Abstract
CellMiner Cross-Database (CellMinerCDB, discover.nci.nih.gov/cellminercdb) allows integration and analysis of molecular and pharmacological data within and across cancer cell line datasets from the National Cancer Institute (NCI), Broad Institute, Sanger/MGH and MD Anderson Cancer Center (MDACC). We present CellMinerCDB 1.2 with updates to datasets from NCI-60, Broad Cancer Cell Line Encyclopedia and Sanger/MGH, and the addition of new datasets, including NCI-ALMANAC drug combination, MDACC Cell Line Project proteomic, NCI-SCLC DNA copy number and methylation data, and Broad methylation, genetic dependency and metabolomic datasets. CellMinerCDB (v1.2) includes several improvements over the previously published version: (i) new and updated datasets; (ii) support for pattern comparisons and multivariate analyses across data sources; (iii) updated annotations with drug mechanism of action information and biologically relevant multigene signatures; (iv) analysis speedups via caching; (v) a new dataset download feature; (vi) improved visualization of subsets of multiple tissue types; (vii) breakdown of univariate associations by tissue type; and (viii) enhanced help information. The curation and common annotations (e.g. tissues of origin and identifiers) provided here across pharmacogenomic datasets increase the utility of the individual datasets to address multiple researcher question types, including data reproducibility, biomarker discovery and multivariate analysis of drug activity.
INTRODUCTION
A critical aim of precision medicine is to match drugs with genomic determinants of response. Identifying tumor molecular features that affect response to specific drug treatments is especially challenging because of patient diversity, incomplete knowledge of the multiple molecular determinants of response and tumor heterogeneity. The relative homogeneity and ease of experimentation of patient-derived cell lines are advantageous, making them widely used model systems for establishing and resolving intrinsic drug response mechanisms as well as performing synthetic lethality screens (1). These features motivated the development of cancer cell line pharmacogenomic databases. Following the omics characterization of a 60-cell line panel developed by the National Cancer Institute (NCI), known as the NCI-60 (2–7), several large cancer cell line sets have been developed with pharmacogenomic data, including the Sanger/Massachusetts General Hospital Genomics of Drug Sensitivity in Cancer (GDSC) (8,9), the Broad/Novartis Cancer Cell Line Encyclopedia (CCLE) (10) and the Broad Cancer Therapeutics Response Portal (CTRP) datasets (11); together they provide information on ∼1400 cancer cell lines. Complementarity and overlap in the strengths of the cancer cell line datasets (e.g. number of drugs or total cell lines) provide the potential for valuable comparisons and integrative analyses in which the total value of the data is greater than the sum of the parts. However, data complexity and sources of inconsistency, such as differences in entity naming (e.g. cell lines, drugs) and data processing, make working across databases challenging; current, source-specific portals do not provide cross-database analyses.
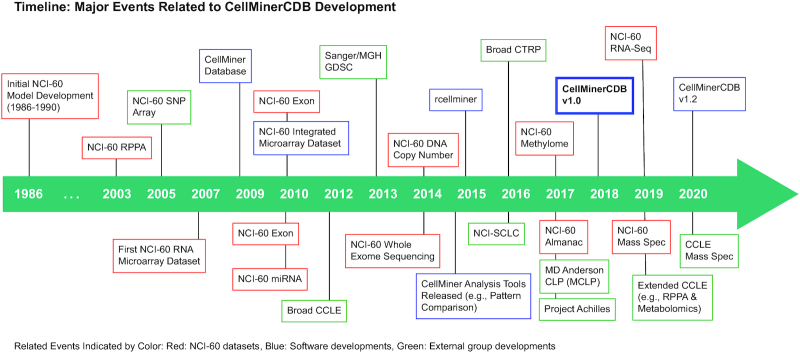
CellMiner Cross-Database (CellMinerCDB) combines the expertise and approaches developed within the Genomics and Pharmacology Facility (discover.nci.nih.gov), Developmental Therapeutics Branch, Center for Cancer Research, National Institutes of Health (NIH). The group has an established history (Figure 1) in the area of bioinformatics and pharmacogenomics, beginning with studies involving the NCI-60 cell line screen developed by the Developmental Therapeutics Program (12). These studies included pioneering efforts to (i) predict drug mechanism of action (MOA) from activity patterns (13), (ii) introduce the now ubiquitously used cluster image maps (i.e. clustered heatmaps) for comparison of molecular alterations and drug activity patterns (14) and (iii) integrate gene expression and compound activity into a single database (3,4,15). CellMiner (discover.nci.nih.gov/cellminer) was set up to allow direct data access and exploration (4,15), and became the platform for additional profiling technologies: protein expression (16,17), transcript microarrays (18,19), microRNA expression (19), DNA methylation (6,20), DNA whole-exome sequencing (5), DNA copy number (21), H2AX protein and phosphorylation levels (22) and RNA sequencing (23). Signatures of each of these data types were developed for integrative analysis (24). The first use of many assaying techniques has occurred on the NCI-60 and then later replicated on other cell line collections such as the GDSC and CCLE (Figure 1) (25–30).
Figure 1.
Timeline of CellMinerCDB development. Related events are indicated by color: NCI-60 datasets (red), software developments (blue) and external group developments (green).
First released in 2018, CellMinerCDB (discover.nci.nih.gov/cellminercdb) is a database accessible through a web interface that enables integrative analyses within and across cancer cell line pharmacogenomic databases (31) (Figure 1). CellMinerCDB integrates pharmacogenomic datasets both generated by the NCI (2–7) and downloaded from other project sites. Those include the CCLE/CTRP (11,32) and GDSC (both GDSC1 and GDSC2) datasets (8,9). CellMinerCDB differs from related cancer genomics aggregation efforts such as cBioPortal (33), a data portal focused on clinical sample cancer genomics. It focuses on cancer patient-derived human cell line molecular and pharmacological data, and it differs from the NCI Genomic Data Commons (34) and the data portals of the CCLE and GDSC in that the data are standardized to allow cross-database analysis; no raw data files are available. CellMinerCDB most closely resembles PharmacoDB (35), which also aggregates pharmacogenomic datasets. However, CellMinerCDB places an emphasis on web-based cross-database analyses of the available datasets. Supplementary Figure S1 showcases the distinctive value of CellMinerCDB to other pharmacogenomic tools (28,33,35–44) in terms of datasets available and capabilities.
In CellMinerCDB, named entities (e.g. drugs and genes) are transparently matched across sources, allowing cell line molecular features and drug responses to be readily compared using bivariate scatter plots and correlation analyses. Multivariate models of factors affecting drug responses or genomic cell line attributes can also be assessed. Analyses can be restricted to tissues of origin, with cell lines across all sources mapped to a uniform tissue type hierarchy. Gene pathway annotations allow assessment and filtering of analysis results. The analyses that CellMinerCDB makes accessible along with the breadth of available data make CellMinerCDB a unique resource for cancer cell line pharmacogenomic data exploration and hypothesis generation. In several cancer types (e.g. pancreatic, prostate, small cell lung cancer, etc.), recent studies have utilized CellMinerCDB to reveal associations between cancer drug sensitivity and (i) gene expression (45–48), (ii) genomic alterations (46) and (iii) cell line subgroups (49).
Here, we present the updates to the data and software infrastructure available in CellMinerCDB (Supplementary Figure S2; discover.nci.nih.gov/cellminercdb). This includes the addition of 12 new datasets and updates to several others. Within these new datasets, new data types are included (e.g. two-drug combinations, CRISPR genetic dependencies, mass spectrometry proteomics and metabolome data) as well as the addition of new gene signatures broadening the scope of CellMinerCDB. Functionally, CellMinerCDB has been improved with features to (i) speed up analysis, (ii) perform a broader range of cross-database analyses, (iii) make it easier to download corresponding data, (iv) present additional drug annotation information, (v) simplify the analysis of subsets of multiple tissue types and (vi) break down univariate associations by tissue type. We provide the reader with use cases highlighting the research potential and complementarity of the composite data.
CELLMINERCDB DATASETS
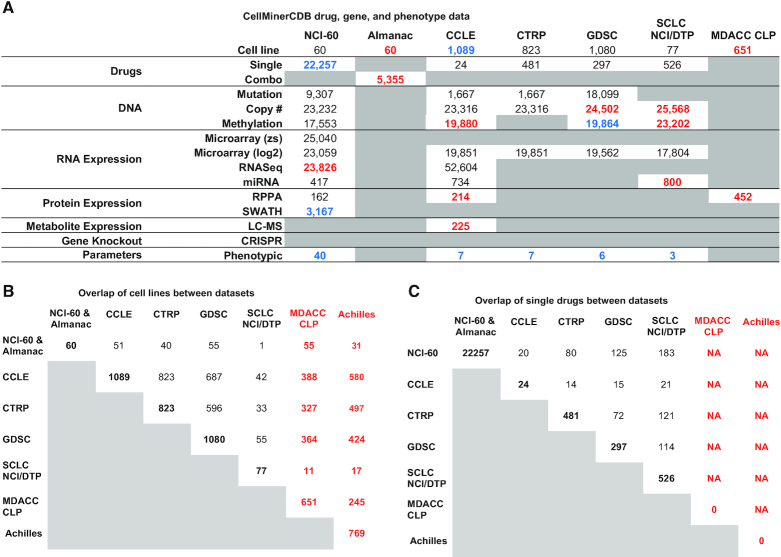
Over the last 2 years, CellMinerCDB (discover.nci.nih.gov/cellminercdb) has been expanded and now includes additional and updated datasets with a focus on genomic, proteomic and metabolomic data (collected prior to treatment), integrated with drug responses and CRISPR-based gene dependency data for these cell lines. Figure 2 summarizes CellMinerCDB content as well as the number of overlaps in cell lines and drugs across the different data sources.
Figure 2.
CellMinerCDB dataset overview. (A) Summary of molecular and drug activity data for the cell line sets included in CellMinerCDB. For the molecular and drug data types, the numbers indicate the number of genes or drugs. Blue numbers indicate a change in the number of features compared to the previous release, red numbers indicate new features and gray boxes denote entries for which there are no data in CellMinerCDB. (B) Number of cell line overlaps between data sources. (C) Number of single-drug activity overlaps between data sources. Abbreviations: datasets: National Cancer Institute (NCI-60), Cancer Cell Line Encyclopedia (CCLE), Cancer Therapeutics Response Portal (CTRP), Genomics of Drug Sensitivity in Cancer (GDSC), the NCI Small Cell Lung Cancer (SCLC) and the MD Anderson Cancer Center (MDACC) Cell Line Project (CLP); other: z-scores (zs), multiplatform microarray average log2 intensities (log2) and liquid chromatography–mass spectrometry (LC–MS).
Updates to existing datasets
In our newest release of the NCI-60 dataset within CellMinerCDB, over 1000 compound activities, including 27 FDA-approved and 412 clinical trial drugs (more than doubled), have been added, with processing as previously reported (3). We removed (curated) drug experiments that had a limited response range or were inconsistent across replicates. We updated the NCI-60 drug MOA and drug names as well as the drug clinical status and synonyms for all datasets. We also reprocessed the existing GDSC methylation data (9).
Newly included datasets
The new release integrates three new cell line sets with new data: the NCI-ALMANAC data for a recent paired drug activity for 105 FDA-approved drugs (50), the RPPA-based protein data from MDACC CLP, which is also known as MCLP (tcpaportal.org/mclp), and the CRISPR–Cas9 gene dependency map with phenotypic data from Project Achilles (28).
Additional data from current cell line sets have been included: (i) RNA-seq and SWATH proteomic data for the NCI-60 cell line set (17,23); (ii) microRNA, methylation and copy number data for the NCI-SCLC cell lines (51,52); (iii) copy number data for GDSC cell lines (9); and (iv) RPPA-based protein data, metabolome data and bisulfite sequencing promoter methylation data for CCLE cell lines (26,29).
Phenotypic gene signatures
Additionally, we have added phenotypic gene signature scores for all data sources for several key cancer phenotypes, including epithelial–mesenchymal transition (EMT) (53), antigen presentation machinery (APM) (54) and neuroendocrine (NE) signatures (55).
SOFTWARE INFRASTRUCTURE
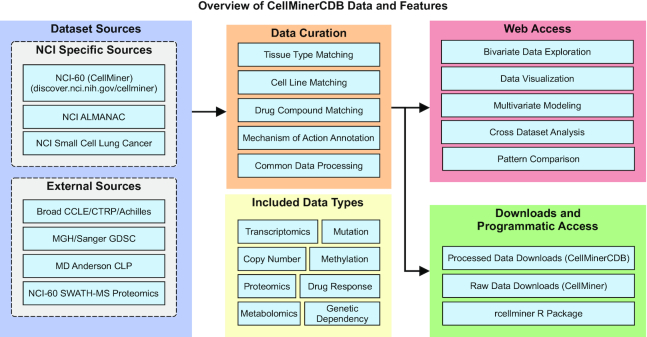
To create CellMinerCDB, cell line datasets are processed and undergo an additional curation step. This allows us to provide cross-database web-accessible analyses and download features (Figure 3). The different components of CellMinerCDB are described in the following sections.
Figure 3.
CellMinerCDB overview: CellMinerCDB integrates cancer cell line information from multiple sources and provides ready-made, user-friendly analysis tools as well as data download features for further analyses.
Data curation
For each data source and cell line, we manually curate a reference table to match cell lines across different sources. Additionally, we assign common terms for the tissue of origin (from the data source) based on the OncoTree (oncotree.mskcc.org; github.com/cBioPortal/oncotree) ontology structure and terms. OncoTree is an attempt to standardize cancer type diagnosis from a clinical perspective. OncoTree was developed and expertly curated by basic researchers and clinicians as a multi-institutional committee including the Memorial Sloan Kettering Cancer Center and Dana-Farber Cancer Institute. We assign OncoTree levels 1–4 (from least to most specific) for each cell line; in cases of ambiguity, resources such as Cellosaurus are consulted during the curation step (56). When available, we record additional cell line annotations such as patient sample information (e.g. gender or age) or curated information such as EMT status (53), SCLC subtypes (57) and triple negative breast cancer status.
For the NCI-60 drug curation, drug information (obtained from the NCI/DTP, dtp.cancer.gov) including National Service Center identifiers is the starting point for drug annotations, including preferred names, aliases, MOAs and FDA approval status. Additional and updated database identifiers are added from multiple sources, including PubChem, the NCI Thesaurus and the scientific literature. We compiled a list of terms that are present in ‘chemical’ names rather than common or generic names (e.g. ‘methyl’, ‘cyclo’, ‘pyrrol’). We scored each drug name on the number of such terms present in it (the ‘chemical name score’) and on the number of characters in the name. In cases where the NCI/DTP provided drug has a chemical name score >3 or the length of the name is >20 characters, we use the PubChem (58) preferred name (if that name has a chemical name score ≤3 and length ≤20). MOAs and clinical status (i.e. FDA approval status) are manually curated with information from NCI/DTP, FDA alerts (fda.gov) and journal articles as new data become available from external data providers. For other drug datasets (e.g. CCLE), we retrieve drug annotation information from our data providers and external sources (e.g. PubChem) including the identifiers, names, synonyms and MOA when available. Using these identifiers, CellMinerCDB matches drugs across data sources.
Data representation and processing
CellMinerCDB datasets are created using R data packages that use two S4 class objects for data representation: molData and drugData as defined by the rcellminer R analysis package (59). The molData object contains results for molecular assays (e.g. genomics, proteomics, etc.) and drugData contains results for drug responses. The core of these objects is a list of ExpressionSet objects. This R data structure is made available by the Biobase Bioconductor package. This data representation allows molecular profiling and drug response data to be conveniently stored with sample metadata using a well-documented and widely available format [details are available in the documentation for the rcellminer and rcellminerData Bioconductor packages (59)].
Each CellMinerCDB data package contains the data from the data provider (i.e. Excel spreadsheets, tab-delimited files, etc.) along with R scripts used to process the data into the molData and drugData objects. In recent work, an instructional data package has been made available at github.com/CBIIT/rcellminerData using a subset of the NCI-60 rcellminerData Bioconductor package data to guide data package developers (59).
Software architecture
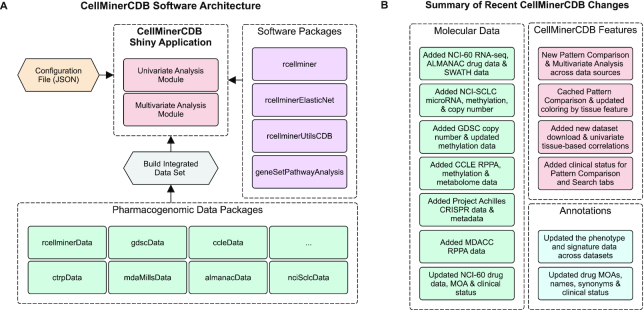
The CellMinerCDB web interface is built using the Shiny framework (shiny.rstudio.com) in the R programming language. This simplifies development, contribution and extension by developers with bioinformatic experience (where R is widely used) (60), in contrast to projects such as cBioPortal that depend on Java and JavaScript knowledge (61). CellMinerCDB architecture (Figure 4A) is based on pharmacogenomic data packages and project-specific R packages, including rcellminer (core data structure functionality), rcellminerUtilsCDB (gene signature and cross-database mapping), rcellminerElasticNet (multivariate analysis) and geneSetPathwayAnalysis (annotation information from GeneCards and Pathway Commons) (62,63). It should be noted that in this update, the rcellminer Bioconductor package removes previous Java code dependencies to simplify installation. Various properties (e.g. datasets to make available and interface properties) of CellMinerCDB are configurable using a JavaScript Object Notation file. CellMinerCDB uses Shiny modules to support independent analysis modules (e.g. the multivariate analysis module) to allow developers to customize the web interface.
Figure 4.
(A) CellMinerCDB software architecture. (B) Summary of recent CellMinerCDB changes. Colors: CellMinerCDB application elements (red), data packages (green), software packages (purple), configuration files (yellow), annotations/metadata (blue) and pre-processing steps (gray).
DATA AVAILABILITY
New functionality allows users to download data from any of the available -omic data via the CellMinerCDB (discover.nci.nih.gov/cellminercdb) Metadata tab as compressed (zip) files. Additionally, users can download drug synonym information or cell line annotation information from this tab. The NCI-60 dataset has an additional method for download via the rcellminerData Bioconductor package.
UPDATES TO CELLMINERCDB FUNCTIONALITY AND USE CASES
In addition to recent dataset updates, CellMinerCDB functionality includes (i) updated drug annotations, (ii) expanded help information, (iii) improved pattern comparison speed, (iv) added functionality that provides correlation results by tissue type, (v) facilitated pattern comparison and (vi) improved multivariate and gene signature-based analyses across data sources (Figure 4B; complete release history: Supplementary Figure S2).
Training materials
CellMinerCDB provides several training materials, including detailed documentation on the CellMinerCDB website (discover.nci.nih.gov/cellminercdb) from the Help tab, as well as a video tutorial (youtube.com/watch?v=XljXazRGkQ8).
Use cases
This section presents several CellMinerCDB use cases, workflow and analysis examples. Figure 5 and Supplementary Figures S4–S6 provide visualizations across multiple use case examples. Each figure presents the input values (in the left-side gray boxes) to allow the exact reproduction of the result shown. Additional use case workflows are in the Help section of the CellMinerCDB website, and we provide a summary of available workflows in Supplementary Figure S3.
Figure 5.
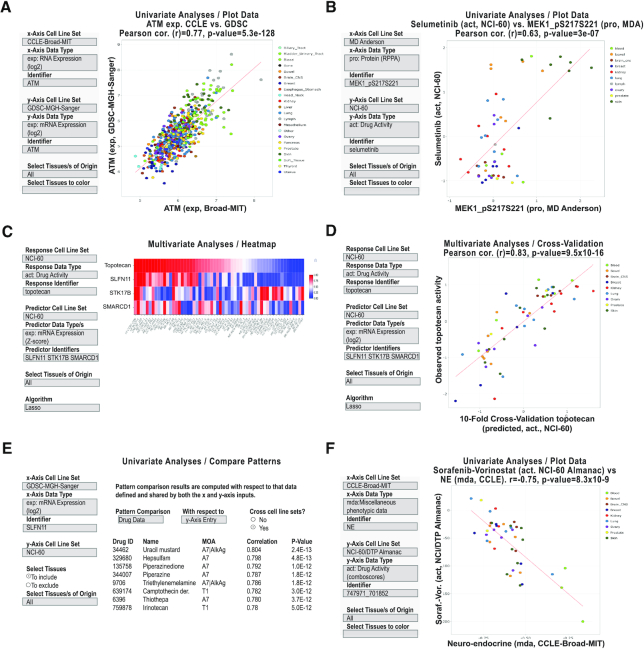
CellMinerCDB analysis examples. (A) Univariate analysis scatter plot of ATM transcript expression levels as measured by CCLE (Broad) versus GDSC (Sanger/MGH). (B) Univariate analysis scatter plot of MAP2K1 (MEK1) phosphoprotein levels versus the MAP2K1 inhibitor selumetinib activity levels. (C) Multivariate analysis and heatmap to identify and visualize molecular predictors for topotecan activity. (D) Multivariate analysis cross-validation scatter plot to visualize the observed versus predicted activities. (E) Univariate analysis ‘Compare Patterns’ output to identify drugs whose activities are most significantly correlated to SLFN11 expression. (F) Univariate analysis scatter plot of the NE transcript expression signature versus the two-drug activity NCI-ALMANAC ComboScore of sorafenib–vorinostat. All examples are captured images from CellMinerCDB (discover.nci.nih.gov/cellminercdb) using the selections detailed in the input box (on the left). Each dot in the scatter plots is a cell line, with tissues of origin indicated in the legend (on right). For the scatter plots, the regression trend line is in red. The x- and y-axes, correlations (Pearson’s r) and P-values are as defined within each panel.
Quality control across database example
Figure 5A provides an example of data reproducibility across datasets. There has been recent discussion regarding the reliability and reproducibility of cell line data, especially when produced at different institutions (32,64,65). Reproducibility issues can arise from multiple factors including variability in what are erroneously perceived to be identical cell lines (66), and from the individuals doing the work and platforms used. CellMinerCDB allows direct comparison of omics profiles or drug activities. The example given shows that ATM expression values as measured by both the CCLE and GDSC are significantly correlated (r = 0.77, P = 5.3e−128). It illustrates an example where users are able to assess the reproducibility of the genomic and pharmacological data across cell lines processed independently with different platforms at different institutions (31). Additional data reproducibility examples are shown for DNA mutation, drug activity, DNA copy number and DNA methylation in Supplementary Figure S4A–E.
Identification of candidate drug biomarkers
Cancer cell lines are the logical starting place for identifying molecular features of relevance as part of precision medicine research. Figure 5B provides an example visualizing the relationship between MAP2K1 (using the name MEK1) phosphorylation status and the MAP2K1 inhibitor selumetinib. MAP2K1 is not presently a qualified biomarker for this FDA-approved drug. Here, we show a significant correlation between the selumetinib drug response in the NCI-60 and phosphorylation status in the MDACC RPPA dataset (r = 0.63, P = 3e−07), suggesting further investigation into patient response predictability. It should be noted that when searching for selumetinib, the drug synonyms (aliases) ‘AZD6244’ and ‘741078’ can alternatively be used; available drug synonyms are downloadable as a table from the Metadata tab of the website. Supplementary Figure S5C and D provides additional examples of the types of analyses to connect drug activity to -omic features with examples using HSP90 and CDK inhibitors, respectively. Correlations for such analyses may not always be extremely high (>0.7) but may nonetheless be informative for further investigations.
Exploration of proteomic complexes
The addition of proteomic data allows users to explore the stoichiometric relationship of protein complex subunits. Supplementary Figure S5A shows the high correlation between protein levels of both subunits of the chromatin remodeling complex FACT (facilitates chromatin transcription): SSRP1 versus SUP16H (r = 0.87, P = 1.6e−19) in the NCI-60 SWATH mass spectrometry data (17,67). Supplementary Figure S5B also shows the correlation between the two components of the catenin complex involved in cell adhesion CTNNA1 and CTNNB1 (α- and β-catenin, respectively) (68).
Multivariate analysis example
In the majority of cases, multiple factors determine drug response in cell lines as well as in patients. Multivariate analysis allows investigation of drug response with respect to multiple -omic features simultaneously. CellMinerCDB allows users to either (i) manually input a multivariate model of their own design or (ii) have the system automatically generate a multivariate model for a molecular entity (i.e. an -omic or a drug response profile); here we have chosen to show the latter.
Figure 5C presents the CellMinerCDB ‘Multivariate Analysis’ functionality using the NCI-60 data. In the example, topotecan activity (a response variable) is analyzed as a function of gene transcript levels (potential response predictors, i.e. features). Using the LASSO (least absolute shrinkage and selection operator) feature selection algorithm (69), which is integrated into CellMinerCDB, three genes are identified automatically: SLFN11, STK17B and SMARCD1. The example puts forward a well-known predictor of topotecan response (SLFN11) (7,70) along with two predictors unknown in the topotecan literature (STK17B and SMARCD1) that may be the genesis of new hypotheses.
The relationship of the drug and selected features (i.e. predictors) of the LASSO-derived regression model is displayed as a heatmap (Figure 5C). The observed experimental topotecan response values and predicted response values (after 10-fold cross-validation) are highly correlated (r = 0.83, P = 9.5e−16; Figure 5D). Predicted response values are obtained (over 10 iterations) by successively holding out 10% of the cell lines and predicting their response using a linear regression model fit to the remaining 90% of the data. After all 10 iterations have been done, each sample has one cross-validated prediction. Details for the LASSO algorithm and the multivariate analysis are in the CellMinerCDB Help section. Biologically, SLFN11 is known to affect topotecan response (7,70). STK17B is a serine/threonine kinase associated with apoptosis, and SMARCD1 is a SWI/SNF subunit of chromatin remodeling factors that could enhance the topoisomerase DNA accessibility necessary for topotecan activity (71,72). While the biological functions of STK17B and SMARCD1 plausibly affect topotecan response, neither has been previously reported in the scientific literature.
It should be noted for users that functionality to fix a model trained on one dataset and apply this to another dataset is not currently available and is better suited for more formal predictive analyses outside of CellMinerCDB to test model generalizability. Also, the interpretation of multivariate models (especially those that are algorithmically generated) requires scrutiny by individual researchers and should be guided by a well-formed understanding of the biological activities of the identified predictors and the MOA of the input drug to guide any follow-up effort. Besides manual and automated model creation, CellMinerCDB can be used to supplement a base model with additional features using partial correlation analysis.
Pattern comparison for the identification of biomarkers and drug targets
Figure 5E shows an example of broad-spectrum biomarker identification; here, a broad-spectrum biomarker is defined as a gene with a molecular profile having predictive strength across cancer types. Using the CellMinerCDB ‘Compare Patterns’ functionality, the transcript expression of SLFN11 from GDSC is compared to the activity profiles of all drugs and compounds in the NCI-60. High correlations are found for multiple DNA-damaging drugs, with TOP1 inhibitors and alkylating agents at the top, as well as TOP2, PARP1 and DNA synthesis inhibitors (not shown in the figure). These results provide unbiased evidence for SLFN11 as a potential biomarker for whole classes of commonly used standard of care chemotherapies (73). This example demonstrates the ability of CellMinerCDB to compare a pattern of interest via a correlation analysis to patterns of other types (transcript expression versus activity in this case). Furthermore, it demonstrates the cross-database capabilities of CellMinerCDB. The values presented here are not corrected for multiple testing though false discovery rate adjusted values are available via the website. Additionally, the pattern comparison feature provides a way to identify co-regulation between genes as previously shown to be involved in EMT (31) and lineage transcription factor pathways in small cell lung cancers (49). Furthermore, the ‘Compare Patterns’ panel includes gene location annotations to help reveal possible gene regulation due to proximity to neighbors and gene copy number alterations. Supplementary Figure S6A provides a visualization of a pattern comparison result (to that in Figure 5E) not employing the cross-cell line sets functionality update, that is GDSC SLFN11 transcription as compared to GDSC FDA-approved drug activities. In both cases, DNA-damaging drugs are recognized.
Exploring drug combinations and gene signatures
Figure 5F provides an example showing the availability of (i) phenotypic gene signatures and (ii) drug combination response data in CellMinerCDB. Correlation of a 50-gene NE status signature (55) with the response of cell lines to a combination of sorafenib and vorinostat shows that NE-like cells (higher NE score) potentially respond less well to the drug combination.
Exploring other data types: genetic dependencies and metabolomic data
The inclusion and integration of the CRISPR–Cas9 knockout data from Project Achilles allow users to search for co-dependent genes across cell lines. For instance, BRCA2 and PALB2 (Partner And Localizer of BRCA2) show high correlation (r = 0.56, P = 7.1e−64; Supplementary Figure S6B). This observation is consistent with the abrogation of homologous recombination (HR) activity that occurs when BRCA1–PALB2 binding is disrupted (74). A second CRISPR–Cas9 example is the correlation between silencing BRAF (on cell survival) and selective activity of vemurafenib (P < 2.9e−15), primarily in the melanoma cell lines, as expected due to their large presence of the V600E mutations (75,76). An example from the CCLE metabolite data, providing a comparison of inosine and guanosine levels (r = 0.55, P = 2.3e−73), is given in Supplementary Figure S6C. Inosine monophosphate (IMP) is converted to xanthosine monophosphate (XMP) by IMP dehydrogenase. XMP is converted to guanosine monophosphate (GMP) by GMP synthase (77).
Exploring drug structures
Currently, CellMinerCDB does not provide access to drug chemical structures. However, structures can be accessed as SMILES representations of NCI-60 compounds via the rcellminer R package on Bioconductor.
CONCLUSION
The past two decades have seen a surge of molecular and pharmacological data for overlapping sets of patient-derived cancer cell lines by multiple institutions (e.g. NCI, Broad Institute and Sanger/MGH). CellMinerCDB (discover.nci.nih.gov/cellminercdb) allows researchers to explore data across institutions using their expertise without bioinformatic support while benefiting from curated genomic and pharmacological annotations. The use case examples presented here are meant to provide a representative selection of explorations and analyses. They include a diverse group of analysis type databases: (i) data reproducibility: the uniquely compiled datasets here allow users to continue the discussion on the reproducibility of pharmacogenomic data (65,78) including the possibility of conducting further systematic analyses; (ii) candidate biomarker discovery; (iii) multivariate analysis of molecular features for drug activity; (iv) exploration of the relationship of composite features (multigene signatures versus response to drug combinations); (v) exploration of protein complexes, genetic dependencies and metabolomic pathways; and (vi) complementary analyses where data from one dataset supplement another (e.g. use of Broad CCLE gene expression data for matching Broad CTRP drug tested cell lines to calculate drug–gene expression correlations). Hence, a wide range of univariate and multivariate explorations can be undertaken. CellMinerCDB seamlessly allows this form of data assessment and integration. It opens the door to hypothesis generation and validation in non-isogenic cancer cell lines across multiple tissues of origin as well as the discovery of potential novel genomic regulatory networks and drug response determinants.
CellMinerCDB supports the analysis of many profiling platforms, including molecular parameters (e.g. gene/protein expression, alteration, metabolomic and genetic dependency), drug compound activity and phenotypic signatures (such as NE status, EMT and subsets of molecularly defined cancer subtypes as in the case of the small cell lung cancer cell lines). Yet, CellMinerCDB does not include drug perturbation analyses, which are provided by other platforms (e.g. the Connectivity Map). CellMinerCDB focuses on the molecular and genomic analyses of cancer cells at their steady-state levels prior to drug response. The goal of CellMinerCDB is to provide accessibility and analytical methods across previously disconnected datasets in a manner that allows its utilization by bench scientists, clinicians and others with domain knowledge, as well as traditional informaticists by combining the strengths and unique features of each dataset.
Going forward, CellMinerCDB will continue to (i) expand datasets [e.g. the PRISM drug repurposing collection (79)] and annotations, (ii) improve analysis speed and (iii) simplify installation for local reuse by other groups. Future software developments are being planned to (i) further advance the accuracy and coverage of annotations, for example through the use of algorithmic prediction [e.g. use of similarity analyses (4) to assess the possible MOA based on the similarity of drug response to known MOA compounds], (ii) use network biology tools and approaches to create models of drug response, (iii) extend analysis features to include comparisons between cell lines and patient cohorts starting with datasets from The Cancer Genome Atlas (TCGA) (80) and (iv) provide future customization by users of available signatures. We anticipate that the modular architecture design of CellMinerCDB will enable these additional analytical features.
CellMinerCDB complements pharmacogenomic data portals through the additional layer of curation and specialized analyses we provide across datasets. This enhances the value of individual pharmacogenomic datasets by its availability for comparison across projects to further our understanding of the combinatorial influences that affect pharmacological outcomes as we move toward personalized medicine.
SOFTWARE AVAILABILITY
All software developed as part of CellMinerCDB is freely available, open source and hosted GitHub in the following repositories: github.com/CBIIT/cellminercdb, github.com/CBIIT/rcellminerUtilsCDB, github.com/CBIIT/geneSetPathwayAnalysis, github.com/CBIIT/rcellminerElasticNet and github.com/CBIIT/rcellminer. Users of CellMinerCDB can provide feedback and ask questions of the development team using webadmin@discover.nci.nih.gov or users can submit developer feedback, file bug reports and request new features using project-specific issue trackers (e.g. github.com/CBIIT/cellminercdb/issues).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the many data providers and collaborators who have made this project possible, specifically Mahmoud Ghandi (CCLE), Paul Clemons (CTRP), Howard Lightfoot, Mathew J. Garnett (GDSC), Beverly Teicher (NCI, CCR), Paul Meltzer (NCI, CCR) and John Minna (UT Southwestern Medical Center). We would like to thank David Goldstein of the NCI Office of Science and Technology Resources for supporting the purchase of software required to enable efficient, multi-user access to the CellMinerCDB site.
Contributor Information
Augustin Luna, cBio Center, Dana-Farber Cancer Institute and Department of Cell Biology, Harvard Medical School, Boston, MA 02215, USA.
Fathi Elloumi, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA; General Dynamics Information Technology Inc., Fairfax, VA 22042, USA.
Sudhir Varma, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA; HiThru Analytics LLC, Princeton, NJ 08540, USA.
Yanghsin Wang, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA; General Dynamics Information Technology Inc., Fairfax, VA 22042, USA.
Vinodh N Rajapakse, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Mirit I Aladjem, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Jacques Robert, Inserm unité 1218, Université de Bordeaux, Bordeaux 33076, France.
Chris Sander, cBio Center, Dana-Farber Cancer Institute and Department of Cell Biology, Harvard Medical School, Boston, MA 02215, USA.
Yves Pommier, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
William C Reinhold, Developmental Therapeutics Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute [Z01 BC006150, ZIC BC011475, ZIA BC010411 and ZIC BC011509]; National Institute of General Medical Sciences [P41 GM103504 to A.L. and C.S.]. Funding for open access charge: National Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F., Pantel S.E. et al.. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014; 1:140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abaan O.D., Polley E.C., Davis S.R., Zhu Y.J., Bilke S., Walker R.L., Pineda M., Gindin Y., Jiang Y., Reinhold W.C. et al.. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013; 73:4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reinhold W.C., Sunshine M., Liu H., Varma S., Kohn K.W., Morris J., Doroshow J., Pommier Y.. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012; 72:3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reinhold W.C., Sunshine M., Varma S., Doroshow J.H., Pommier Y.. Using CellMiner 1.6 for systems pharmacology and genomic analysis of the NCI-60. Clin. Cancer Res. 2015; 21:3841–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reinhold W.C., Varma S., Sousa F., Sunshine M., Abaan O.D., Davis S.R., Reinhold S.W., Kohn K.W., Morris J., Meltzer P.S. et al.. NCI-60 whole exome sequencing and pharmacological CellMiner analyses. PLoS One. 2014; 9:e101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reinhold W.C., Varma S., Sunshine M., Rajapakse V., Luna A., Kohn K.W., Stevenson H., Wang Y., Heyn H., Nogales V. et al.. The NCI-60 methylome and its integration into CellMiner. Cancer Res. 2017; 77:601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zoppoli G., Regairaz M., Leo E., Reinhold W.C., Varma S., Ballestrero A., Doroshow J.H., Pommier Y.. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl Acad. Sci. U.S.A. 2012; 109:15030–15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W., Greninger P., Thompson I.R., Luo X., Soares J. et al.. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012; 483:570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Gonçalves E., Barthorpe S., Lightfoot H. et al.. A landscape of pharmacogenomic interactions in cancer. Cell. 2016; 166:740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rees M.G., Seashore-Ludlow B., Cheah J.H., Adams D.J., Price E.V., Gill S., Javaid S., Coletti M.E., Jones V.L., Bodycombe N.E. et al.. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol. 2016; 12:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stinson S.F., Alley M.C., Kopp W.C., Fiebig H.H., Mullendore L.A., Pittman A.F., Kenney S., Keller J., Boyd M.R.. Morphological and immunocytochemical characteristics of human tumor cell lines for use in a disease-oriented anticancer drug screen. Anticancer Res. 1992; 12:1035–1053. [PubMed] [Google Scholar]
- 13. Weinstein J.N., Kohn K.W., Grever M.R., Viswanadhan V.N., Rubinstein L.V., Monks A.P., Scudiero D.A., Welch L., Koutsoukos A.D., Chiausa A.J.. Neural computing in cancer drug development: predicting mechanism of action. Science. 1992; 258:447–451. [DOI] [PubMed] [Google Scholar]
- 14. Weinstein J.N., Myers T.G., O’Connor P.M., Friend S.H., Fornace A.J. Jr, Kohn K.W., Fojo T., Bates S.E., Rubinstein L.V., Anderson N.L. et al.. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997; 275:343–349. [DOI] [PubMed] [Google Scholar]
- 15. Scherf U., Ross D.T., Waltham M., Smith L.H., Lee J.K., Tanabe L., Kohn K.W., Reinhold W.C., Myers T.G., Andrews D.T. et al.. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000; 24:236–244. [DOI] [PubMed] [Google Scholar]
- 16. Nishizuka S., Charboneau L., Young L., Major S., Reinhold W.C., Waltham M., Kouros-Mehr H., Bussey K.J., Lee J.K., Espina V. et al.. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl Acad. Sci. U.S.A. 2003; 100:14229–14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo T., Luna A., Rajapakse V.N., Koh C.C., Wu Z., Liu W., Sun Y., Gao H., Menden M.P., Xu C. et al.. Quantitative proteome landscape of the NCI-60 cancer cell lines. iScience. 2019; 21:664–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reinhold W.C., Mergny J.-L., Liu H., Ryan M., Pfister T.D., Kinders R., Parchment R., Doroshow J., Weinstein J.N., Pommier Y.. Exon array analyses across the NCI-60 reveal potential regulation of TOP1 by transcription pausing at guanosine quartets in the first intron. Cancer Res. 2010; 70:2191–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu H., D’Andrade P., Fulmer-Smentek S., Lorenzi P., Kohn K.W., Weinstein J.N., Pommier Y., Reinhold W.C.. mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol. Cancer Ther. 2010; 9:1080–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reinhold W.C., Reimers M.A., Lorenzi P., Ho J., Shankavaram U.T., Ziegler M.S., Bussey K.J., Nishizuka S., Ikediobi O., Pommier Y.G. et al.. Multifactorial regulation of E-cadherin expression: an integrative study. Mol. Cancer Ther. 2010; 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C.. High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner. PLoS One. 2014; 9:e92047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ji J., Zhang Y., Redon C.E., Reinhold W.C., Chen A.P., Fogli L.K., Holbeck S.L., Parchment R.E., Hollingshead M., Tomaszewski J.E. et al.. Phosphorylated fraction of H2AX as a measurement for DNA damage in cancer cells and potential applications of a novel assay. PLoS One. 2017; 12:e0171582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reinhold W.C., Varma S., Sunshine M., Elloumi F., Ofori-Atta K., Lee S., Trepel J.B., Meltzer P.S., Doroshow J.H., Pommier Y.. RNA sequencing of the NCI-60: integration into CellMiner and CellMinerCDB. Cancer Res. 2019; 79:3514–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gmeiner W.H., Reinhold W.C., Pommier Y.. Genome-wide mRNA and microRNA profiling of the NCI 60 cell-line screen and comparison of FdUMP[10]with fluorouracil, floxuridine, and topoisomerase 1 poisons. Mol. Cancer Ther. 2010; 9:3105–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J. et al.. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005; 436:117–122. [DOI] [PubMed] [Google Scholar]
- 26. Li H., Ning S., Ghandi M., Kryukov G.V., Gopal S., Deik A., Souza A., Pierce K., Keskula P., Hernandez D. et al.. The landscape of cancer cell line metabolism. Nat. Med. 2019; 25:850–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S. et al.. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017; 49:1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M. et al.. Defining a cancer dependency map. Cell. 2017; 170:564.e16–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H. et al.. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nusinow D.P., Szpyt J., Ghandi M., Rose C.M., McDonald E.R. 3rd, Kalocsay M., Jané-Valbuena J., Gelfand E., Schweppe D.K., Jedrychowski M. et al.. Quantitative proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020; 180:387.e16–402.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rajapakse V.N., Luna A., Yamade M., Loman L., Varma S., Sunshine M., Iorio F., Sousa F.G., Elloumi F., Aladjem M.I. et al.. CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience. 2018; 10:247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cancer Cell Line Encyclopedia Consortium and Genomics of Drug Sensitivity in Cancer Consortium Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015; 528:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. et al.. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jensen M.A., Ferretti V., Grossman R.L., Staudt L.M.. The NCI Genomic Data Commons as an engine for precision medicine. Blood. 2017; 130:453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smirnov P., Kofia V., Maru A., Freeman M., Ho C., El-Hachem N., Adam G.-A., Ba-Alawi W., Safikhani Z., Haibe-Kains B.. PharmacoDB: an integrative database for mining in vitro anticancer drug screening studies. Nucleic Acids Res. 2018; 46:D994–D1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R. et al.. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013; 41:D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caroli J., Sorrentino G., Forcato M., Del Sal G., Bicciato S.. GDA, a web-based tool for genomics and drugs integrated analysis. Nucleic Acids Res. 2018; 46:W148–W156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. et al.. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017; 171:1437.e17–1452.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahmed J., Meinel T., Dunkel M., Murgueitio M.S., Adams R., Blasse C., Eckert A., Preissner S., Preissner R.. CancerResource: a comprehensive database of cancer-relevant proteins and compound interactions supported by experimental knowledge. Nucleic Acids Res. 2011; 39:D960–D967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cokelaer T., Chen E., Iorio F., Menden M.P., Lightfoot H., Saez-Rodriguez J., Garnett M.J.. GDSCTools for mining pharmacogenomic interactions in cancer. Bioinformatics. 2018; 34:1226–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smirnov P., Safikhani Z., El-Hachem N., Wang D., She A., Olsen C., Freeman M., Selby H., Gendoo D.M.A., Grossmann P. et al.. PharmacoGx: an R package for analysis of large pharmacogenomic datasets. Bioinformatics. 2016; 32:1244–1246. [DOI] [PubMed] [Google Scholar]
- 42. Zagidullin B., Aldahdooh J., Zheng S., Wang W., Wang Y., Saad J., Malyutina A., Jafari M., Tanoli Z., Pessia A. et al.. DrugComb: an integrative cancer drug combination data portal. Nucleic Acids Res. 2019; 47:W43–W51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu H., Zhang W., Zou B., Wang J., Deng Y., Deng L.. DrugCombDB: a comprehensive database of drug combinations toward the discovery of combinatorial therapy. Nucleic Acids Res. 2020; 48:D871–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seo H., Tkachuk D., Ho C., Mammoliti A., Rezaie A., Madani Tonekaboni S.A., Haibe-Kains B.. SYNERGxDB: an integrative pharmacogenomic portal to identify synergistic drug combinations for precision oncology. Nucleic Acids Res. 2020; 48:W494–W501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Perkail S., Andricovich J., Kai Y., Tzatsos A.. BAP1 is a haploinsufficient tumor suppressor linking chronic pancreatitis to pancreatic cancer in mice. Nat. Commun. 2020; 11:3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hsieh Y.-Y., Liu T.-P., Chou C.-J., Chen H.-Y., Lee K.-H., Yang P.-M.. Integration of bioinformatics resources reveals the therapeutic benefits of gemcitabine and cell cycle intervention in SMAD4-deleted pancreatic ductal adenocarcinoma. Genes. 2019; 10:766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cheteh E.H., Sarne V., Ceder S., Bianchi J., Augsten M., Rundqvist H., Egevad L., Östman A., Wiman K.G.. Interleukin-6 derived from cancer-associated fibroblasts attenuates the p53 response to doxorubicin in prostate cancer cells. Cell Death Discov. 2020; 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kriegsman B.A., Vangala P., Chen B.J., Meraner P., Brass A.L., Garber M., Rock K.L.. Frequent loss of IRF2 in cancers leads to immune evasion through decreased MHC class I antigen presentation and increased PD-L1 expression. J. Immunol. 2019; 203:1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tlemsani C., Pongor L., Girard L., Roper N., Elloumi F., Varma S., Luna A., Rajapakse V.N., Sebastian R., Kohn K.W. et al.. SCLC_CellMiner: integrated genomics and therapeutics predictors of small cell lung cancer cell lines based on their genomic signatures. Cell Rep. 2020; 33:108296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sidorov P., Naulaerts S., Ariey-Bonnet J., Pasquier E., Ballester P.J.. Predicting synergism of cancer drug combinations using NCI-ALMANAC data. Front. Chem. 2019; 7:509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Polley E., Kunkel M., Evans D., Silvers T., Delosh R., Laudeman J., Ogle C., Reinhart R., Selby M., Connelly J. et al.. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J. Natl Cancer Inst. 2016; 108:djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krushkal J., Silvers T., Reinhold W.C., Sonkin D., Vural S., Connelly J., Varma S., Meltzer P.S., Kunkel M., Rapisarda A. et al.. Epigenome-wide DNA methylation analysis of small cell lung cancer cell lines suggests potential chemotherapy targets. Clin. Epigenetics. 2020; 12:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kohn K.W., Zeeberg B.M., Reinhold W.C., Pommier Y.. Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS One. 2014; 9:e99269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang S., He Z., Wang X., Li H., Liu X.-S.. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. eLife. 2019; 8:e49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang W., Girard L., Zhang Y.-A., Haruki T., Papari-Zareei M., Stastny V., Ghayee H.K., Pacak K., Oliver T.G., Minna J.D. et al.. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl. Lung Cancer Res. 2018; 7:32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bairoch A. The Cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 2018; 29:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rudin C.M., Poirier J.T., Byers L.A., Dive C., Dowlati A., George J., Heymach J.V., Johnson J.E., Lehman J.M., MacPherson D. et al.. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019; 19:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. et al.. PubChem substance and compound databases. Nucleic Acids Res. 2016; 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Luna A., Rajapakse V.N., Sousa F.G., Gao J., Schultz N., Varma S., Reinhold W., Sander C., Pommier Y.. rcellminer: exploring molecular profiles and drug response of the NCI-60 cell lines in R. Bioinformatics. 2016; 32:1272–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pittard W.S., Li S.. The essential toolbox of data science: Python, R, Git, and Docker. Methods Mol. Biol. 2020; 2104:265–311. [DOI] [PubMed] [Google Scholar]
- 61. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al.. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al.. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.1–1.30.33. [DOI] [PubMed] [Google Scholar]
- 63. Rodchenkov I., Babur O., Luna A., Aksoy B.A., Wong J.V., Fong D., Franz M., Siper M.C., Cheung M., Wrana M. et al.. Pathway Commons 2019 update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020; 48:D489–D497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Safikhani Z., El-Hachem N., Quevedo R., Smirnov P., Goldenberg A., Juul Birkbak N., Mason C., Hatzis C., Shi L., Aerts H.J. et al.. Assessment of pharmacogenomic agreement. F1000Res. 2016; 5:825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haibe-Kains B., El-Hachem N., Birkbak N.J., Jin A.C., Beck A.H., Aerts H.J.W.L., Quackenbush J.. Inconsistency in large pharmacogenomic studies. Nature. 2013; 504:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ben-David U., Siranosian B., Ha G., Tang H., Oren Y., Hinohara K., Strathdee C.A., Dempster J., Lyons N.J., Burns R. et al.. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018; 560:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huang H., Santoso N., Power D., Simpson S., Dieringer M., Miao H., Gurova K., Giam C.-Z., Elledge S.J., Zhu J.. FACT proteins, SUPT16H and SSRP1, are transcriptional suppressors of HIV-1 and HTLV-1 that facilitate viral latency. J. Biol. Chem. 2015; 290:27297–27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Debruyne P., Vermeulen S., Mareel M.. The role of the E-cadherin/catenin complex in gastrointestinal cancer. Acta Gastroenterol. Belg. 1999; 62:393–402. [PubMed] [Google Scholar]
- 69. Tibshirani R. Regression shrinkage and selection via the Lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996; 58:267–288. [Google Scholar]
- 70. Reinhold W.C., Thomas A., Pommier Y.. DNA-targeted precision medicine; have we been caught sleeping?. Trends Cancer Res. 2017; 3:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doherty G.A., Byrne S.M., Austin S.C., Scully G.M., Sadlier D.M., Neilan T.G., Kay E.W., Murray F.E., Fitzgerald D.J.. Regulation of the apoptosis-inducing kinase DRAK2 by cyclooxygenase-2 in colorectal cancer. Br. J. Cancer. 2009; 101:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Priam P., Krasteva V., Rousseau P., D’Angelo G., Gaboury L., Sauvageau G., Lessard J.A.. SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPϵ dependent mechanism. Nat. Genet. 2017; 49:753–764. [DOI] [PubMed] [Google Scholar]
- 73. Murai J., Thomas A., Miettinen M., Pommier Y.. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol. Ther. 2019; 201:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Foo T.K., Tischkowitz M., Simhadri S., Boshari T., Zayed N., Burke K.A., Berman S.H., Blecua P., Riaz N., Huo Y. et al.. Compromised BRCA1–PALB2 interaction is associated with breast cancer risk. Oncogene. 2017; 36:4161–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Patrawala S., Puzanov I.. Vemurafenib (RG67204, PLX4032): a potent, selective BRAF kinase inhibitor. Future Oncol. 2012; 8:509–523. [DOI] [PubMed] [Google Scholar]
- 76. Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M. et al.. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011; 364:2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gu J.J., Tolin A.K., Jain J., Huang H., Santiago L., Mitchell B.S.. Targeted disruption of the inosine 5′-monophosphate dehydrogenase type I gene in mice. Mol. Cell. Biol. 2003; 23:6702–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Safikhani Z., Smirnov P., Freeman M., El-Hachem N., She A., Rene Q., Goldenberg A., Birkbak N.J., Hatzis C., Shi L. et al.. Revisiting inconsistency in large pharmacogenomic studies. F1000Res. 2016; 5:2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Corsello S.M., Nagari R.T., Spangler R.D., Rossen J., Kocak M., Bryan J.G., Humeidi R., Peck D., Wu X., Tang A.A. et al.. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer. 2020; 1:235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S. et al.. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018; 173:321.e10–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New functionality allows users to download data from any of the available -omic data via the CellMinerCDB (discover.nci.nih.gov/cellminercdb) Metadata tab as compressed (zip) files. Additionally, users can download drug synonym information or cell line annotation information from this tab. The NCI-60 dataset has an additional method for download via the rcellminerData Bioconductor package.