Abstract
Infertility is a complex multifactorial disease that affects up to 10% of couples across the world. However, many mechanisms of infertility remain unclear due to the lack of studies based on systematic knowledge, leading to ineffective treatment and/or transmission of genetic defects to offspring. Here, we developed an infertility disease database to provide a comprehensive resource featuring various factors involved in infertility. Features in the current IDDB version were manually curated as follows: (i) a total of 307 infertility-associated genes in human and 1348 genes associated with reproductive disorder in 9 model organisms; (ii) a total of 202 chromosomal abnormalities leading to human infertility, including aneuploidies and structural variants; and (iii) a total of 2078 pathogenic variants from infertility patients’ samples across 60 different diseases causing infertility. Additionally, the characteristics of clinically diagnosed infertility patients (i.e. causative variants, laboratory indexes and clinical manifestations) were collected. To the best of our knowledge, the IDDB is the first infertility database serving as a systematic resource for biologists to decipher infertility mechanisms and for clinicians to achieve better diagnosis/treatment of patients from disease phenotype to genetic factors. The IDDB is freely available at http://mdl.shsmu.edu.cn/IDDB/.
Graphical Abstract
Graphical Abstract.
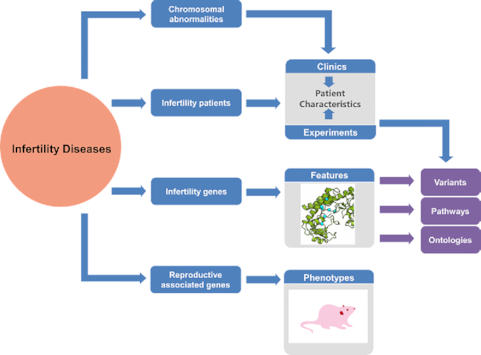
The major components of Infertility Disease Database and their combinations.
INTRODUCTION
Infertility is a diverse multifactorial disease affecting approximately 10% of couples worldwide, and it includes disorders of the female reproductive tract, ovulation defects, inferior quality of spermatozoa, etc. linked with infertility (1). For many affected couples, molecular infertility mechanisms remain unclear, and a considerable number of couples suffer from misdiagnosis and ineffective treatment of infertility or bypass the issues with assisted reproductive technologies (ART) leading to transmission of genetic defects to offspring. The identification of molecular infertility mechanisms enables the selection of the most appropriate management strategy for each couple (2).
Previous studies of human infertility revealed that a large number of pathogenic variations and chromosomal abnormalities play essential roles in patients with infertility and could be responsible for causing human infertility (3–6). Patients with infertility are at a high risk of being carriers of genetic anomalies (7,8). For example, mutations of PANX1 in sporadic female infertility cause oocyte death and infertility (9). An extra X chromosome is commonly found in men with Klinefelter syndrome (47,XXY), characterized as eunuchoid body proportions, gynecomastia, small firm testes and azoospermia (10). Meanwhile, gene-targeted model organisms have advantages in the study of risk factors in infertility (11,12). Several model organisms, such as Mus musculus (mouse), Drosophila melanogaster (fruit fly), Caenorhabditis elegans (nematode) and Danio rerio (zebrafish), have been widely used to assess reproduction and infertility (13–16). Recently, piwi mutants were found to have spermatogenic arrest in the mouse model, a finding that was quickly followed by confirmation in patients with azoospermia (17). Despite growing awareness of genetic causes of infertility, progress in diagnosis and treatment has remained ineffective due to the lack of systematic data and analyses in infertility diseases.
To facilitate the understanding of the pathology and molecular mechanisms underlying infertility, we present the Infertility Disease Database (IDDB), a novel and curated gene database related to infertility for studying the biology of infertility. Manually curated information on associated gene mutations, chromosomal abnormalities, patients’ characteristics, diseases/gene ontology terms and pathways along with supporting reference literature has been collected and included in the IDDB. The IDDB also collected findings from model organisms, and a great many entries represent human homologs of genes shown to affect reproduction in model organisms.
DATA COLLECTION AND ORGANIZATION
A search for original articles published through May 2020 was performed using PubMed to identify studies on genetic variants and chromosomal abnormalities that cause infertility; the data pertaining to the single-nucleotide polymorphisms (SNPs) and irrelevant references were therefore excluded. We curated the data manually and retrieved gene–disease relationships according to the ClinGen clinical validity classification (18). Hyperlinks to the original articles in the PubMed database were provided. We also annotated the gene (genetic variants, gene function and pathway annotation), disease and infertility patients’ samples.
Similarly, relevant information on chromosomal abnormalities causing infertility was also sorted out. The key messages were linked to online existing database browsers, such as National Center for Biotechnology Information (NCBI), GeneCards (19), HUGO Gene Nomenclature Committee (HGNC) (20), Kyoto Encyclopedia of Genes and Genomes (KEGG) (21), UniProt (22) and Gene Ontology (GO) (23). Moreover, we also collected model organism genes involved in reproductive progress, which were annotated with corresponding infertile status and infertile phenotypic information.
Standardization of disease names in infertility was the first step for data integration, sharing and exchange, and it facilitates better communication and diagnosis of infertility diseases. We integrated the disease list from the following sources: (i) Online Mendelian Inheritance in Man (OMIM) (24); (ii) Unified Medical Language System (UMLS); and (iii) NCI Thesaurus (NCIt). We considered the term usage variations in disease names and identifiers (IDs). We mapped all the disease categories among the currently controlled disease ontology databases, including international classification of diseases-version 10/11 (ICD10/11), Mondo Disease Ontology (MONDO) (25), Human Phenotype Ontology (HPO) (26) and ORPHANET (27). As a result, 60 infertility diseases and their identifiers (IDs) were archived in the IDDB, comprising the Infertility Disease Ontology (IDO).
Collectively, all data on infertility were ultimately organized in a unified way into four data entities in the IDDB: genes, diseases, regions (chromosomal abnormalities) and model organisms.
DATA ARCHITECTURE AND WEB INTERFACE
The main page of the IDDB is divided into two main sections: infertility-associated genes and regions in humans and reproduction-associated genes in model organisms. For each human infertility-associated gene, the details were subdivided into six main sections, i.e., infertility pathway maps, pathogenic variants, patient characteristics, regions and chromosomes, disease and gene ontology. The comprehensive descriptions of these sections and the relationship between them are shown in Supplementary Figure S1.
The current version of the IDDB offers multiple web interfaces and data visualizations to facilitate a better understanding of the relationships between the phenotype and the infertility disease genotype. The following five browsing tools were designed under the web site ‘BROWSE’ and ‘ORGANISM’ menus: (i) Disease Browser: infertility diseases are arranged and depicted in interactive Sankey diagrams, and clicking each node displays the disease description and genes and regions records; (ii) Gene Browser: this tool links to infertility-associated genes in the IDDB; at the first level, users can preview the list of genes and filter the records with the names of infertility diseases in the left tree panel; at the second level, prosperity of details for the gene–disease relationships is viewed in multiple sections, as illustrated in Supplementary Figure S1. When studying a single gene, if it has a residual mutation that can be mapped to its experimental crystal structures or computational modeled structures predicted by the ITASSER server (28), the mutation will be shown in the PDB loading file by the JSMol panel under the mutation section; (iii) Region Browser: this page shows all regions of the disorder found in human chromosomes. The region list and details are shown with the first and second level pages similar to those for Gene Browser; (iv) Chromosome Browser: all human genes in the IDDB are labeled in an ideogram based on their chromosome location, and users can select a single chromosome and search the genes in the Gene Browser section; and (v) Model Organism Browser: users can easily explore genes of a species by selecting the options under the ‘ORGANISM’ menu. All entities stored in the IDDB web site are efficiently cross-linked and annotated. Users can easily navigate between genes and their associated, corresponding model organisms, gaining a current and composite snapshot of interdisciplinary research on infertility. Users can query, create subsets and download a single source—the module-specific dataset—for further analysis.
In addition to the navigation features described above, the IDDB also supports flexible queries for diseases, genes, regions and their annotations. With the three search options under the ‘SEARCH’ menu, users can (i) query any field by Global Search, (ii) extract gene records by inputting a list of gene symbol (e.g., CPEB1 LHCGR ZP1) or NCBI Gene ID (e.g., 64506 3973 22917) by Gene Search or (iii) choose Chromosome Search and query disorder regions by chromosome number (e.g., 1–22, X, Y) or chromosome details (e.g., 47, XXY). To facilitate use, the IDDB provides the list and corresponding links in the ‘MISC’ menu for ontological infertility. The data statistics and data download can be executed under the ‘DOWNLOAD’ menu. The help documents and detailed tutorials are readily accessible in the ‘HELP’ menu.
DATA CONTENT AND ANALYSIS
The IDDB comprises a total of 307 human infertility-associated genes, 202 chromosomal abnormalities in the human infertility-associated genes and regions section and 1348 model organism genes associated with infertility (Table 1).
Table 1.
Summary statistics for infertility genes and chromosomal abnormalities in the IDDB 2020
| Diseases/Phenotypes | No. of genes | No. of CAa |
|---|---|---|
| 1. Female infertility | 131 | 53 |
| Endocrine diseases | 47 | 1 |
| Ovarian dysfunction | 60 | 12 |
| Infertility due to oocyte factors | 10 | 0 |
| Unspecified syndrome with female infertility | 19 | 19 |
| Female infertility, unspecified | 3 | 21 |
| 2. Male infertility | 209 | 136 |
| Endocrine diseases | 58 | 5 |
| Abnormal spermatogenesis | 96 | 56 |
| Abnormal sperm morphology | 51 | 0 |
| Genital tract obstruction | 2 | 0 |
| Unspecified syndrome with male infertility | 26 | 35 |
| Male infertility, unspecified | 8 | 40 |
| 3. Recurrent pregnancy loss/miscarriage | 25 | 13 |
| 4. Reproductive disorder in model organisms | 1348 | |
| Mouse | 1076 | |
| Fruit fly | 168 | |
| Nematode | 68 | |
| Zebrafish | 29 | |
| Rat/Rabbit/Sheep/Pig/Chicken | 7 |
aChromosomal ‘abnormalities’ means visible structural changes in karyotype that are sufficiently large to cause clinical abnormalities. Genetic variants (e.g., frameshifts) are not included; CA, chromosomal abnormalities.
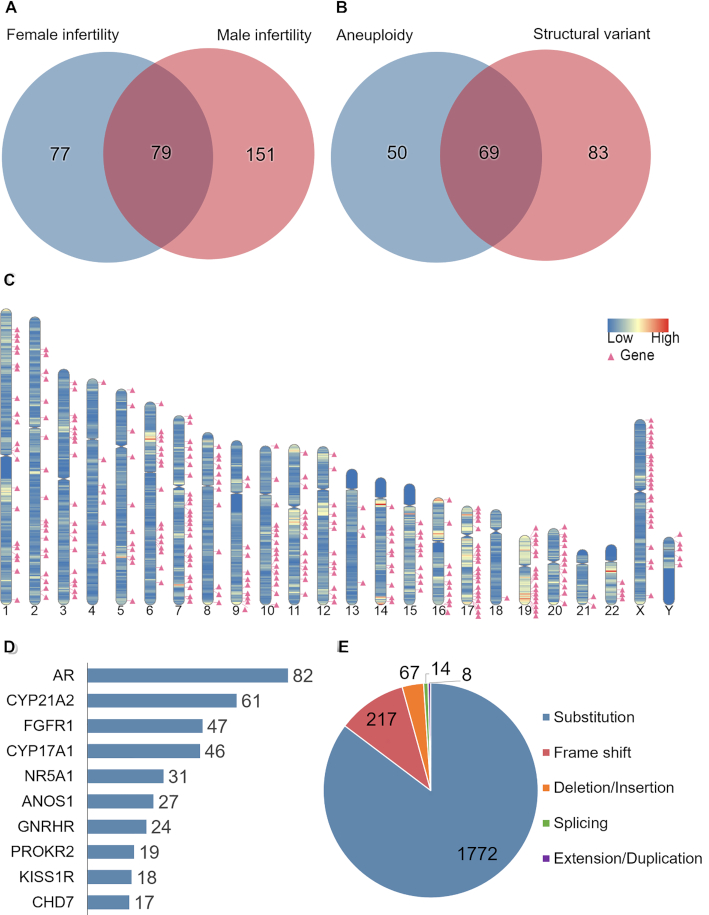
The human infertility-associated genes and regions section is described in Figure 1. There is a total of 79 genes shared by both sexes causing infertility in human (Figure 1A). Most of the 79 genes (e.g., GNRHR, CYP21A2, FSHB) are involved in signal transduction, steroidogenic pathways, etc. (29), and loss-of-function of these genes may cause endocrine infertility. We further annotated 202 cases of human infertility-associated chromosomal abnormalities (e.g., Klinefelter's syndrome, as known as 47, XXY), including 69 cases of both numerical and structural chromosomal abnormalities (Figure 1B). These results demonstrate that the IDDB is a valuable source of information related to common genetic causes of human infertility.
Figure 1.
Statistics of human infertility genes and chromosomal abnormalities in the IDDB 2020. (A) The intersection of infertility genes between females and males. (B) The intersection of chromosomal abnormalities between aneuploidies and structural variants. (C) Schematic representation of the chromosomal location of human infertility-associated genes leads to human infertility disease. (D) Top 10 mutated human infertility-associated genes calculated by unique pathogenic variants for each gene. (E) Statistics of protein variants collected from infertility patients’ samples in the IDDB 2020.
A total of 307 genes causing human infertility were visually represented according to their gene locations on high-resolution chromosome ideograms to allow clinical and laboratory geneticists and clinical pharmacists to access convenient visual images of the position and distribution of human infertility-associated genes. The detected human infertility-associated genes were unevenly distributed across chromosomes (Figure 1C). Specifically, the percentage of human infertility-associated genes ranged from 0.11% to 1.22% on chromosomes. The largest percentage (1.22%) of human infertility-associated genes was discovered on Chromosome X. This suggests that Chromosome X is the most pathogenic chromosome in human infertility, which is consistent with an earlier study (30). Next, these genes were rated by the number of unique pathogenic variants and the 10 most frequently mutated genes in human infertility were identified, including AR, CYP21A2, FGFR1, CYP17A1, NR5A1, ANOS1, GNRHR, PROKR2, KISS1R and CHD7 (Figure 1D). All of the top 10 mutated genes are associated with three main categories of human infertility in the IDDB, including androgen-insensitive syndrome (causative gene: AR), hypogonadotropic hypogonadism (causative genes: FGFR1, NR5A1, ANOS1, GNRHR, PROKR2, KISS1R and CHD7) and congenital adrenal hyperplasia (causative genes: CYP21A2, CYP17A1). Androgen-insensitive syndrome (AIS) is the most common type of 46, XY sex development disorder (DSD) and has a wide range of clinical symptoms, ranging from hypospadias, completely normal female external genitals to male infertility. Mutation of AR is the leading cause of AIS (31). The current version of our developed database includes a total of 82 different pathogenic variants of AR in various AIS patients. We also compiled a total of 2078 pathogenic variants from infertility patients’ samples across 60 different types of human infertility diseases obtained from published literature. We classified the mutations into five categories, including substitution, frameshift, deletion or insertion, splicing and duplication, and we generated a pie chart showing the proportion in each category (Figure 1E).
Additionally, a total of 1348 genes that had been genetically modulated and resulted in significant changes in reproductive phenotype (e.g., azoospermia in mice) in model organisms were included. Most observations were from the four most popular biomedical model organisms: mouse, fruit fly, nematode and zebrafish. However, several results from other model organisms, such as rat, rabbit, sheep, pig and chicken were also included (Table 1). All of the 1348 candidate genes associated with reproductive disorders in model organisms were mapped to their human orthologs using the comparison orthology predictor search engine HCOP (http://www.genenames.org/cgi-bin/hcop) and vice versa; subsequently, as shown in supplementary Figure S2A, approximately 57.2% (176/307) of human infertility genes were found to be associated with reproductive disorder in model organisms. However, 131 human infertility genes had no overlap with the reproduction associated genes, including three genes without homologous gene found in model organisms (TUBB8, SHOX, RMST). Moreover, 1219 reproductive associated genes found in model organisms had no mutation found in infertility patients. The distribution of infertility genes (were listed into six categories) is associated with infertility diseases/phenotypes classified by species and sexes (only in humans), as shown in Supplementary Figure S2B.
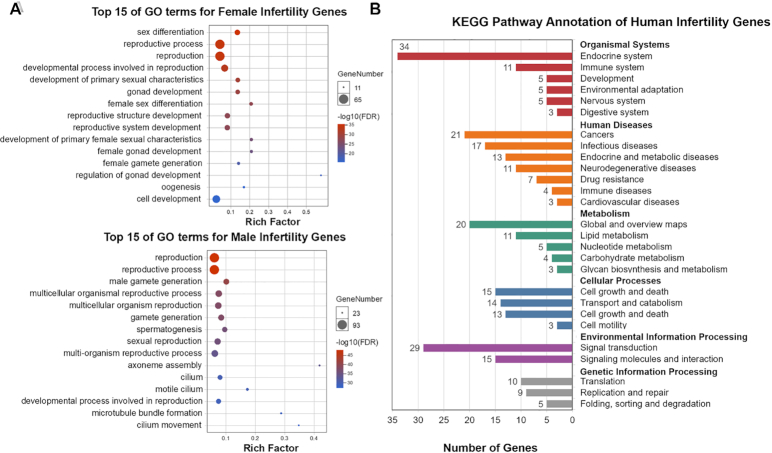
To identify the primary functions of human infertility-associated genes, we performed GO enrichment analysis using DAVID (32) and the R package Bioconductor (33). The bubble chart of GO enrichment analysis depicted in Figure 2A lists the top 15 GO functions for both male and female gene sets. Compared to the context of all protein-coding genes, approximately 46% of genes from both female (65/131) and male (93/209) gene sets are enriched in ‘reproductive process’ and ‘reproduction’ functions, indicating that the collected data in the current version of the IDDB are significantly related to infertility mechanisms. Notably, there are 167 (54%) human genes that were not covered by the ‘reproduction’ or ‘reproductive process’ GO terms in IDDB. Among the 167 genes, 87 infertility related genes are enriched in ‘response to stimulus’, 74 in ‘developmental process’, 53 in ‘cilium movement of cell or subcellular component’ and 27 in ‘cilium assembly’. The genes were reported to be closely related to either endocrine system and development (e.g. KISS1R, FGFR1, GNRHR) or sperm motility (e.g. DNAAF5, ARMC4,DNAH6). Therefore, the infertility diseases in IDDB are not only caused by the genes in ‘reproduction’ or ‘reproductive process’ GO terms, but also associated with genes that play important roles in endocrine, development and sperm motility functions. Comparing the enriched GO function terms, we found that both male and female genes are expected to have become distinct from one another. For females, the genes are associated with female-specific functions, including female sex differentiation, development of primary female sexual characteristics, generation of the female gamete, oogenesis, etc. Specific GO terms such as axoneme assembly, motile cilium, male gamete generation and spermatogenesis are focused primarily on males. Accordingly, these results suggest that human infertility is a complex disease that can be influenced by a multitude of mechanisms. Sex-related gene disorders might indeed cause potential reproductive issues for a particular sex, i.e. dysfunction of GDF9 affects important reproductive-related functions in females including female gamete generation and oocyte growth that could result in primary ovarian insufficiency (IDO:01020100). Another example is AR, which plays an important role in the androgen receptor signaling pathway, and its dysregulation leads to male androgen-insensitivity syndrome (IDO:02010300).
Figure 2.
Enrichment analysis for human infertility gene sets. (A) Gene Ontology (GO)-based functional annotation of female/male infertility gene sets and enrichment analysis. The X-axis represents the rich factor corresponding to the GO term, and the Y-axis represents the name of the GO term. The color of the dot represents the size of the q-value. The smaller the q-value, the closer the color is to blue. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway annotation of human infertility gene sets: for each gene in each disease category, records with a total frequency >2 are displayed in this histogram.
We also conducted KEGG pathway enrichment analysis (29) using Omicshare CloudTools (http://www.omicshare.com/tools/?l=en-us) to recognize the human genes included in the current version of the IDDB from a cellular network perspective. The human infertility-associated genes are involved in 27 pathway subcategories from 6 KEGG categories, namely, organismal systems, human diseases, metabolism, cellular processes, environmental information processing and genetic information processing (Figure 2B). Over 20 infertility-associated genes are enriched in the pathway categories of the endocrine system (34 genes), signal transduction (29 genes) and cancer (21 genes); therefore, dysregulation of these physiological processes may induce infertility. Subsequently, in the top 5 enriched pathways, we found that 3 of 5 belong to the endocrine system category, namely, ovarian steroidogenesis (hsa04913, 10 genes), prolactin signaling pathway (hsa04917, 8 genes) and cortisol synthesis and secretion (hsa04927, 8 genes), as listed in supplementary Table S1. The CYP17A1 gene is common to each of the endocrine pathways above. In addition, numerous different receptor genes also play a significant role in two endocrine pathways, i.e., STAR in ovarian steroidogenesis and cortisol synthesis and secretion. Indeed, the current version of the IDDB has integrated infertility disease-causing genes reported in numerous references with a large-scale gene collection. In addition to extracting the disordered pathways associated with infertility diseases, we can also decode new target genes from the pathways in our future work.
CONCLUSION AND FUTURE DIRECTION
The IDDB platform provides information about pathogenic variants and chromosomal abnormalities associated with human infertility. We also identified reproduction-associated genes in 9 different model organisms. This integrated platform will facilitate the discovery of genes or clusters of genes, construction of networks and study of pathways to decipher mechanisms of infertility and identify potential targets for therapeutic intervention. Additionally, clinicians may also benefit from the accumulated clinical and molecular information in the IDDB, leading to more accurate diagnoses and therapy for infertility diseases. In the future, the IDDB database will be continually updated with new information on human infertility-associated genes and regions and reproduction-associated genes in model organisms.
In the future, we will provide at least one updated release of IDDB per half of year to include more infertility patients’ samples and maintain it as a useful resource for the research and physician community. Future work will also include the integration of additional genetic features associated with infertility into the database.
Supplementary Material
Contributor Information
Jing Wu, Department of Assisted Reproduction, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200011, China; Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Danjun Li, Department of Assisted Reproduction, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200011, China; Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Xinyi Liu, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Qian Li, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Xinheng He, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Jiale Wei, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Xinyi Li, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Mingyu Li, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Ashfaq Ur Rehman, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Yujia Xia, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Chengwei Wu, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China.
Jian Zhang, Medicinal Bioinformatics Center, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200025, China; School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou 450001, China.
Xuefeng Lu, Department of Assisted Reproduction, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (SJTU-SM), Shanghai 200011, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81873856, 81925034, 21778037, 81721004, 81603023, in part]; ‘Two-hundred Talent’ [20191814]; Shanghai Health and Family Planning Commission [20184Y0268]; Subgrant [2017YFC0908502] of the Chinese National Precise Medical Research key project [2017YFC0908500]; Shanghai Municipal Education Commission [2019-01-07-00-01-E00036]; Shanghai Science and Technology Innovation [19431901600]; Shanghai Health and Family Planning System Excellent Subject Leader and Excellent Young Medical Talents Training Program [2018BR12]; Subgrant [2018ZX09711002-001-006] of the National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ [2018ZX09711002]; Clinical Rese Clinical Research Program of 9th People's Hospital, Shanghai Jiao Tong University School of Medicine [JYLJ035]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCE
- 1. Vander Borght M., Wyns C.. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018; 62:2–10. [DOI] [PubMed] [Google Scholar]
- 2. Devroey P., Fauser B.C., Diedrich K. Evian Annual Reproduction Workshop, G. . Approaches to improve the diagnosis and management of infertility. Hum. Reprod. Update. 2009; 15:391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krausz C., Riera-Escamilla A.. Genetics of male infertility. Nat. Rev. Urol. 2018; 15:369–384. [DOI] [PubMed] [Google Scholar]
- 4. Colaco S., Modi D.. Genetics of the human Y chromosome and its association with male infertility. Reprod. Biol. Endocrinol. 2018; 16:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Group E.C.W. Genetic aspects of female reproduction. Hum. Reprod. Update. 2008; 14:293–307. [DOI] [PubMed] [Google Scholar]
- 6. Alhathal N., Maddirevula S., Coskun S., Alali H., Assoum M., Morris T., Deek H.A., Hamed S.A., Alsuhaibani S., Mirdawi A. et al.. A genomics approach to male infertility. Genet. Med. 2020; 22:1–9. [DOI] [PubMed] [Google Scholar]
- 7. Evian Annual Reproduction Workshop, G. Fauser B.C., Diedrich K., Bouchard P., Dominguez F., Matzuk M., Franks S., Hamamah S., Simon C., Devroey P. et al.. Contemporary genetic technologies and female reproduction. Hum. Reprod. Update. 2011; 17:829–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barratt C.L.R., Björndahl L., De Jonge C.J., Lamb D.J., Osorio Martini F., McLachlan R., Oates R.D., van der Poel S., St John B., Sigman M. et al.. The diagnosis of male infertility: an analysis of the evidence to support the development of global WHO guidance-challenges and future research opportunities. Hum. Reprod. Update. 2017; 23:660–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sang Q., Zhang Z., Shi J., Sun X., Li B., Yan Z., Xue S., Ai A., Lyu Q., Li W. et al.. A pannexin 1 channelopathy causes human oocyte death. Sci. Transl. Med. 2019; 11:eaav8731. [DOI] [PubMed] [Google Scholar]
- 10. Groth K.A., Skakkebæk A., Høst C., Gravholt C.H., Bojesen A.. Clinical review: Klinefelter syndrome–a clinical update. J. Clin. Endocrinol. Metab. 2013; 98:20–30. [DOI] [PubMed] [Google Scholar]
- 11. Weinberg-Shukron A., Rachmiel M., Renbaum P., Gulsuner S., Walsh T., Lobel O., Dreifuss A., Ben-Moshe A., Zeligson S., Segel R. et al.. Essential role of BRCA2 in ovarian development and function. N. Engl. J. Med. 2018; 379:1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gou L.T., Kang J.Y., Dai P., Wang X., Li F., Zhao S., Zhang M., Hua M.M., Lu Y., Zhu Y. et al.. Ubiquitination-Deficient mutations in human piwi cause male infertility by impairing Histone-to-Protamine exchange during spermiogenesis. Cell. 2017; 169:1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matzuk M.M., Lamb D.J.. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008; 14:1197–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alphey L., Jimenez J., White-Cooper H., Dawson I., Nurse P., Glover D.M.. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell. 1992; 69:977–988. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z., Lau S.W., Zhang L., Ge W.. Disruption of zebrafish Follicle-Stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology. 2015; 156:3747–3762. [DOI] [PubMed] [Google Scholar]
- 16. Xu X.Z., Sternberg P.W.. A C. elegans sperm TRP protein required for sperm-egg interactions during fertilization. Cell. 2003; 114:285–297. [DOI] [PubMed] [Google Scholar]
- 17. Gou L.T., Kang J.Y., Dai P., Wang X., Li F., Zhao S., Zhang M., Hua M.M., Lu Y., Zhu Y. et al.. Ubiquitination-Deficient mutations in human piwi cause male infertility by impairing Histone-to-Protamine exchange during spermiogenesis. Cell. 2017; 169:1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strande N.T., Riggs E.R., Buchanan A.H., Ceyhan-Birsoy O., DiStefano M., Dwight S.S., Goldstein J., Ghosh R., Seifert B.A., Sneddon T.P. et al.. Evaluating the clinical validity of Gene-Disease Associations: An Evidence-Based framework developed by the clinical genome resource. Am. J. Hum. Genet. 2017; 100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al.. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 2016; 54:1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- 20. Bruford E.A., Lush M.J., Wright M.W., Sneddon T.P., Povey S., Birney E.. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008; 36:D445–D448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M.. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016; 44:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019; 47:D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A.. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019; 47:D1038–D1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shefchek K.A., Harris N.L., Gargano M., Matentzoglu N., Unni D., Brush M., Keith D., Conlin T., Vasilevsky N., Zhang X.A. et al.. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020; 48:D704–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Köhler S., Carmody L., Vasilevsky N., Jacobsen J.O.B., Danis D., Gourdine J.P., Gargano M., Harris N.L., Matentzoglu N., McMurry J.A. et al.. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019; 47:D1018–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rath A., Olry A., Dhombres F., Brandt M.M., Urbero B., Ayme S.. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum. Mutat. 2012; 33:803–808. [DOI] [PubMed] [Google Scholar]
- 28. Yang J., Zhang Y.. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015; 43:W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M.. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014; 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Butler M.G., Rafi S.K., McGuire A., Manzardo A.M.. Currently recognized clinically relevant and known genes for human reproduction and related infertility with representation on high-resolution chromosome ideograms. Gene. 2016; 575:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hughes I.A., Davies J.D., Bunch T.I., Pasterski V., Mastroyannopoulou K., MacDougall J.. Androgen insensitivity syndrome. Lancet. 2012; 380:1419–1428. [DOI] [PubMed] [Google Scholar]
- 32. Jiao X., Sherman B.T., Huang da W., Stephens R., Baseler M.W., Lane H.C., Lempicki R.A.. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012; 28:1805–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T. et al.. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015; 12:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.