Abstract
The current state of the COVID-19 pandemic is a global health crisis. To fight the novel coronavirus, one of the best-known ways is to block enzymes essential for virus replication. Currently, we know that the SARS-CoV-2 virus encodes about 29 proteins such as spike protein, 3C-like protease (3CLpro), RNA-dependent RNA polymerase (RdRp), Papain-like protease (PLpro), and nucleocapsid (N) protein. SARS-CoV-2 uses human angiotensin-converting enzyme 2 (ACE2) for viral entry and transmembrane serine protease family member II (TMPRSS2) for spike protein priming. Thus in order to speed up the discovery of potential drugs, we develop DockCoV2, a drug database for SARS-CoV-2. DockCoV2 focuses on predicting the binding affinity of FDA-approved and Taiwan National Health Insurance (NHI) drugs with the seven proteins mentioned above. This database contains a total of 3,109 drugs. DockCoV2 is easy to use and search against, is well cross-linked to external databases, and provides the state-of-the-art prediction results in one site. Users can download their drug-protein docking data of interest and examine additional drug-related information on DockCoV2. Furthermore, DockCoV2 provides experimental information to help users understand which drugs have already been reported to be effective against MERS or SARS-CoV. DockCoV2 is available at https://covirus.cc/drugs/.
INTRODUCTION
From December 2019 to June 2020, SARS-CoV-2 has infected over 10 million people, and caused >500 thousand deaths worldwide (1). As a result, the last few months has seen a rapid expansion of COVID-19 research (2–7) including the discovery of potential inhibitors and drugs against SARS-CoV-2 (8–12).
The SARS-CoV-2 genome contains ∼30 000 nucleotides that encode about 29 proteins such as spike protein, 3C-like protease (3CLpro, also called main protease, Mpro), Papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp, also named nsp12), and nucleocapsid (N) protein (9,13–15). These five proteins are all known to play important roles in the viral replication process in some way. For example, the surface spike glycoprotein is known to be critical for virus entry through binding to the major antigens of host receptors such as angiotensin-converting enzyme 2 (ACE2) (16,17). The 3CLpro is a key enzyme which digests pp1a and pp1ab, two proteins that yield functional viral proteins in the virus' life cycle (10,18). Blocking the activity of this enzyme would therefore suppress viral replication, making it an efficient therapeutic strategy. Likewise, PLpro is responsible for the cleavage of the viral polypeptide (19), and RdRp catalyzes viral RNA synthesis and plays an important role in the replication and transcription machinery of SARS-CoV-2 (20). Remdesivir, the potent inhibitor against SARS-CoV-2, is effective because of how it targeted the RdRp protein (20,21). N protein is an RNA-binding protein necessary for viral RNA transcription and replication (15). In addition to the viral proteins, SARS-CoV-2 uses human ACE2 for viral entry and human transmembrane serine protease family member II (TMPRSS2) for spike protein priming (16). We therefore hypothesize that these five SARS-CoV-2 proteins, spike, 3CLpro, PLpro, RdRp, N protein, and two host proteins, ACE2 and TMPRSS2, can be potential druggable targets.
Previously, researchers have targeted these viral proteins for drug discovery by using structure-assisted design, virtual and high-throughput screening (8,11). Similarly, these approaches can also be used for SARS-CoV-2 research (22). For example, Jin et al. used a computer-aided drug design and virtual screening to identify the mechanism-based inhibitor (N3) and the antineoplastic drug carmofur targeting Mpro (10,11). They further determined the crystal structures of the N3- and carmofur-Mpro complexes and confirmed the antiviral activities of N3 and carmofur using the plaque-reduction assays (10,11).
Drug repositioning is the process of finding new uses for existing approved drugs, and is believed to offer great benefits over de novo drug discovery, as well as enable rapid clinical trials and regulatory review for COVID-19 therapy (23). Here we develop the database, DockCoV2, by performing molecular docking analyses of seven proteins including spike, 3CLpro, PLpro, RdRp, N protein, ACE2 and TMPRSS2 with 2285 FDA-approved and 1478 NHI drugs. DockCoV2 also provides appropriate experimental information with literature support. Several databases focus on delivering repurposing drugs against SARS-CoV-2. For example, Excelra's COVID-19 Drug Repurposing Database includes 128 small molecules and biologics (https://www.excelra.com/covid-19-drug-repurposing-database/) and the OpenData collected the actionable data and validated methodologies (https://doi.org/10.1101/2020.06.04.135046). Additionally, Covid19_DB lists the curated data of clinical trials in Covid-19/2019-nCoV (http://www.redo-project.org/covid19db/). To our knowledge, no database provides a more up-to-date and comprehensive resource with drug-target docking results for repurposed drugs against SARS-CoV-2.
DATA SOURCES
For drug repositioning, we selected FDA and Taiwan NHI (National Health Insurance Administration) as our list of approved drugs. FDA-approved drugs were retrieved from the ZINC15 database (24), and Taiwan NHI-approved drugs were downloaded from the website of NHI. Drug names were extracted from the top five ingredients of each product, and drug structures were obtained from the PubChem Compound database (25). Due to the limitation of the ligand preparation pipeline, we dropped some compounds with ions which could not be converted properly by the gen3d operation of OpenBabel (version 3.0.0) (26). Note that we also added some additional FDA-approved drugs which are not in the ZINC15 database. In total, 2285 FDA-approved drugs and 1478 Taiwan NHI-approved drugs were recruited as our candidates for virtual screening.
Seven target proteins were selected because of their involvement in viral entry and replication. The protein structures, including 3CLpro (PDB ID: 6LU7) (10), PLpro (PDB ID: 6WX4) (https://doi.org/10.1101/2020.04.29.068890), RdRp (PDB ID: 7BV2) (21), spike receptor-binding domain (RBD) (PDB ID: 6M0J) (27), N protein (PDB ID: 6M3M) (15) and ACE2 (PDB ID: 1R42) (28) were retrieved from Protein Data Bank. To ensure the availability of the binding sites of the proteins, we removed the compounds added for protein structure determination such as ligands, cofactors, and ions. For the spike protein, only the RBD was used for molecular docking and for 7BV2, only the nsp12 was used out of the nsp7–nsp8–nsp12 complex. The structure of human TMPRSS2 is currently not available in Protein Data Bank, so we used homology modeling on the sequence of TMPRSS2 from Uniprot (ID: O15393) (29) to build an approximate structure based on the template of the human serine protease hepsin (PDB ID: 5CE1) using SWISS-MODEL (30).
DATABASE CONTENT
DockCov2 aims to speed up the process of finding potential drugs by providing a computational representation of molecular docking with the most commonly-used drug databases (31–35). Through our website, researchers can query the docking scores of candidate drugs with essential drug-related information from the database to identify which drugs could be potential drugs. We designed a virtual screening pipeline, and docked 2285 FDA approved drugs and 1478 Taiwan National Health Insurance (NHI) approved drugs with the seven main proteins of the SARS-CoV-2 infection mechanism (36–38). All of our docking results are saved in the formats of Protein Data Bank, Partial Charge (Q) and Atom Type (T) (PDBQT), and can be viewed either directly on the website through NGLView (39) or downloaded for further docked structure refinement.
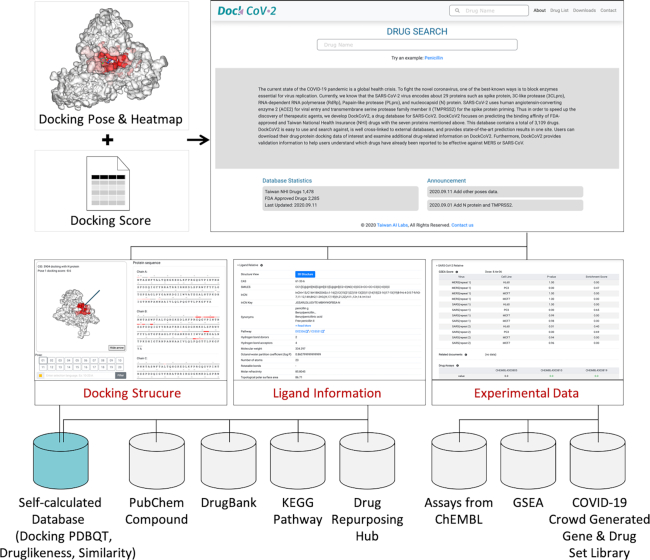
The overview of the database is shown in Figure 1. In addition to the molecular docking score, the joint panel section has three tabs: docking structure, ligand information and experimental data. The docking structure panel visualizes the docking PDBQT from the self-calculated database, which also contains druglikeness information and compound similarity. The ligand information panel contains (a) structural information from PubChem Compound database (25); (b) drug information from DrugBank (33); (c) pathway information from Kyoto Encyclopedia of Genes and Genomes (KEGG) (40); (d) clinical-related information from Repurposing Hub (34); and (e) other chemical information. In the experimental data panel, we collected experimental and literary evidence from ChEMBL (35), a study of gene set enrichment analysis (GSEA) from Zhou et al. (41) and COVID-19 Crowd Generated Gene and Drug Set Library (https://amp.pharm.mssm.edu/covid19).
Figure 1.
The overview of the database content. In addition to the docking scores, DockCoV2 designed a joint panel section to provide the following related information: docking structure, ligand information and experimental data.
DATA PROCESSING AND INTEGRATION
Identifier conversion
We chose PubChem compound identifiers (CIDs) as the key identifier in DockCoV2 because it is unique and widely used and can be easily linked to other external databases. For drugs retrieved from NHI, drug names were translated into CIDs using whole source mappings provided by UniChem (31) and PubChem Identifier Exchange Service (https://pubchem.ncbi.nlm.nih.gov/idexchange/). Specifically, multiple CIDs may be returned by PubChem Identifier Exchange Service for each drug name. We canonically chose the first returned record in order to be consistent with the result by manual search via the graphical interface of PubChem. Once we have the PubChem CIDs of our compound library, simplified molecular-input line-entry system (SMILES) (42), international chemical identifier (InChI), hashed values of international chemical identifiers (InChIKey), and synonyms were fetched through the PubChem PUG API (43).
Ligand information
For drug development, Lipinski's rule of five (44), Ghose filter (45), Veber filter (46), rapid elimination of swill (REOS) filter (47) are several available indicators to evaluate the druglikeness. Therefore, components of these rules were calculated by RDKit (http://www.rdkit.org). Moreover, fragment screening is able to find a sensible starting point for the evolution of a new molecule in modern drug discovery. Previously, a large crystallographic fragment screen (48) was performed against 3CLpro by the Diamond Light Source group. Seventy-eight hits were released to the public in March 2020. In DockCoV2, we performed the substructure matching of these fragments toward the drug library we used here. To let the user easily fetch structurally similar compounds, similarity search (49) strategies were also adopted. Similarity search within the DockCoV2 and toward whole PubChem compound databases are both supported. To generate similarity values between any two compounds in DockCoV2, a 512-bit fingerprint of each compound is calculated using the Morgan algorithm (50). Each bit represents the presence or absence of a particular structural feature. From here, similarity values between any compound pairs are represented by the Dice coefficient. In the current version of DockCoV2, the rule of five, substructure matching, and the calculation of similarity values were all done by using the RDKit 2020.03.2 release.
Pathway information can serve as the basis of hypothesis in drug discovery. Therefore, two databases of KEGG (namely, KEGG Drug and KEGG Compound) were integrated into DockCoV2. PubChem CIDs were converted into DrugBank IDs first, then further converted into KEGG Drug IDs and KEGG Compound IDs. Alternatively, PubChem CIDs were directly converted to KEGG Drug IDs and KEGG Compound IDs by PubChem Identifier Exchange Service. Clinically related information including approval information, clinical phase, mechanism of action (MoA), target, disease area, and indication were also retrieved from the metadata file from the Drug Repurposing Hub.
Experimental data
To provide useful validation information for drug repurposing and development, chemical information was aggregated either from external databases or directly calculated from structures. Three biological assays CHEMBL4303805, CHEMBL4303810, CHEMBL4303819 were fetched from ChEMBL (35). All of these three assays are functional assays. For CHEMBL4303805, Antiviral activities were determined as inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells at 10 uM after 48 h by high content imaging. Ratios of inhibition of each compound were reported. For CHEMBL4303810, Antiviral activities against SARS-CoV-2 (USA-WA1/2020 strain) were measured by imaging in HRCE cells at MOI 0.4 after 96 h. Hit scores were reported from 0 to 1 for on-disease versus off-disease activity: scores >0.6 are considered hits. For CHEMBL4303819, Inhibition of cell viability relative to arbidol control (inhibition index) was measured by fluorescence (OD590nm) in Vero E6 cells, which were infected with SARS-CoV-2 (strain BavPat1) at MOI 0.002 after 72 h. Inhibition index with a value over 1 indicates higher activity. ChEMBL IDs were translated into CIDs using whole source mappings provided by UniChem (31) and PubChem Identifier Exchange Service as well.
Gene Set Enrichment Analysis (GSEA) is a method that determines whether an a priori defined set of genes show statistically significant expression differences between two biological states (51). Here, three NCBI Gene Expression Omnibus (GEO) data, GSE1739, GSE33267 and GSE122876 were used to identify sets of differentially expressed genes post SARS or MERS infection. Gene expression profiles of drug treatment were retrieved from the Connectivity Map (CMAP) database. The GSEA enrichment scores indicate the potential effectiveness of candidate drugs to reverse the gene expression signature of HCoV infection. For detailed information, please see Zhou et al. (41). Literature was also fetched from COVID 19 Coward Generated Gene and Drug Set Library to assist the user to validate the hypothesis of an interaction between a drug and its target. Only compounds that were reported in experiments were collected.
VIRTUAL SCREENING
There are many softwares that exist for virtual screening such as VSDK (52), AUDocker (53), DockoMatic (54), PyRx (55), etc. However, the efficiency of these tools may not be applicable on large drug banks such as the complete FDA-approved drug list. Therefore, we used AutoDock Vina (version 1.1.2) (56) as the core docking utility to reconstruct our virtual screening pipeline, and ran it on a Kubernetes server with parallel computing (Figure 2). Kubernetes is an engine server, which can automatically deploy, scale and manage the containerized tasks to clusters. In practice, we uploaded the FDA-approved and Taiwan NHI-approved drug lists to Redis data storage, and generated 128 Kuberenetes pods in parallel. Each pod continuously takes up a new docking task of a drug until the Redis drug list is completed. Based on our implementation, the virtual screening for our drug list only costs 3 days per protein.
Figure 2.
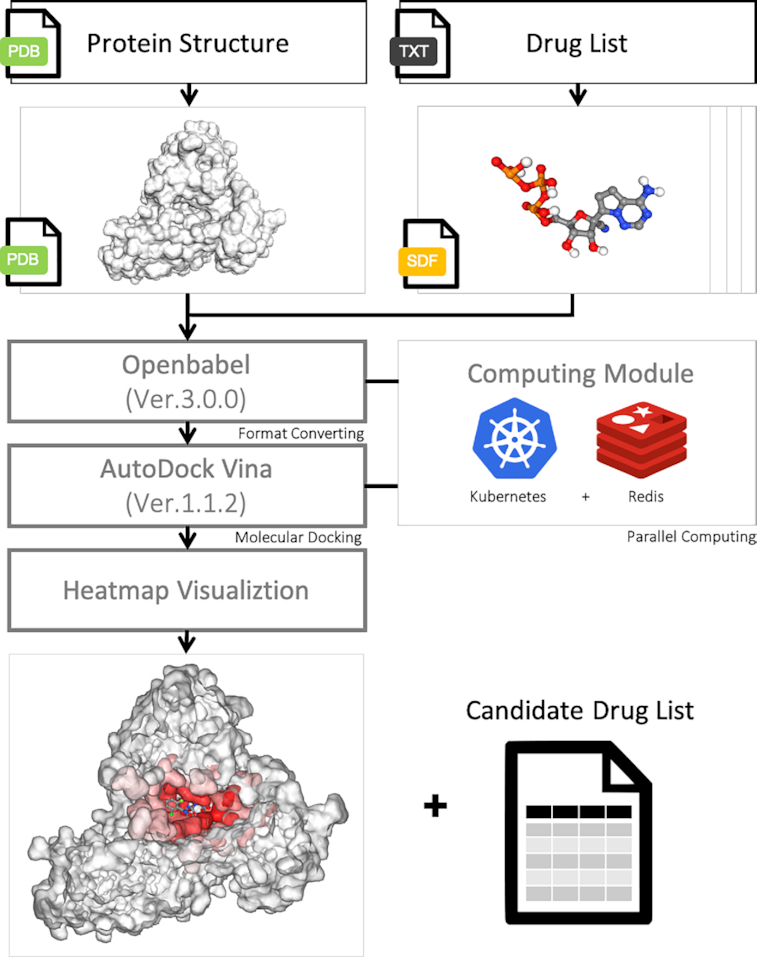
The virtual screening pipeline. We used AutoDock Vina as the core docking utility to reconstruct the virtual screening pipeline. In addition, the Kubernetes server and Redis data store were adopted for parallel computing. After finishing docking, the top 20 docking poses were taken to build a protein heatmap for visualization.
To simulate binding affinity between protein and ligands, each of the drug's 3D structure was obtained from the PubChem database in structure-data file (SDF) format. The gen3d operation of OpenBabel (version 3.0.0) (26) was used for energy minimization. This operation iterated 500 cycles of performing the geometry optimization with the MMFF94 force field and conducting the weighted rotor conformational search, in order to generate a likely global minimum energy conformer in MOL2 file format. Since AutoDock Vina only takes the PDBQT format as input, we used AutoDockTool 1.5.7 (http://autodock.scripps.edu/resources/adt) to convert the file format from MOL2 to PDBQT with default parameter settings excluding a parameter -A, which would add hydrogens to structure only if there are none already. After that, we applied rigid-body docking on these converted files using AutoDock Vina. In order to consider all of the potential docking poses, the entire protein is taken as the search space for blind search. We noted that the number of runs of the docking simulation should be adjusted accordingly by considering the variety of protein sizes. In AutoDock Vina, the number of runs is set by the exhaustiveness parameter, and the default setting of exhaustiveness is set at eight for a search space below 30 × 30 × 30 Å (56). We proportionally scaled the exhaustiveness depending on the size of the protein by a factor of 2. For example, if the size of a protein is 60 × 60 × 60 Å, the exhaustiveness would be 8 × (60/30) × (60 /30) × (60 /30) × 2 = 128. AutoDock Vina reported multiple docking scores for each run, and the top score was selected as the final result shown on the website. The distribution of the final scores of each protein is shown in Figure 3. By comparing the scores between systems (different proteins), there is no bias of docking scores toward any protein we are interested in. By examining the intra-system scores, we observed that for each protein if there is a group or few ligands that are with particularly good docking scores.
Figure 3.
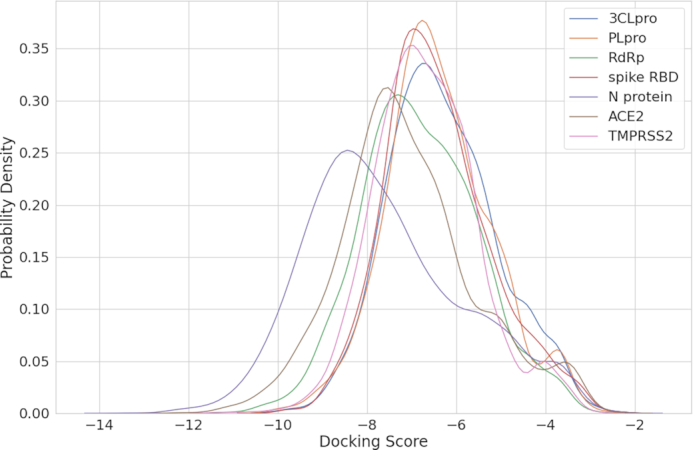
The docking score distribution of each protein. The x-axis is the minimum docking score of each protein–ligand pair, and the y-axis is the probability density estimated by kernel density estimation.
After finishing docking, AutoDock Vina will generate multiple docking poses for each ligand–protein pair. For the sake of a straightforward representation of the docking results, the top 20 docking poses from AutoDock Vina were taken to build a protein heatmap. For each pose, a sphere centered at the centroid of ligand with a radius of 5 Å is highlighted, and the frequency of residues inside the sphere is incremented. The protein heatmap and the ligand docking position with the best affinity score are shown on the main page of each entry (a ligand–protein pair) in DockCoV2, where the red scale of the protein heatmap denotes the frequency of residues over twenty trials. Figure 4 is a demonstration of the structure of 3CLpro (PDB ID: 6LU7) (10) with molecular docking. In order to validate our virtual screening pipeline, we separated the original structure into a ligand and a receptor, and docked them with the proposed pipeline. The left-hand side of Figure 4 shows that eight out of the top 20 docking poses (colored in blue) are located in the binding pocket of the protein. This figure also demonstrates that users can explore the binding pocket through the hot spot of our heatmap. The right-hand side zooms in the binding pocket and shows that the top docking pose (colored in blue) of our pipeline is close to the ligand (colored in red) in the crystal structure. For reproducing our results or repurposing the proposed pipeline for other drugs or proteins, we released our scripts and the configuration file for docking procedures on GitHub (https://github.com/ailabstw/DockCoV2).
Figure 4.
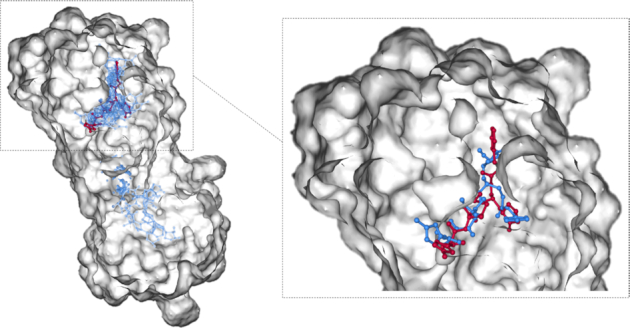
A demonstration of the bound structure of 3CLpro (PDB ID: 6LU7, where the ligand was colored in red) with molecular docking. The left-hand side shows the results of the top 20 docking poses (colored in blue). Eight out of 20 docking poses are located in the binding pocket of the protein. The right-hand side zooms in the binding pocket and shows that the top docking pose (colored in blue) is close to the bound ligand (colored in red).
INTERFACE
In order to provide an efficient searching interface for users, we built a website with a search engine based on Django 3.0 (https://djangoproject.com) and Bootstrap 4.3.1 (https://getbootstrap.com/). The search bar, which can be found either in the index page or on navbar, allows the user to input the drug name, PubChem CID or synonym to explore the docking score with all proteins. The details of each ligand are shown after choosing a specific ligand–protein pair, including docking structure, ligand information and experimental data. PDBQT files of ligand and protein from AutoDock Vina are also provided on the main page of each ligand-protein pair. In the docking structure panel, both protein heatmap and sequences are displayed on the right-side of the detail page. Protein heatmap is interactively demonstrated with the NGL viewer tool, which is a WebGL-based molecular viewer (57). To easily visualize the targeted binding sites on the proteins, not only basic operations on protein heatmap, but also selecting or limiting atoms/residues through the selection language is available on the docking structure panel. The ligand information panel is divided into ligand relative, clinical relative, druglikeness, other relative info and drug similarity. Ligand relative part consists of basic compound Information, including 3D structure view, CAS number, SMILES, InChI, InChI key, synonyms and pathway from KEGG Drug and KEGG Compound databases. Clinical Relative consists of approval information, which denotes whether or not this ligand has been approved by the Taiwan NHI or FDA, as well as other corresponding data from Drug Repurposing Hub, including clinical phase, mechanism of action (MoA), target, disease area, and indication. Results and components of Lipinski's rule of five, Ghose filter, Veber filter and REOS filter are shown in druglikeness section. External links to PubChem and Drugbank are also provided in the other relative info section. SARS-CoV-2 relative, related documents and drug assays are included in experimental data.
CONCLUSION
Drug repositioning, the exploration of repurposing existing drugs that have already passed the safety screening for new indications, is a relatively quick therapeutic solution for emerging infectious diseases like SARS-CoV-2. For the purpose of drug repositioning, molecular docking is a well-known virtual screening method to evaluate a great amount of compounds automatically. Based on the urgent need to find effective drugs, we established DockCoV2, the first virtual screening database for SARS-CoV-2. DockCoV2 focuses on predicting 2285 FDA-approved and 1478 Taiwan NHI-approved drugs targeting five proteins relating to the mechanism of viral entry and replication. Those docked structures could also be further utilized and get more accurate binding poses and the correct ranking of the binding affinity through methods such as molecular mechanics Poisson-Boltzman surface area (MM/PBSA) and molecular mechanics generalized Boltzman surface area (MM/GBSA) calculations (22,58). In addition, DockCoV2 also provides experimental data, including biological assays, pathway information, and gene set enrichment analysis recruited from other validated databases. Future versions will include the prediction of binding affinity with other potentially druggable targets against SARS-CoV-2 and the reported experimental assays and high-throughput screening results to support our prediction.
ACKNOWLEDGEMENTS
We are pleased to thank Professor Chun-Hua Hsu for the useful discussion and Tzu-Jung Hung for help on the website layout design.
Contributor Information
Ting-Fu Chen, Taiwan AI Labs, Taipei 10351, Taiwan.
Yu-Chuan Chang, Taiwan AI Labs, Taipei 10351, Taiwan.
Yi Hsiao, Taiwan AI Labs, Taipei 10351, Taiwan.
Ko-Han Lee, Taiwan AI Labs, Taipei 10351, Taiwan.
Yu-Chun Hsiao, Taiwan AI Labs, Taipei 10351, Taiwan.
Yu-Hsiang Lin, Taiwan AI Labs, Taipei 10351, Taiwan.
Yi-Chin Ethan Tu, Taiwan AI Labs, Taipei 10351, Taiwan.
Hsuan-Cheng Huang, Institute of Biomedical Informatics, National Yang-Ming University, Taipei 11221, Taiwan.
Chien-Yu Chen, Taiwan AI Labs, Taipei 10351, Taiwan; Department of Biomechatronics Engineering, National Taiwan University, Taipei 10617, Taiwan.
Hsueh-Fen Juan, Taiwan AI Labs, Taipei 10351, Taiwan; Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei 10617, Taiwan; Department of Life Science, National Taiwan University, Taipei 10617, Taiwan.
FUNDING
Ministry of Science and Technology, Taiwan [MOST 108-2221-E-002-079-MY3, MOST 109-2221-E-002-161-MY3, MOST109-2327-B-002-009, MOST 109-3114-Y-001-001]; Higher Education Sprout Project [NTU-109L8837A]. Funding for open access charge: Ministry of Science and Technology, Taiwan [MOST 109-3114-Y-001-001]
Conflict of interest statement. None declared.
REFERENCES
- 1. Dong E., Du H., Gardner L.. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020; 20:533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. et al.. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020; 181:1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. et al.. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020; 182:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S. et al.. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020; 181:1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H. 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M. et al.. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020; 182:429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park A., Iwasaki A.. Type I and Type III interferons - induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe. 2020; 27:870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. et al.. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020; 26:1070–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dai W., Zhang B., Jiang X.-M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F. et al.. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020; 368:1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. et al.. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020; 583:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. et al.. Structure of M from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020; 582:289–293. [DOI] [PubMed] [Google Scholar]
- 11. Jin Z., Zhao Y., Sun Y., Zhang B., Wang H., Wu Y., Zhu Y., Zhu C., Hu T., Du X. et al.. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020; 27:529–532. [DOI] [PubMed] [Google Scholar]
- 12. Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C. et al.. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020; 182:417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. et al.. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579:265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. et al.. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X., He S., Zhou Z., Zhou Z., Chen Q. et al.. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B. 2020; 10:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. et al.. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020; 181:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A.. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020; 368:630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R.. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020; 368:409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D.. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect Dis. 2020; 6:2099–2109. [DOI] [PubMed] [Google Scholar]
- 20. Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. et al.. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020; 368:779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M. et al.. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020; 368:1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.-G., Mishra Y.K., Suar M., Ahuja R.. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020; 6:eabb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guy R.K., Kiplin Guy R., DiPaola R.S., Romanelli F., Dutch R.E.. Rapid repurposing of drugs for COVID-19. Science. 2020; 368:829–830. [DOI] [PubMed] [Google Scholar]
- 24. Sterling T., Irwin J.J.. ZINC 15 – ligand discovery for everyone. J. Chem. Inf. Model. 2015; 55:2324–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open Babel: an open chemical toolbox. J. Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. et al.. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020; 581:215–220. [DOI] [PubMed] [Google Scholar]
- 28. Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A. et al.. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J. Biol. Chem. 2004; 279:17996–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. et al.. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018; 46:W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chambers J., Davies M., Gaulton A., Hersey A., Velankar S., Petryszak R., Hastings J., Bellis L., McGlinchey S., Overington J.P.. UniChem: a unified chemical structure cross-referencing and identifier tracking system. J. Cheminform. 2013; 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corsello S.M., Bittker J.A., Liu Z., Gould J., McCarren P., Hirschman J.E., Johnston S.E., Vrcic A., Wong B., Khan M. et al.. The Drug Repurposing Hub: a next-generation drug library and information resource. Nat. Med. 2017; 23:405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M. et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li G., De Clercq E.. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat. Rev. Drug Discov. 2020; 19:149–150. [DOI] [PubMed] [Google Scholar]
- 37. Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y.. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016; 15:327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y. et al.. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020; 181:894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nguyen H., Case D.A., Rose A.S.. NGLview–interactive molecular graphics for Jupyter notebooks. Bioinformatics. 2018; 34:1241–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F.. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988; 28:31–36. [Google Scholar]
- 43. Kim S., Thiessen P.A., Cheng T., Zhang J., Gindulyte A., Bolton E.E.. PUG-View: programmatic access to chemical annotations integrated in PubChem. J. Cheminform. 2019; 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J.. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001; 46:3–26. [DOI] [PubMed] [Google Scholar]
- 45. Ghose A.K., Viswanadhan V.N., Wendoloski J.J.. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999; 1:55–68. [DOI] [PubMed] [Google Scholar]
- 46. Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D.. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002; 45:2615–2623. [DOI] [PubMed] [Google Scholar]
- 47. Walters W.P., Namchuk M.. Designing screens: how to make your hits a hit. Nat. Rev. Drug Discov. 2003; 2:259–266. [DOI] [PubMed] [Google Scholar]
- 48. Hubbard R.E. Hubbard R.E. Fragment Screening: An Introduction. Structure-Based Drug Discovery. 2006; Royal Society of Chemistry; 142–172. [Google Scholar]
- 49. Willett P., Barnard J.M., Downs G.M.. Chemical similarity searching. J. Chem. Inf. Comput. Sci. 1998; 38:983–996. [Google Scholar]
- 50. Rogers D., Hahn M.. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010; 50:742–754. [DOI] [PubMed] [Google Scholar]
- 51. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Baba N., Akaho E.. VSDK: virtual screening of small molecules using AutoDock Vina on Windows platform. Bioinformation. 2011; 6:387–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sandeep G., Nagasree K.P., Hanisha M., Kumar M.M.K.. AUDocker LE: a GUI for virtual screening with AUTODOCK Vina. BMC Res. Notes. 2011; 4:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bullock C., Cornia N., Jacob R., Remm A., Peavey T., Weekes K., Mallory C., Oxford J.T., McDougal O.M., Andersen T.L.. DockoMatic 2.0: high throughput inverse virtual screening and homology modeling. J. Chem. Inf. Model. 2013; 53:2161–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dallakyan S., Olson A.J.. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015; 1263:243–250. [DOI] [PubMed] [Google Scholar]
- 56. Trott O., Olson A.J.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlic A., Rose P.W.. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018; 34:3755–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang E., Sun H., Wang J., Wang Z., Liu H., Zhang J.Z.H., Hou T.. End-Point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 2019; 119:9478–9508. [DOI] [PubMed] [Google Scholar]