Abstract
CRISPR–Cas is an anti-viral mechanism of prokaryotes that has been widely adopted for genome editing. To make CRISPR–Cas genome editing more controllable and safer to use, anti-CRISPR proteins have been recently exploited to prevent excessive/prolonged Cas nuclease cleavage. Anti-CRISPR (Acr) proteins are encoded by (pro)phages/(pro)viruses, and have the ability to inhibit their host's CRISPR–Cas systems. We have built an online database AcrDB (http://bcb.unl.edu/AcrDB) by scanning ∼19 000 genomes of prokaryotes and viruses with AcrFinder, a recently developed Acr-Aca (Acr-associated regulator) operon prediction program. Proteins in Acr-Aca operons were further processed by two machine learning-based programs (AcRanker and PaCRISPR) to obtain numerical scores/ranks. Compared to other anti-CRISPR databases, AcrDB has the following unique features: (i) It is a genome-scale database with the largest collection of data (39 799 Acr-Aca operons containing Aca or Acr homologs); (ii) It offers a user-friendly web interface with various functions for browsing, graphically viewing, searching, and batch downloading Acr-Aca operons; (iii) It focuses on the genomic context of Acr and Aca candidates instead of individual Acr protein family and (iv) It collects data with three independent programs each having a unique data mining algorithm for cross validation. AcrDB will be a valuable resource to the anti-CRISPR research community.
INTRODUCTION
Prokaryotes (bacteria and archaea) are constantly attacked by viruses (1). According to the red queen hypothesis, prokaryotes and their viruses have been under endless arms race for billions of years (2). To avoid viral infections, prokaryotes have evolved an arsenal of anti-viral defense mechanisms (3) encoded in their genomes, e.g., restriction–modification (RM) systems, CRISPR–Cas systems and toxin–antitoxin (TA) systems (4,5). To overcome these defense systems, viruses have developed anti–anti-viral (or anti-defense) strategies (6), among which anti-CRISPRs specifically inhibit CRISPR–Cas systems of their hosts (7).
In addition, the current CRISPR–Cas genome editing tools, which are widely employed in numerous research labs and companies worldwide, are not perfectly safe to use in humans (8,9). As the naturally occurring inhibitors of CRISPR–Cas, anti-CRISPRs have a great application in the development of safer and more controllable CRISPR–Cas genome editing tools. Since 2017, over 15 cases have already been published to apply Acrs to finely control CRISPR–Cas gene editing, gene regulation, epigenetic modification, DNA imaging, and gene drive (recently reviewed in (10)). It is certain that more Acr applications will happen with acceleratingly more Acrs being discovered.
Anti-CRISPR (Acr) proteins were first discovered in 2013 in Pseudomonas phages and prophages (11). Acr encoding genes often form operons with putative transcription regulator genes that encode Acr-associated (Aca) proteins (12,13). Since 2013, 65 experimentally characterized Acr proteins (43 since 2018 and 18 already in 2020) have been published, and most of them had been identified with the help of bioinformatics (7,14,15). Notably, very little sequence similarity is found between characterized Acrs, and most of them do not have any conserved Pfam domains and have no homology to known proteins. Given that CRISPR–Cas is so broadly distributed in prokaryotes (16) and the genomic diversity of their viruses is astronomically high (17), it is believed that the 65 experimentally characterized anti-CRISPRs only represent the tiny tip of an iceberg of the possible anti-CRISPR diversity in nature (7,12,18). This means that more Acr subtypes are waiting to be discovered in various taxonomic groups, and more advanced computational approaches are needed other than the classical sequence homology-based approach.
Bioinformatics prescreening genomes has been a critical step in characterization of new anti-CRISPRs (7,14,15). However, compared to experimental studies, relatively fewer bioinformatics tool development has been published for anti-CRISPR research. Table 1 lists all bioinformatics resources that are currently online, among which two are databases for experimentally characterized Acrs and their homologs (19,20), and one (21) is a database for Acrs predicted with machine learning.
Table 1.
Online bioinformatics tools for Acr research
| Name (ref.) | Year | Resource provided | Features |
|---|---|---|---|
| Anti-CRISPRDB (19) | 2018 | Database | Experimentally characterized Acrs and their homologs and BLAST search |
| CRISPRminer (20) | 2018 | Database | Experimentally characterized Acrs and their homologs and genomic context |
| Acr nomenclature (14) | 2018 | Google spreadsheets | Experimentally characterized Acrs and Acas nomenclature |
| Self-Targeting Spacer Searcher (27) | 2018 | Standalone package | Workflow for self-targeting spacer identification |
| AcrCatalog (21) | 2020 | Database and model code | Predicted Acrs from decision tree ML classifier + heuristic filtering |
| AcRanker (23) | 2020 | Web server and standalone package | XGBoost classifier using AA biases |
| AcrFinder (22) | 2020 | Web server and standalone package | Workflow combining Homology + GBA + self-targeting and user-friendly website |
| PaCRISPR (24) | 2020 | Web server | SVM classifier using PSSMs to capture evolutionary features |
| AcrDetector (29) | 2020 | Model code | Decision tree classifier using six sequence features |
Additionally, in the year 2020, four standalone/webserver bioinformatics tools become available for automated Acr discovery (Table 1). Three of these tools were developed by authors of this paper: AcrFinder (22), AcRanker (23) and PaCRISPR (24). AcrFinder implemented a bioinformatics pipeline that was reported in (25), where we developed a bioinformatics pipeline to identify genomic operons containing Acr homologs and/or Aca homologs by combining three computational approaches: homology, GBA (guilt-by-association), and STS (self-targeting spacer) (26). AcRanker uses amino acid compositions of known Acr and non-Acr proteins to train an Extreme Gradient Boosting (XGBoost) ranking algorithm to rank putative Acr proteins in a given test proteome, and has been used to assist experimental discovery of novel Acr proteins (23). PaCRISPR builds position-specific scoring matrices (PSSMs) to capture evolutionary features in Acr alignments, and then trains a support vector machine (SVM) for Acr classification (24). All these three recent tools have their strengths and weaknesses, and we were motivated to combine predictions from three independent tools to create a comprehensive Acr-Aca operon database with a user-friendly website.
Briefly, combining AcrFinder (22), AcRanker (23), and PaCRISPR (24), we have performed a large-scale bioinformatics data mining for putative Acr-Aca operons in all sequenced genomes of bacteria, archaea, and their viruses. As a result, a new pre-computed Acr-Aca operon database AcrDB was developed. As described in our previous papers (22) and (25), putative Acr-Aca operons are defined as genomic loci meeting the following criteria: (i) all genes on the same strand (running in the same direction); (ii) all intergenic distance <150 bp; (iii) all genes encode proteins <200 aa; (iv) at least one gene homologous to known Acr or Aca proteins.
The only three published online Acr databases (Table 1) include: anti-CRISPRDB (19), CRISPRminer (20), and AcrCatalog (21). Anti-CRISPRDB and CRISPRminer focus on experimentally characterized Acr proteins and their homologs. AcrCatalog provides predicted Acrs by using a decision tree-based machine learning algorithm but has very minimum web utilities.
DATA COLLECTION
We have scanned 15 203 complete bacterial genomes, 961 (complete and draft) archaeal genomes and 2658 complete prokaryotic viral genomes of the NCBI RefSeq database to identify potential Acrs and their associated operons. Here ‘draft’ means the genomes contain contigs or scaffolds with gaps, while ‘complete’ means the genomes are fully assembled into chromosomes without gaps. The scanning process (Figure 1) was started by running our standalone AcrFinder (https://github.com/HaidYi/acrfinder) program on all the genomes, followed by running AcRanker on genomes containing predicted Acr-Aca operons. We have also run PaCRISPR only on proteins in AcrFinder predicted Acr-Aca operons, as PaCRISPR is much slower than AcrFinder and AcRanker and not suitable for genome-scale Acr predictions. In other words, data in AcrDB were primarily generated by AcrFinder, but were further scored and ranked by AcRanker and PaCRISPR.
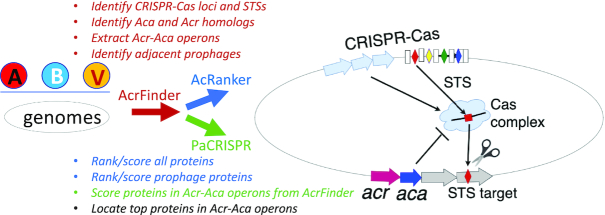
Figure 1.
Overview of data collection for AcrDB. Genomes of A (Archaea), B (Bacteria), and V (Viruses) are used as input for AcrFinder. Within AcrFinder, Acr homologs, Aca homologs, CRISPR–Cas loci, self-targeting spacers (STSs) and their targets (red diamonds) are identified. Acr and Aca homologs are analyzed to see if they form operons, if they are adjacent to or within a prophage region and/or to an STS target. Genomes with AcrFinder result are further analyzed by AcRanker to receive a rank and score, by PaCRISPR to receive a score, and lastly to locate the top ranked proteins in Acr-Aca operons.
The detailed algorithm and methodology of the three tools, as well as their prediction evaluation results have been described in our previous papers (22–25). Briefly, AcrFinder integrates a multi-step genome processing pipeline, which includes identifications of CRISPR–Cas loci, self-targeting spacers (STSs) (27), Acr and Aca homologs, Acr-Aca operons, prophages, and examination of gene neighborhood of these genetic elements (Figure 1). According to the presence/absence of STSs and the proximity to STSs in the genome, Acr-Aca operons are also classified into three groups: high (presence of STSs nearby), medium (presence of STSs but not nearby), or low confidence (absence of STSs in the genome) groups (22). After the AcrFinder step, 419, 5850 and 2044 genomes of archaea, bacteria and viruses, respectively, were found to have Acr-Aca operons each containing at least one Acr homolog or Aca homolog. For these genomes, two AcRanker runs were made (Figure 1): one to rank all the proteins in the genome (i.e. the complete proteome), while the other only to rank all the prophage proteins in the genome (a subset of the proteome). The top 10% ranked proteins in the two runs were then intersected with the Acr-Aca operons from AcrFinder. Acr-Aca operons containing top 10% ranked proteins are considered to be more confident candidates according to the AcRanker paper (23). Meanwhile, after the AcrFinder step, all the 57 879 proteins (unique IDs) of the predicted Acr-Aca operons were processed by PaCRISPR to receive a prediction score (Figure 1) and those with a score >0.5 were considered as more confident candidates according to the PaCRISPR paper (24).
DATABASE CONTENT
As shown in Table 2, Acr-Aca operons with homologs of known Acr proteins are found in only a small percentage of genomes: 2.8% (27/961) Archaea, 7.4% Bacteria and 3.4% Viruses. This is expected as most known Acr proteins tend to have very few homologs in the public databases. These percentages, however, are much higher for Acr-Aca operons with Aca homologs (all confidence levels, defined in (22)): 43.5% (418/961) Archaea, 33.0% Bacteria and 76.9% Viruses, consistent with the fact that Aca proteins are much more conserved than Acr proteins. Furthermore, although genomes without CRISPR self-targeting spacers (STSs) can also have anti-CRISPRs (25), genomes with STSs are more likely to encode Acrs to avoid autoimmunity. When only considering operons found in genomes with STSs, as only a small number of genomes have STSs, a much lower percentage of genomes is found to have both STSs and Acr-Aca operons (high and medium confidence levels): 8.8% (85/961) Archaea and 7.2% Bacteria.
Table 2.
Statistics of data in AcrDB
| # of Genomes, genera and Acr-Aca operons | Archaea | Bacteria | Viruses |
|---|---|---|---|
| Total genomes searched | 961 | 15,203 | 2,659 |
| Genomes (genera) with Acr homologs in Acr-Aca operonsb | 27 (2) | 1127 (97) | 91(20) |
| Genomes (genera) with Aca homologs in Acr-Aca operons | 418 (94) | 5014 (603) | 2043 (424) |
| Acr-Aca operons of high, medium, and low confidence levels | 1481 | 31 683 | 4125 |
| Genomes (genera) with Aca homologs in Acr-Aca operons and STSs | 85 (36) | 1101 (244) | NAa |
| Acr-Aca operons of high and medium confidence levels | 361 | 7,889 | NAa |
| Genomes (genera) with Aca homologs in Acr-Aca operons and candidate Acrs ranked in 10% by AcrRanker | 293 (79) | 4753 (609) | 1330 (314) |
| Acr-Aca operons of high, medium, and low confidence levels | 634 | 17 208 | 1,857 |
| Genomes (genera) with Aca homologs in Acr-Aca operons and STSs and candidate Acrs ranked in 10% by AcrRanker | 67 (29) | 1045 (236) | NAa |
| Acr-Aca operons of high and medium confidence levels | 75 | 2,565 | NAa |
| Genomes (genera) with Aca homologs in Acr-Aca operons and candidate Acrs verified by PaCRISPR (score > 0.5) | 247 (59) | 2799 (446) | 1542 (330) |
| Acr-Aca operons of high, medium, and low confidence levels | 359 | 5,706 | 2455 |
| Genomes (genera) with Aca homologs in Acr-Aca operons and STSs and candidate Acrs verified by PaCRISPR (score > 0.5) | 53 (25) | 796 (185) | NAa |
| Acr-Aca operons of high and medium confidence levels | 64 | 1869 | NAa |
aNA because viruses were not analyzed for the presence of CRISPR–Cas and STSs.
bThese include single gene operon (i.e. only the Acr homolog).
As mentioned above, proteins were also ranked/scored by AcRanker and PaCRISPR (Figure 1) in terms of the likelihood of sharing similar amino acid compositions or sharing sequence profiles with known Acr proteins. When counting Acr-Aca operons encoding at least one protein ranked in the top 10% of all the proteins of the genome, 30.5% (293/961) archaeal, 31.3% bacterial and 50.0% of viral genomes remained to have Acr-Aca operons. Applying all the conditions (with Aca homologs in the operon, with STSs in the genome, and with proteins ranked in the top 10% by AcRanker) as the filters found only 75 and 2565 Acr-Aca operons from 67 (7.0%) and 1045 (6.9%) genomes of Archaea and Bacteria, respectively. Similarly, when counting Acr-Aca operons encoding at least one protein with PaCRISPR score >0.5, 25.7% (247/961) archaeal, 18.4% bacterial and 58.0% of viral genomes remained to have Acr-Aca operons. Applying all the conditions (with Aca homologs in the operon, with STSs in the genome, and with proteins scored >0.5 by PaCRISPR) as the filters found only 64 and 1869 Acr-Aca operons from 53 (5.5%) and 796 (5.2%) genomes of Archaea and Bacteria, respectively.
Altogether AcrFinder identified 1481 (Archaea) + 31 683 (Bacteria) + 4125 (Viruses) = 37 289 Acr-Aca operons (Table 1 and Figure 2A). These operons include 25,353 (68.0%) operons that are from genomes with STSs, or contain at least one protein meeting the rank/score thresholds of AcRanker and PaCRISPR (Figure 2A). Operons in the intersection of the three circles (STS, AcRanker and PaCRISPR) in Figure 2A only make up a very small percentage (1458/37 289 = 3.9%) of operons, but obviously represent the most confident predictions (Supplementary Table S1) in AcrDB. The taxonomy distribution of Acr-Aca operons of different confidence levels indicates that anti-CRISPRs are widely distributed in 41 phyla of prokaryotes and viruses (Figure 2B and Supplementary Table S2). Specifically, Acr-Aca operons with Acr homologs are found in 12 phyla (first column of Figure 2B), Acr-Aca operons with Aca homologs are found in 41 phyla (second column of Figure 2B), and even the most accurate Acr-Aca operons in the intersection of STS, AcRanker and PaCRISPR are also found in 17 phyla (eighth column of Figure 2B). The taxonomy distribution of Acr-Aca operons at the class level is available in Supplementary Figure S1 and Supplementary Table S3.
Figure 2.
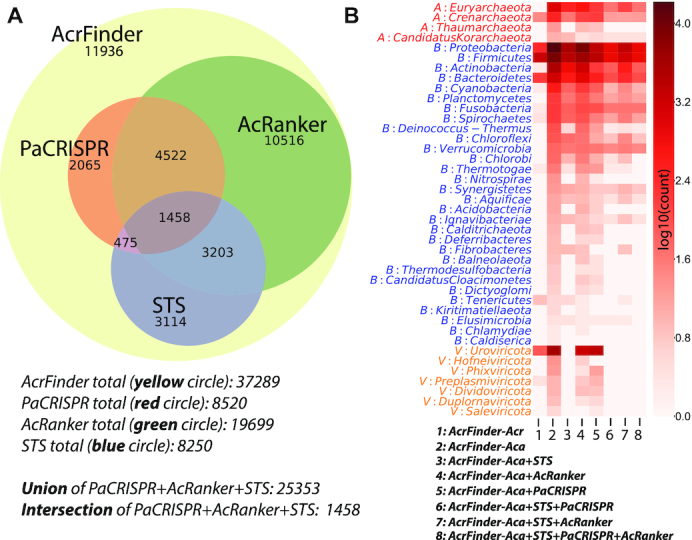
Overview of Acr-Aca operons in AcrDB. (A) Venn diagram of the three circles (STS, AcRanker, PaCRISPR) within the AcrFinder circle (37 289 operons containing Aca homologs). (B). Heatmap of the taxonomy distribution of Acr-Aca operons meeting different criteria. Each row is a taxonomic phylum (V: Virus, B: Bacteria, A: Archaea). There are eight columns: 1. Acr-Aca operons containing Acr homologs; 2. Acr-Aca operons containing Aca homologs; 3. Acr-Aca operons containing Aca homologs and in genomes with STSs; 4. Acr-Aca operons containing Aca homologs and candidate Acrs ranked in top 10% by AcrRanker; 5. Acr-Aca operons containing Aca homologs and candidate Acrs scored by PaCRISPR >0.5; 6. Acr-Aca operons containing Aca homologs and candidate Acrs scored by PaCRISPR >0.5 and in genomes with STSs; 7. Acr-Aca operons containing Aca homologs and candidate Acrs ranked in top 10% by AcrRanker and in genomes with STSs; 8. Acr-Aca operons containing Aca homologs and candidate Acrs scored by PaCRISPR >0.5 and ranked in top 10% by AcrRanker and in genomes with STSs.
WEB DESIGN
AcrDB is powered by PHP+MySQL+Apache2+JavaScript. The web interface provides access to all the pre-computed data that were collected for Acr-Aca operons from 419, 5850 and 2044 genomes of Archaea, Bacteria, and Viruses, respectively. To browse the data (Figure 3A), users have three options:
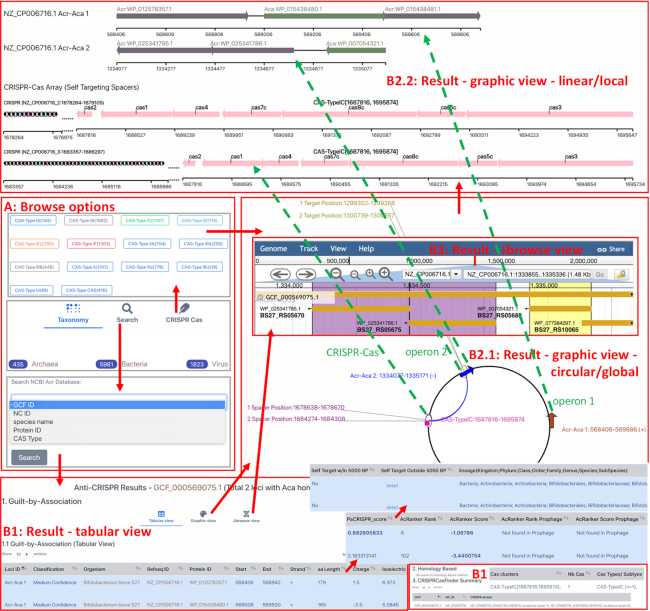
Figure 3.
Screenshots of AcrDB website to demonstrate its utilities. (A) Browse page allows three options to enter the database. The default is entry by CRISPR Cas Type. One can also click on Taxonomy for browsing by the NCBI Taxonomy tree, and click on Search to type in keywords and search different data fields. (B) Result page has different components to better organize the genome-specific results. Example screenshots shown here are taken from this URL: http://bcb.unl.edu/AcrDB/anti_crispr_results.php?type=ncbi&organism=GCF_000569075.1. (B1) shows the default tabular view, where a large table is displayed. The table has 23 columns and one can use mouse to move to other parts of the table. The bottom of the result page also shows a CRISPRCasFinder summary Table. B2.1 and B2.2 are on the same page and can be seen by clicking on Graphic view in B1. (B2.1) shows the graphic global view of the genome in a circular representation. Different features are shown as different shapes, and the size of shapes do not indicate their real size in the genome: the Acr-Aca operon positions (as arrows), the STS (purple ovals) and their target positions (yellow green ovals) connected with blue arcs, and the Cas loci positions (pink squares). The green dotted lines point from these Acr-Aca operons (arrows) to their corresponding local view (B2.2) or Jbrowse view (B3). In B2.2, each Acr-Aca operon is shown with component genes (directional arrows) together with a ruler to indicate its position in the genome. CRISPR–Cas arrays are shown if they contain an STS and have an adjacent Cas locus (<10 kb). The size of the CRISPR array and Cas loci is proportional to their real lengths.
By CRISPR–Cas types. This is for Bacteria and Archaea only. According to AcrFinder, Acr-Aca operons of prokaryotes must be from genomes with complete CRISPR–Cas systems (levels 3 and 4 of CRISPRCasFinder (28)). Therefore, CRISPR–Cas types of the genome were assigned to the Acr-Aca operons (see (22)).
By Taxonomy. This is for Bacteria, Archaea and Viruses. Taxonomy lineages of all the genomes was retrieved from NCBI Taxonomy database.
By Keyword search. This is for Bacteria, Archaea, and Viruses. Different query fields (e.g. RefSeq GCF ID or species name) are allowed to change and search. The input query can be autocompleted while the user is typing.
The result page contains three pieces (Figure 3B1) of data from AcrFinder: guilt-by-association (GBA) result (operons containing Aca homologs), homology based result (operons containing Acr homologs), and CRISPRCasFinder result (to infer subtypes for Acr-Aca operons, and to identify STSs within AcrFinder pipeline, see (22)). The GBA data has been further organized to have three different views for better presentation:
Tabular view (Figure 3B1) provides a table with 23 columns including various information of the identified operons. Pre-computed data are gathered with different tools and organized for biologists to quickly look for interested targets, such as protein sequence, sequence length, isoelectric point, Aca HTH domain match, PaCRISPR score (bold if >0.5), AcRanker rank/score (bold if score > –5), phage protein homologs, proximity to STSs, and taxonomy lineages.
Graphic view (Figure 3B2) is designed only for Bacteria, because all bacterial genomes are completely assembled. Two component graphs are provided, one for a global view of the entire genome (shown as a circle as most bacterial genomes are circular (Figure 3B2.1)), and the other for a local view of Acr-Aca operons and CRISPR–Cas loci in a linear representation (Figure 3B2.2). For the global view, in the circular genome, various important sites/loci are indicated as different shapes (size not proportional to the real base pair length). It should be mentioned that a genome can contain multiple CRISPR–Cas loci and genomes do not contain STS are not shown in this global view. For the local view, in each Acr-Aca operon, each arrow represents a gene, and the arrow direction represents the strand, and the size of arrows is proportional to the real length of the gene. The CRISPR–Cas loci that contain self-targeting spacers (STSs) are also shown in this local view. To find the CRISPR array and Cas locus pair, the CRISPR array that contains STSs is located first, then look for neighboring Cas loci. If the distance between the CRISPR array and the neighboring Cas locus is larger than 10 kb, we do not show them.
Jbrowse view (Figure 3B3) shows the genomic context of each Acr-Aca operon. Unlike the static global and local views, the Jbrowse view is dynamic, meaning one can use the menu and navigation icons of Jbrowse to zoom in or out and move the window to either directions. Nucleotide sequence of each gene and other RefSeq annotation data can be retrieved by clicking on each gene in this view.
Additionally, we also provide a very detailed Help page to explain the website, and a Statistics page to allow users to quickly grab the data summary and navigate to important dataset (e.g. link to the most confident Acr-Aca operon or certain taxonomy groups). The Download page provides the flat files that are organized as different folders, as compressed archives, Fasta sequence files, or TSV text files, and can be batch downloaded conveniently. The links to our AcrFinder web server, PaCRISPR server, AcRanker server, and other related bioinformatics resources are also provided.
CONCLUSIONS
AcrDB provides a collection of computationally predicted Acr-Aca operons that are present in >7000 RefSeq genomes of prokaryotes and their viruses. Among three existing databases (anti-CRISPRDB (19), CRISPRminer (20) and AcrCatalog (21)), anti-CRISPRDB and CRISPRminer collect experimentally characterized Acr proteins and their BLAST homologs, equivalent to AcrDB’s very small Acr homolog dataset (Table 2). AcrCatalog is more similar to AcrDB, as both focus on computationally predicted data.
However, AcrDB differs from AcrCatalog in the following aspects, which can be considered as advantages of AcrDB: (i) AcrDB offers a more user-friendly web interface with various browsing, viewing in graphical representations, searching, and downloading options, while AcrCatalog does not provide any of these functions except for tabular browsing (http://acrcatalog.pythonanywhere.com/catalog/); (ii) AcrDB contains a much larger data (37 289 Acr-Aca operons [total 99 648 proteins and 56 851 unique IDs] with Aca homologs, and 2477 Acr-Aca operons [total 4359 proteins and 1567 unique IDs] with Acr homologs) than AcrCatalog (16 919 Acr proteins of 2500 Acr families (21)); (iii) AcrDB features the genomic context of Acr and Aca homologs, while AcrCatalog focuses on clustering Acr proteins into sequence similarity-based families; and (iv) AcrDB collects data with three independent tools each having a unique data mining algorithm (AcrFinder: guilt-by-association, STS, and gene neighborhood-based pipeline; AcRanker: amino acid k-mer composition-based XGBoost classifier; PaCRISPR: evolutionary conservation in a form of Acr sequence alignment-based SVM classifier), while AcrCatalog uses a random forest (RF) classifier capturing eight Acr sequence features. The idea of using multiple tools for cross validation and performance improvement has been commonly used in genomic data science, and future development of AcrDB will consider further incorporate AcrCatalog standalone package and other new tools in our data collection pipeline.
To conclude, AcrDB focuses on providing user-friendly access to computationally predicted Acr-Aca operons rather than experimentally characterized Acrs and homologs. All the three tools used for AcrDB data collection have been extensively evaluated in terms of their prediction performances in our recent papers (22–25). Various types of computational evidence are provided in a tabular view, e.g. Acr homology, Aca homology, AcRanker rank and score, PaCRISPR score, neighboring prophage genes and presence/absence of STSs in the genome. Graphical representation is also developed to view identified Acr-Aca operons, CRISPR–Cas loci, STS and their targets in the genome presented as a circle or as individual zoomed-in gene clusters. All pre-computed data are available through a batch download page, including AcrFinder GBA, Acr homology, AcRanker, PaCRISPR, and CRISPR–Cas results.
FUTURE WORK
As acknowledged in our recent paper (23), AcrFinder has two limitations: (i) it requires Acrs be in the gene neighborhood of HTH-containing Acas, which will miss Acr proteins that do not need Aca regulators; and (ii) it requires Acrs be in prokaryotic genomes with complete CRISPR–Cas systems, which will fail to identify anti-CRISPRs in genomes with decayed/incomplete CRISPR–Cas systems or without CRISPR–Cas systems at all (possible according to (25)). Although we have purposely imposed these requirements in AcrFinder algorithm in order to reduce false positives, clearly these limitations will lead to false negatives and have been inherited by the current version of AcrDB. Both AcRanker and PaCRISPR have no such limitations; however, PaCRISPR is not designed for genome-scale Acr discovery due to its time-consuming PSSM building step, and AcRanker's scores/ranks are not a good indicator of prediction confidence. For future development, we will continue to improve these programs by considering more Acr sequence features (e.g. protein 3D structure, protein charge and isoelectric point), and explore new machine learning-based algorithms that will address limitations of current programs.
We plan to update AcrDB annually with newly characterized Acr and Aca proteins as well as our improved computational tools (AcrFinder, AcRanker, PaCRISPR and others). We will also include more genomes (not only complete but also draft genomes) and metagenomes in the discovery of Acr-Aca operons. AcrDB will complement existing anti-CRISPR databases (Table 1), provide the largest collection of genome-wide Acr-Aca operons, and facilitate the experimental characterization of new Acr proteins and the development of safer CRISPR–Cas genome editing technologies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was partially completed utilizing the Holland Computing Center of the University of Nebraska, which receives support from the Nebraska Research Initiative.
Contributor Information
Le Huang, Department of Genetics, University of North Carolina at Chapel Hill, NC, USA.
Bowen Yang, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska - Lincoln, Lincoln, NE 68588, USA.
Haidong Yi, Department of Computer Science, University of North Carolina at Chapel Hill, NC, USA.
Amina Asif, Pakistan Institute of Engineering and Applied Sciences (PIEAS), PO Nilore, Islamabad, Pakistan; Department of Computer Science, National University of Computer and Emerging Sciences (NUCES), Islamabad, Pakistan.
Jiawei Wang, Infection and Immunity Program, Biomedicine Discovery Institute and Department of Microbiology, Monash University, VIC 3800, Australia.
Trevor Lithgow, Infection and Immunity Program, Biomedicine Discovery Institute and Department of Microbiology, Monash University, VIC 3800, Australia.
Han Zhang, College of Artificial Intelligence, Nankai University, Tianjin, China.
Fayyaz ul Amir Afsar Minhas, Department of Computer Science, University of Warwick, Coventry, UK.
Yanbin Yin, Nebraska Food for Health Center, Department of Food Science and Technology, University of Nebraska - Lincoln, Lincoln, NE 68588, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) CAREER award [DBI-1933521], United States Department of Agriculture (USDA) award [58-8042-9-089]; start-up grant of UNL [2019-YIN to Y.Y.]. Funding for open access charge: NSF CAREER award [DBI-1933521].
Conflict of interest statement. None declared.
REFERENCES
- 1. Rostol J.T., Marraffini L.. (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe. 2019; 25:184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stern A., Sorek R.. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011; 33:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernheim A., Sorek R.. The pan-immune system of bacteria: antiviral defence as a community resource. Nat. Rev. Microbiol. 2020; 18:113–119. [DOI] [PubMed] [Google Scholar]
- 4. Hampton H.G., Watson B.N.J., Fineran P.C.. The arms race between bacteria and their phage foes. Nature. 2020; 577:327–336. [DOI] [PubMed] [Google Scholar]
- 5. Dy R.L., Richter C., Salmond G.P., Fineran P.C.. Remarkable mechanisms in microbes to resist phage infections. Annu Rev Virol. 2014; 1:307–331. [DOI] [PubMed] [Google Scholar]
- 6. Samson J.E., Magadan A.H., Sabri M., Moineau S.. Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 2013; 11:675–687. [DOI] [PubMed] [Google Scholar]
- 7. Pawluk A., Davidson A.R., Maxwell K.L.. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 2018; 16:12–17. [DOI] [PubMed] [Google Scholar]
- 8. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P et al.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li H., Yang Y., Hong W., Huang M., Wu M., Zhao X.. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther. 2020; 5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marino N.D., Pinilla-Redondo R., Csorgo B., Bondy-Denomy J.. Anti-CRISPR protein applications: natural brakes for CRISPR–Cas technologies. Nat. Methods. 2020; 17:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bondy-Denomy J., Pawluk A., Maxwell K.L., Davidson A.R.. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013; 493:429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Borges A.L., Davidson A.R., Bondy-Denomy J.. The discovery, mechanisms, and evolutionary impact of Anti-CRISPRs. Annu. Rev. Virol. 2017; 4:37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bondy-Denomy J. Protein inhibitors of CRISPR–Cas9. ACS Chem. Biol. 2018; 13:417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bondy-Denomy J., Davidson A.R., Doudna J.A., Fineran P.C., Maxwell K.L., Moineau S., Peng X., Sontheimer E.J., Wiedenheft B.. A unified resource for tracking Anti-CRISPR names. CRISPR J. 2018; 1:304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stanley S.Y., Maxwell K.L.. Phage-Encoded Anti-CRISPR defenses. Annu. Rev. Genet. 2018; 52:445–464. [DOI] [PubMed] [Google Scholar]
- 16. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H et al.. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dion M.B., Oechslin F., Moineau S.. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020; 18:125–138. [DOI] [PubMed] [Google Scholar]
- 18. Davidson A.R., Lu W.T., Stanley S.Y., Wang J., Mejdani M., Trost C.N., Hicks B.T., Lee J., Sontheimer E.J.. Anti-CRISPRs: Protein inhibitors of CRISPR–Cas systems. Annu. Rev. Biochem. 2020; 89:309–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dong C., Hao G.F., Hua H.L., Liu S., Labena A.A., Chai G., Huang J., Rao N., Guo F.B.. Anti-CRISPRdb: a comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 2018; 46:D393–D398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang F., Zhao S., Ren C., Zhu Y., Zhou H., Lai Y., Zhou F., Jia Y., Zheng K., Huang Z.. CRISPRminer is a knowledge base for exploring CRISPR–Cas systems in microbe and phage interactions. Commun Biol. 2018; 1:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gussow A.B., Park A.E., Borges A.L., Shmakov S.A., Makarova K.S., Wolf Y.I., Bondy-Denomy J., Koonin E.V.. Machine-learning approach expands the repertoire of anti-CRISPR protein families. Nat. Commun. 2020; 11:3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yi H., Huang L., Yang B., Gomez J., Zhang H., Yin Y.. AcrFinder: genome mining anti-CRISPR operons in prokaryotes and their viruses. Nucleic Acids Res. 2020; 48:W358–W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eitzinger S., Asif A., Watters K.E., Iavarone A.T., Knott G.J., Doudna J.A., Minhas F.. Machine learning predicts new anti-CRISPR proteins. Nucleic Acids Res. 2020; 48:4698–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Dai W., Li J., Xie R., Dunstan R.A., Stubenrauch C., Zhang Y., Lithgow T.. PaCRISPR: a server for predicting and visualizing anti-CRISPR proteins. Nucleic Acids Res. 2020; 48:W348–W357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yin Y., Yang B., Entwistle S.. Bioinformatics identification of Anti-CRISPR loci by using homology, Guilt-by-Association, and CRISPR Self-Targeting spacer approaches. mSystems. 2019; 4:e00455-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rauch B.J., Silvis M.R., Hultquist J.F., Waters C.S., McGregor M.J., Krogan N.J., Bondy-Denomy J.. Inhibition of CRISPR–Cas9 with bacteriophage proteins. Cell. 2017; 168:150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Watters K.E., Fellmann C., Bai H.B., Ren S.M., Doudna J.A.. Systematic discovery of natural CRISPR–Cas12a inhibitors. Science. 2018; 362:236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Couvin D., Bernheim A., Toffano-Nioche C., Touchon M., Michalik J., Neron B., Rocha E.P.C., Vergnaud G., Gautheret D., Pourcel C.. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018; 46:W246–W251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dong C., Pu D.-K., Ma C., Wang X., Wen Q.-F., Zeng Z., Guo F.-B.. Precise detection of Acrs in prokaryotes using only six features. 2020; bioRxiv doi:26 May 2020, preprint: not peer reviewed 10.1101/2020.05.23.112011. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.