P., Young P.E., Mccorkindale A.L., Dang T.H.Y., Clancy J.L., Humphreys D.T., Preiss T., Hutvagner G., Martin D.I.K., Cropley J.E. et al.. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014; 42:8984–8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar P., Mudunuri S.B., Anaya J., Dutta A.. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015; 43:D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gupta N., Singh A., Zahra S., Kumar S.. PtRFdb: a database for plant transfer RNA-derived fragments. Database (Oxford). 2018; 2018:bay063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thompson A., Zielezinski A., Plewka P., Szymanski M., Nuc P., Szweykowska-Kulinska Z., Jarmolowski A., Karlowski W.M.. tRex: a web portal for exploration of tRNA-derived fragments in Arabidopsis thaliana. Plant Cell Physiol. 2018; 59:e1. [DOI] [PubMed] [Google Scholar]
- 33. Pliatsika V., Loher P., Magee R., Telonis A.G., Londin E., Shigematsu M., Kirino Y., Rigoutsos I.. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects. Nucleic Acids Res. 2018; 46:D152–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng L.L., Xu W.L., Liu S., Sun W.J., Li J.H., Wus J., Yang J.H., Qu L.H.. tRF2Cancer: A web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016; 44:W185–W193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. La Ferlita A., Alaimo S., Veneziano D., Nigita G., Balatti V., Croce C.M., Ferro A., Pulvirenti A.. Identification of tRNA-derived ncRNAs in TCGA and NCI-60 panel cell lines and development of the public database tRFexplorer. Database (Oxford). 2019; 2019:baz115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yao D., Sun X., Zhou L., Amanullah M., Pan X., Liu Y., Liang M., Liu P., Lu Y.. OncotRF: an online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 2020; 17:1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Helwak A., Kudla G., Dudnakova T., Tollervey D.. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leinonen R., Sugawara H., Shumway M.. The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10. [Google Scholar]
- 41. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 43. Hasler D., Lehmann G., Murakawa Y., Klironomos F., Jakob L., Grässer F.A., Rajewsky N., Landthaler M., Meister G. et al.. The lupus autoantigen La prevents Mis-channeling of tRNA fragments into the human MicroRNA pathway. Mol. Cell. 2016; 63:110–124. [DOI] [PubMed] [Google Scholar]
- 44. Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R.. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1404–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kruger J., Rehmsmeier M.. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006; 34:W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng W., Nickle D.C., Learn G.H., Maust B., Mullins J.I.. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user's datasets. Bioinformatics. 2007; 23:2334–2336. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
tsRBase is freely available at http://www.tsrbase.org.