Abstract
Pharmacotranscriptomics has become a powerful approach for evaluating the therapeutic efficacy of drugs and discovering new drug targets. Recently, studies of traditional Chinese medicine (TCM) have increasingly turned to high-throughput transcriptomic screens for molecular effects of herbs/ingredients. And numerous studies have examined gene targets for herbs/ingredients, and link herbs/ingredients to various modern diseases. However, there is currently no systematic database organizing these data for TCM. Therefore, we built HERB, a high-throughput experiment- and reference-guided database of TCM, with its Chinese name as BenCaoZuJian. We re-analyzed 6164 gene expression profiles from 1037 high-throughput experiments evaluating TCM herbs/ingredients, and generated connections between TCM herbs/ingredients and 2837 modern drugs by mapping the comprehensive pharmacotranscriptomics dataset in HERB to CMap, the largest such dataset for modern drugs. Moreover, we manually curated 1241 gene targets and 494 modern diseases for 473 herbs/ingredients from 1966 references published recently, and cross-referenced this novel information to databases containing such data for drugs. Together with database mining and statistical inference, we linked 12 933 targets and 28 212 diseases to 7263 herbs and 49 258 ingredients and provided six pairwise relationships among them in HERB. In summary, HERB will intensively support the modernization of TCM and guide rational modern drug discovery efforts. And it is accessible through http://herb.ac.cn/.
INTRODUCTION
Having a complete understanding of the molecular effects of chemical compounds facilitates strategic selection of candidates for advancement in modern drug discovery (1). To evaluate the molecular effects of active compounds, functional assays using cell lines and animal models are often utilized to study whole-transcriptomic changes by using high-throughput technologies to identify the effects of various treatments or perturbations (2). The largest such database for modern drugs, called Connectivity Map (CMap), includes transcriptomic-level perturbation datasets for thousands of well-annotated small molecules profiled in a core set of nine cell lines (3). Furthermore, there are other similar data available from databases storing functional genomics datasets, e.g. the Gene Expression Omnibus (GEO) repository (4). The recent explosion of availability of such transcriptomics perturbation datasets has transformed the field of pharmacology and helped researchers rapidly identify promising chemical compounds for various diseases (5). For example, researchers recently identified a compound, celastrol, which acts as a leptin sensitizer to treat obesity, by mapping the gene expression profile of celastrol to that of reduced endoplasmic reticulum (ER) stress, a condition tightly linked to obesity (6). Similarly, withaferin A, another leptin sensitizer for the treatment of obesity, was discovered through a CMap library analysis of those small molecules that have gene expression profiles similar to that of celastrol (7).
Traditional Chinese medicine (TCM) provides a rich basis for modern drug discovery and development. To date, 97 FDA-approved drugs have been derived from TCM, as recorded by DrugBank (Supplementary Table S1). For example, ephedrine, which is derived from the TCM herb Ma Huang (Herba ephedrae), is used as an anti-asthmatic drug (8). Similarly, artemisinin (qinghaosu), derived from the Chinese herb qinghao (Herba Artemisiae annuae), is now a first-line drug for malaria (9). The traditional knowledge of Chinese herbs, as well as their combinations as prescriptions, is built upon thousands of years of folk testing, including iteratively identifying natural products with improved clinical efficacy for the treatment of a wide range of diseases. Therefore, isolating the active ingredients in TCM and further dissecting their mechanisms of action provides a promising starting point for new therapeutics (10,11).
With the advent of next-generation sequencing technology, an increasing number of TCM studies have focused on identifying the molecular effects of active herbs and ingredients using high-throughput techniques (12). For example, microarray and RNA sequencing approaches have been used in TCM research since 2006 and 2011, respectively. Through 2019, there were >6000 samples in the GEO database on cell lines, animals, or patients that were treated with TCM herbs/ingredients and then analyzed using these transcriptomic technologies. The rapid accumulation of omics data related to natural products has provided new opportunities to understand the transcriptional responses and regulatory changes resulting from the activity of each herb/ingredient (13). Importantly, these data can be mapped to CMap to systematically evaluate the similarities between TCM components and modern drugs, providing objective, data-driven support for the investigation of TCM therapies as candidates for novel therapeutics, such as TCM-HUB (14). However, due to the diversity of data resulting both from varying production platforms and experimental conditions, there is currently no systematic database organizing the data and findings from these high-throughput TCM experiments.
There have been a large number of studies recently published aiming to decode gene targets of the active components of TCM, and link them to modern diseases. The most recent efforts to curate these references were the HIT (15) and TCMID (16) databases published in 2011 and 2012, respectively. HIT contains 1,301 gene targets related to 586 herbal ingredients gathered from PubMed mining, and TCMID contains 680 herbal targets from Chinese articles mined from the WeiPu database. However, the typical sets of associations between herb-target, herb-disease, and ingredient-disease, have not previously been collected and curated from published references. Curated information is of significantly higher value than previous efforts that indirectly linked them using intermediate components, such as targets. Thus, the bulk of studies published within the last decade required manual curation in order to develop a high-confidence database linking targets, diseases, and TCM herbs/ingredients. Such a system would offer high-quality, evidence-based connections between TCM and modern drugs.
Therefore, we built HERB (BenCaoZuJian as its Chinese name), a high-throughput experiment- and reference-guided database of TCM. HERB integrates multiple TCM databases and thus contains the most comprehensive list of herbs and ingredients created to date. We gathered and uniformly reanalyzed public data from all available high-throughput experiments (i.e. microarrays and RNA-seq experiments) that tested herbs or their active ingredients. In this manner, we generated a library containing 6164 gene expression profiles from 1037 experiments evaluating herbs or ingredients and built data-driven connections among herbs, active ingredients, and 2837 modern drugs with pharmacotranscriptomics datasets in CMap. Furthermore, we collected 17,886 TCM-related papers that were published since 2011 by PubMed text mining and manually connected 1241 gene targets and 494 modern diseases for 473 herbs/ingredients from 1966 of those references. This newly curated target and disease information for TCM is of high quality due to its manual confirmation, and was cross-referenced to databases for modern drugs, including the TTD (17), DisGeNet (18), HPO (19), and Disease Ontology (20) databases. Together with database mining and statistical inference, we linked 12 933 targets and 28 212 diseases to 7263 herbs and 49 258 ingredients within HERB. The objective, data-based connections between TCM and modern medicines described in HERB provide strong support for further pharmacological studies of TCM as a fundamental arm of modern drug discovery efforts.
MATERIALS AND METHODS
Catalogs of TCM herbs and ingredients
In HERB, we first prepared a list of herbs and ingredients and determined their relationships by integrating multiple TCM databases including SymMap (21), TCMID 2.0 (22), TCMSP 2.3 (23) and TCM-ID (24). To obtain a non-redundant list, we merged herbs/ingredients with the same IDs, names, or aliases. In case of discrepancies across databases, we selected the entry with the most recent publication date. Note that the molecule formula and molecule smile information for all ingredients were standardized according to SciFinder (25) and PubChem (26), two authoritative chemical databases. We then mapped the TCM ingredients to the DrugBank database (27) and labeled a subset of TCM ingredients as approved drugs. Last, we searched an authoritative database of active herbal ingredients, the ‘National Database for Chemical Composition in TCM’ (http://cintmed.cintcm.com/cintmed/), which has been continuously maintained for several decades by the Institute of Information on Traditional Chinese Medicine, China Academy of Chinese Medical Sciences.
High-throughput experimental data for TCM herbs and ingredients
We gathered public data from GEO high-throughput experiments that studied treatments with herbs or ingredients in multiple cell types or animal models. We only retained those datasets that were generated in Homo sapiens or Mus musculus and that were obtained by a typical bulk RNA sequencing platform (Illumina) or microarray studies. Then, we built different pipelines for uniformly reanalyzing RNA-seq and microarray data.
For RNA-seq data, we first downloaded the raw reads in fastq format for each sample (with unique GSM numbers in GEO) using fasterq-dump 2.9.2 (28). Then, we filtered adaptor sequences and low-quality reads by using Trim Galore 0.6.4 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with the parameters ‘–quality 20 –length 20 –consider_already_trimmed 10’. Next, we mapped the filtered reads to the genome of Homo sapiens (assembly hg38 and annotation v100) or Mus musculus (assembly mm10 and annotation v100) using STAR 2.7.3a (29). Samples with a mapping rate lower than 60% were discarded. Finally, we quantified the read counts for each gene in each sample using featureCounts 2.0.0 (30), preprocessed and quantile-normalized the data into a gene expression matrix using the ‘voom’ function in the R package limma 3.42.2 (31).
For microarray data, we reanalyzed the samples with raw data provided. We first extracted the probe expression profile from the raw data using R package oligo 1.50.0 (32) for the Affymatrix platform, or using R package limma 3.42.2 (31) for the Agilent platform. Next, we normalized the probe expression profile with the robust multichip average algorithm (rma). For other samples without accessible raw data, we directly used their normalized expression profile for probes. Finally, we transformed the probe profile into the normalized gene expression matrix by systematically constructing the conversion relationships between probes from diverse array platforms to genes. When multiple probes could be converted to the same gene, the probe with the median expression level was selected to represent that gene. To that end, we provide an automatic conversion script named probe2gene, which embeds conversion files in SOFT format for 105 platforms downloaded from NCBI and automatically conducts the probe-to-gene conversion according to the SOFT files.
Differential expression analysis and functional enrichment analysis
We conducted differential expression analysis for each TCM herb/ingredient. First, we manually extracted the comparative relationship between samples. In NCBI-GEO, an experiment with a unique GSE number always contains several samples (with unique GSM numbers), and the samples included control samples as well as samples treated with each herb/ingredient in varying biological/physiological conditions. We defined a comparison as a set of control samples and treatment samples that were performed in the same condition, called an HERB experiment (EXP) hereafter. To ensure reproducibility, we required at least two biological replicates of treatment samples (≥2 GSMs) in each EXP. As there can be one or many EXPs for each herb/ingredient, we performed downstream analysis individually for each EXP and then merged the analyzed results from multiple EXPs later. Note that data from EXPs performed in Homo sapiens or Mus musculus were merged and displayed separately.
For each EXP, we performed differential expression analysis using the R package limma 3.42.2. In brief, we first fit linear models from the normalized gene expression matrix using the ‘lmFit’ function and then computed the empirical Bayes statistics using the ‘eBayes’ function. Finally, we selected genes with sufficient expression difference, i.e. |log2(fold change)| ≥ 0.5 and P ≤ 0.05, as differentially expressed genes (DEGs). Based on the DEG list for each EXP, we conducted further functional enrichment analysis for each EXP using the R package clusterProfiler 3.14.3 (33). We used the ‘enrichGO’ function for GO enrichment and the ‘enrichKEGG’ function for KEGG enrichment. Enriched GO terms and KEGG pathways were selected when P ≤ 0.05. Of note, we conducted separate GO/KEGG analyses in each EXP for all genes, up-regulated genes, and down-regulated genes, respectively.
Then, we merged the analyzed results for each herb/ingredient that had multiple EXPs. We firstly transformed the initial two-tailed P-values for EXPs to two one-tailed P-values, P/2 and 1 −P/2, for considering up-regulation and down-regulation. Then, we merged two unified probabilities for each gene, one for up-regulation (P-up), and the other for down-regulation (P-down) using Fisher's method (34). The test statistic (X2) following a chi-square distribution is shown below.
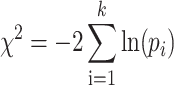 |
We used the ‘fisher.method’ function in the R package metaseqR (35) to compute the statistics and the corresponding P-values. Genes with only one significant P-value (either P-up < 0.05 or P-down < 0.05) were retained in the DEG list, and the directions of their dysregulations were determined by assessing which P-value was significant. Similarly, we also required an average |log2(fold change)| ≥ 0.5 based on those EXPs with significant differences in the individual tests. Based on the merged set of differentially expressed genes for each herb/ingredient, we performed separate GO and KEGG enrichment analyses for all, up and down-regulated genes, respectively. Furthermore, we adjusted up and down P-values separately to account for false discovery rates (FDRs) using the BH method (36).
Data-driven mapping of TCM herbs/ingredients with modern drugs
Once we established the gene expression profiles for TCM herbs/ingredients, we further evaluated the similarities between TCM herbs/ingredients and modern drugs by mapping HERB-EXP to CMap, which contains pharmacotranscriptomics datasets for thousands of well-annotated small molecules. We thus built the data-driven connections by first mapping the DEG list derived from each EXP to CMap and then merged the mapped results from all EXPs from a given herb or ingredient. Note that data from EXPs performed in H. sapiens or M. musculus were mapped and displayed separately.
For each EXP, the list of differentially expressed genes was submitted to the CMap website (https://clue.io/query) in batch query mode. Of note, a maximum of the top 300 genes in the DEG list were retained (5). It is noteworthy that the DEG genes from mouse EXPs were firstly converted to their human orthologs by using the R package homologene with the conversion table from NCBI (ftp://ftp.ncbi.nih.gov/pub/HomoloGene/build68/) (Supplementary Table S2), and then the human genes for each mouse EXP were submitted for CMap calculation. The CMap dataset we queried, called Touchstone, consisted of perturbation data for 2837 compounds that were tested in nine cell lines. Using the connectivity map method (3), the similarities between the input DEG list and the gene expression profiles for all these 2837 perturbagens were evaluated and ranked by their connectivity map scores, τ, ranked from −100 to 100. A τ of +90/−90 indicates that only 10% of random perturbations showed stronger connectivity to the input DEG list, and ‘plus’ or ‘minus’ represents a positive or negative correlation, respectively.
Then, we merged the connectivity results for each herb/ingredient with multiple EXPs. We followed the method of maximum quantile statistic in CMap (3) to merge the connectivity map scores derived from each comparison (EXP). The summarized connectivity map score for each herb/ingredient was calculated according to the following formula,
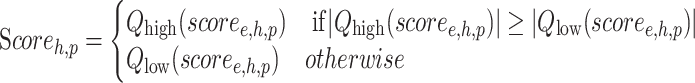 |
where the final scoreh,p stands for the overall similarity measure between a herb/ingredient h in HERB and a perturbagen p in the CMap database, and the scoree,h,p indicates the individual similarity measure between each EXP e related to the herb/ingredient h and the perturbagen p. This procedure compares the Qhigh and Qlow quantiles of scoree,h,p and retains whichever is of higher absolute magnitude. The scores of Qhigh and Qlow were set to be 0.67 and 0.33 respectively, as suggested by CMap.
Manually curated reference data for TCM herbs and ingredients
We collected TCM-related papers from 2011 to 2020 through PubMed text mining. The names and aliases of herbs and ingredients were used as key words, and the key words were required to be present in the title or abstract. In order to narrow down the initial paper set, we first required that references should be published in journals with an impact factor above five according to the Journal Citation Report 2019, and we only selected from articles classified in the biomedical-related categories according to the classification system of the National Science Library, Chinese Academy of Sciences (http://www.fenqubiao.com/). Then, we restricted our search to research articles that were not labeled as review papers in PubMed. Third, we only kept those papers that were related to active herbal ingredients that were included in the National Database for Chemical Composition in TCM (see the first section in Materials and Methods). After PubMed mining and hierarchical filtering, we obtained a list of 17 886 references for manual curation.
Ten Ph.D. candidates were recruited to read, select, and extract information from this set of 17 886 references, with senior students performing a secondary verification. This effort resulted in a set of usable information on gene targets and diseases related to TCM herbs/ingredients from 1966 of these references, called HERB-REF, most of which had evidence from low-throughput experiments. Thus, the resulting data included information not only on herb/ingredient and target/disease relationships, but also on the cell line, animal model, or patients used in the experiments. We further standardized gene names for the newly curated TCM targets according to the NCBI nomenclature and GeneCards database (37), and then cross-referenced them with the TTD database (17), a widely used therapeutic target database for modern compounds. Then, we standardized the disease information to MeSH terms (https://www.nlm.nih.gov/mesh/meshhome.html) and the DisGeNet database (18), and cross-referenced them with the HPO and Disease Ontology (20) databases. Further, we curated a number of clinically relevant phenotypes in HERB-REF for users’ convenience. Finally, we transferred the relationships between targets and diseases from DisGeNet (18) to HERB. As a result of this approach, HERB offers a set of data-based connections linking TCM and modern drugs, built on basis of high-quality manual curation of references.
Other associations among HERB components
Prior to HERB-REF, databases like SymMap (21), HIT (15), TCMSP (23) and TCMID (22) have also curated ingredient-target relationships. Therefore, we merged together the ingredient−target pairs from reference and database mining. Other relationships, e.g. ingredient−disease, herb−target and herb−disease, were generally indirectly linked by combining two or more direct relationships. For example, the indirect relationship between ingredient and disease can be obtained using the gene targets as a middle component (Supplementary Figure S1A). As HERB contains the most comprehensive list of herbs and targets, we calculated the indirect associations for these three relationships again using Fisher's exact test, which is called statistical inference and adopted from SymMap. Taking the ingredient-disease relationship as an example, we first acquired all indirect associations by linking ingredient-target and target-disease relationships together and then selected reliable associations (FDR < 0.01) from them using Fisher's exact test followed by multiple test corrections using the BH method (36). This strategy was also used to infer the herb−target and herb−disease relationships by using ingredient and target as the middle components, respectively (Supplementary Figures S1B, C). Finally, we combined the associated pairs from statistical inference and reference mining together.
Implementation of HERB
In summary, HERB provides information about herbs, ingredients, their gene targets, diseases in modern terms, and the relevant high-throughput experimental data and manually curated references. HERB provides a convenient web interface for users to browse, search, visualize, and download data. HERB is freely accessible at http://herb.ac.cn without a need for user registration. The HERB website was built using the Python-Flask, Nginx and React JavaScript frameworks and is compatible with most major browsers. The HERB data are stored in a MySQL database.
RESULTS
Data contents in HERB
HERB contains a comprehensive list of TCM herbs (7263) and ingredients (49 258) by integrating multiple TCM databases. HERB was created through reanalysis of 6164 gene expression profiles from 1037 high-throughput experiments of herbs/ingredients and a collection of 1966 TCM-related references, from which 1241 gene targets and 494 modern diseases for 473 herbs/ingredients, as well as 709 clinical-relevant phenotypes were curated. Further, HERB includes information on targets and diseases gathered by database mining and statistical inference, resulting in a final total of 12 933 targets and 28 212 diseases related to herbs and ingredients. These data statistics are shown in Table 1. We also provided the statistics of six pairwise relationships among herbs, ingredients, targets, and diseases that were gathered from reference mining, database mining, and/or indirect association through statistical inference (Supplementary Table S3). HERB contains two important novel features lacking from previous TCM databases. First, HERB presents a comprehensive pharmacotranscriptomics data set for TCM herbs and ingredients, allowing mapping to modern drugs with data in CMap. Second, HERB provides manually curated data of targets and diseases mined from recent literature, providing high-confidence information for ranking TCM herbs and ingredients as promising candidates for therapeutic development.
Table 1.
Overview of the data curated in HERB
| Components | Amount | Data source |
|---|---|---|
| Herbs | 7263 | Integrated from SymMap, TCMID, TCMSP and TCM-ID databases |
| Ingredients | 49 258 | Integrated from SymMap, TCMID, TCMSP and TCM-ID databases |
| Experiments | 1037 | Downloaded, manually curated, and automatically analyzed from the NCBI GEO database |
| References | 1966 | Downloaded, manually curated, and extracted information from the NCBI PubMed database |
| Targets | 12 933 | Partially from the manual curation from PubMed references and others from previous databases including SymMap, HIT, TCMSP, and TCMID |
| Diseases | 28 212 | Partially from the manual curation from PubMed references and others from the DisGetNet database |
High-throughput data analyzed in HERB
Through GEO mining using herbs and ingredients as key words, we gathered 472 high-throughput GEO datasets containing 6164 GEO samples. Each GEO dataset performed by a specific lab had a unique GSE number. Each GEO sample conducted in a particular biological/physiological condition also had a unique GSM number. We manually obtained the herb/ingredient-centric classification of these data by defining a HERB experiment (EXP), as a set of control and treatment GEO samples related to a herb/ingredient in one GEO dataset. This resulted in 1037 EXPs in HERB, with 83.9% of them consisting of microarray data and the remaining 16.1% as RNA-seq data; 10.4% of HERB-EXPs are related to herbs and 89.6% of them are related to ingredients (Figure 1A-1). The average number of biological replicates for control and treatment samples in HERB-EXPs were 4.0 ± 2.9 and 3.7 ± 2.6 respectively. Of note, 83.2% and 74.1% of control and treatment samples had at least three replicates, respectively (Figure 1A-2). Sufficient numbers of replicates facilitate downstream analysis of these high-throughput data.
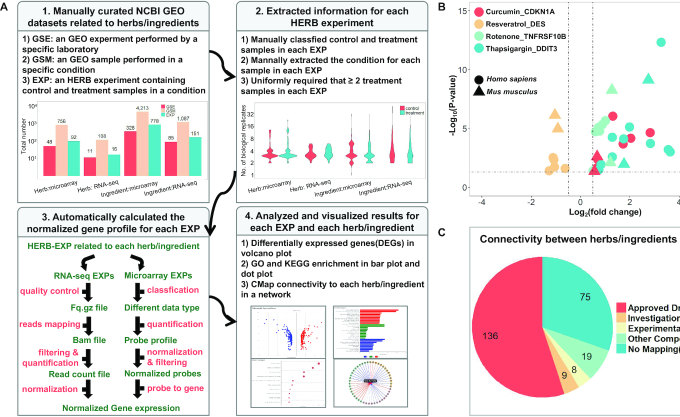
Figure 1.
Construction and characterization of HERB-EXP. (A) A schematic diagram of the data processing framework in HERB-EXP in four consecutive steps: 1. download data from GEO experiments (GSE) and samples (GSM), extract HERB experiments (EXP) for each herb/ingredient, and show the number of GSE/GSM/EXPs for each data type in bar plots. 2. For each EXP, illustrate the number of biological replicates for control and treatment samples in violin plots. 3. Automatically analyze RNA-seq and microarray data according to different pipelines for each EXP first, and then merge results from multiple EXPs together for each herb/ingredient. 4. Visualize the results of differential expression analysis, GO and KEGG enrichment, or CMap connectivity for each EXP, and then for each herb/ingredient, in volcano plot, bar plot, dot plot or network respectively. (B) The differential expression of four known targets (CDKN1A, DES, TNFRSF10B and DDIT3) for four TCM ingredients (curcumin, resveratrol, rotenone, and thapsigargin) in multiple EXPs related to these ingredients are shown in volcano plots. The X-axis shows the expression change compared to control samples and the Y-axis shows statistical significance. The four colors correspond to four target-ingredient pairs, and the two shapes indicate the species information for each EXP (mouse/human). (C) Classification of all herbs/ingredients with HERB-EXP data, according to their mapped compounds in CMap.
Then, we built automatic pipelines to analyze the microarray and RNA-seq data of each individual EXP (Figure 1A-3). To process the RNA-seq data, we used raw data as an input, mapped the filtered reads to the genome, and got the normalized read count for each gene. For microarray raw data, we first processed the data into a probe profile and then converted it into normalized gene expression data. If no raw data were available, the former step was skipped. Based on the normalized gene expression matrix for control and treatment samples in each EXP, we identified DEGs for each EXP and then merged the results into a ranked DEG list for each herb/ingredient. We visualized the DEG list for herbs and ingredients by volcano plot (Figure 1A-4). Based on the DEG list, we visualized the enriched GO terms and KEGG pathways in bar plots and dot plots, for all, up and down-regulated genes (Figure 1A-4), respectively. Moreover, we queried the CMap data containing pharmacotranscriptomics datasets for 2837 modern drugs while using the DEG lists of each herb/ingredient as the query. The connections between the TCM herb/ingredienst and their map-able modern drugs were visualized with a interactive network (Figure 1A-4).
In summary, there were an average of 1801 DEGs upon EXP-level analysis (Supplementary Figure S2A) and 2864 DEGs, on average, for herbs/ingredients after merging the results from multiple EXPs (Supplementary Figure S2B). For GO terms, the average numbers were 1083 and 1260 for the EXP-level and herb/ingredient-level results (Supplementary Figure S3). For KEGG pathways, the two numbers were 44 and 50, respectively (Supplementary Figure S4). We also illustrated the GO/KEGG enrichment results for up-regulated and down-regulated genes (Supplementary Figures S3 and S4). Next, we selected all DEGs related to TCM ingredients that were described in previously published papers and explored their expression changes in the HERB experiments. Across 325 such ingredient-target pairs, 208 of them (64%) showed consistent patterns of differential expression (Supplementary Table S4). Other inconsistencies may be caused by post-transcriptional regulation issues, as evidences from references are mainly based on protein expression levels. We also showed 4 distinguished ingredient-gene pairs in Figure 1B. The direction of regulation for them, including the curcumin-CDKN1A (38), resveratrol-DES (39), rotenone-TNFRSF10B (40) and thapsigargin-DDIT3 (41), were the same as identified in their corresponding references, and cross-validated by both human and mouse data that are merged separately. Thus, they are most likely to be the bona fide targets that are highly related to the downstream pathways affected by these ingredients.
Next, we connected TCM herbs and ingredients to modern pharmaceutic compounds using connectivity mapping between HERB-EXP data and CMap data. In total, 20 herbs and 152 ingredients could be mapped to 978 CMap compounds using a cutoff of absolute connectivity score above 95. The clinical status for 383 out of 978 CMap compounds could be classified as approved drugs, investigational drugs, experimental drugs, or other compounds, by connecting their IDs (InChiKey or PubChem ID) documented in CMap to DrugBank. Thus, we further classified herbs/ingredients according to the type of compounds to which they were mapped. Out of 247 herbs/ingredient with high-throughput data in HERB, there were 136 (55.1%) herbs and ingredients that could be mapped to approved drugs. Another 9 (3.6%) and 8 (3.2%) herbs/ingredients could be mapped to investigational drugs that were in clinical trials and experimental drugs that were under preclinical research, respectively (Figure 1C). Taken together, the gene expression patterns for >60% of herbs/ingredients were similar or antagonistic to modern drugs. These findings illustrate the potential for other TCM herbs/ingredients to be investigated in pharmaceutic development. We have provided a detailed list of the mapped drugs for the TCM herbs/ingredients in Supplementary Table S5. Note that for mouse EXP, a total of 15 123 mouse DEG genes can be converted to their human orthologs, from which 15 060 mouse genes were converted in a one-versus-one manner. On average, 65.3% of mouse DEG genes in these EXPs can be mapped (Supplementary Figure S5). As a result, the CMap mapping results based on mouse data maybe incomplete. So the mouse data was just provided as an auxiliary information.
References curated in HERB
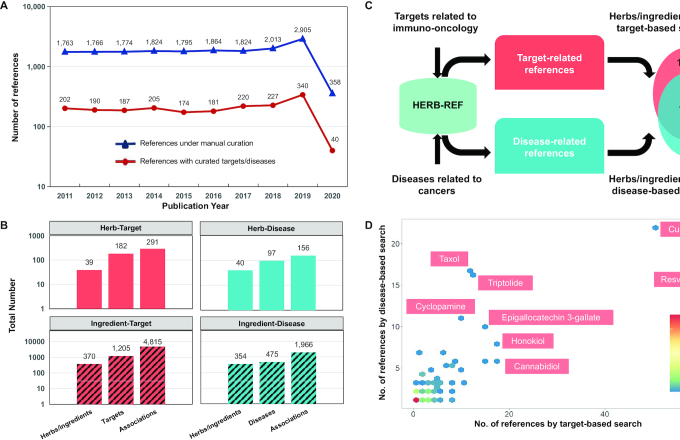
We collected TCM-related papers from 2011 to 2020 by PubMed text mining and obtained a list of 17 886 references for manual curation after hierarchical filtering (Figure 2A, blue line). The number of published papers related to TCM herbs/ingredients has increased dramatically in the past three years, with 2905 relevant papers published in 2019 at the top (with partial data for 2020). After manually reading, selecting, and extracting information from the initial 17 886 references, 1966 references were selected, from which we collected a large amount of high-quality data on gene targets and modern diseases for TCM herbs/ingredients. The pattern of the final set of references with curated targets/diseases (Figure 2A, red line) was quite similar with the initial set of references under manual curation (Figure 2A, blue line), suggesting that the process of manual curation by 10 Ph.D. candidates was unbiased. Only 10.9% of the initial set of references were selected, because we carefully filtered out a large number of references without relevant or reliable information. Finally, we manually extracted 1241 gene targets of 39 herbs and 370 ingredients and manually connected 494 modern diseases with 40 herbs and 354 active ingredients (Figure 2B). The number of newly curated targets was similar to that of HIT 2011 (1301), and larger than TCMID 2012 (680), demonstrating that a continuous number of recent TCM studies have focused on molecular and mechanistic studies. This new set of curated disease information included direct experimental data, and is thus likely of external validity than prior approaches that indirectly linked herbs/ingredients to diseases using gene targets as intermediates. Further, 709 clinically relevant phenotypes, although not diseases themself, were curated and presented for users’ convenience.
Figure 2.
Overview of HERB-REF. (A) Statistics on the number of herb/ingredient-related references published in each year. The blue line represents the total references from manual curation, and the red line represents the references with curated targets/diseases. (B) The number of curated herbs/ingredients, targets/diseases, and their associations from HERB-REF are shown in bar plots, with the four facets showing four types of associations: herb-target, herb-disease, ingredient–target, and ingredient–disease. (C) HERB-REF datasets were used to search and rank promising herbs or ingredients for cancer-related diseases that could be connected to immuno-oncology related targets. The number of herbs/ingredients obtained by target-based search and/or disease-based search was shown in a Venn plot. (D) The selected herbs/ingredients that were supported by both target-based and disease-based search were illustrated in a dot plot, with the X- and Y-axis showing the number of references from each search, respectively. The density of dots representing the number of dots at the same point are represented in color scales.
Next, we ranked herbs or ingredients that were promising for a particular disease with a set of candidate targets, based on the newly curated HERB-REF data. For example, as TCM has been shown to have clinical benefits for regulating the immunity of cancer patients (42), we used the targets and diseases related to immuno-oncology as the input to query HERB-REF. We first collected a list of 400 target genes related to immuno-oncology, according to external published papers (43–45). Then, we searched the 1,966 references in HERB-REF for 53 cancer-related diseases using the key words ‘neoplasm’, ‘tumor’, ‘cancer’ or ‘carcinoma’. We intersected the two lists with the target-related or disease-related herbs or ingredients (Figure 2C). This resulted in 282 herbs/ingredients that had targets in the immuno-oncology related list and were supported by 745 of the references in HERB-REF, and obtained 178 herbs/ingredients that could be connected to immuno-oncology related diseases and supported by 396 references. When we required both target and disease relationships, we were left with a final list of 9 herbs and 137 ingredients that were highly related to immuno-oncology. The list of 146 herbs/ingredients is provided in Supplementary Table S6.
We further visualized the 146 herbs/ingredients related to immuno-oncology in Figure 2D. Most of these herbs/ingredients were supported by a small number of references from either target-based search or disease-based search, as indicated by the high density of dots in the bottom-left corner. The dots in the upper-right corner represent higher confidence estimates with large numbers of supporting references. For example, curcumin, an active ingredient produced by the Curcuma longa plant, inhibits the COP9 signalosome 5 (CSN5) and diminishes the expression of PD-L1 in cancer cells, which is a well-known target for cancer immune therapies (46). As a result, curcumin may represent a promising candidate for combination therapy in cancer. In our HERB-REF search, the connection of curcumin to immuno-oncology was supported by 65 references, with 51 target-based references, 22 disease-based references, and 8 references from both searches, demonstrating the power of HERB-REF for searching and ranking TCM solutions for a given disease. Cannabidiol, a phytocannabinoid from cannabis plants, has long been shown to have anti-tumor activity (47). In both cell lines and an animal model of triple-negative breast cancer, cannabidiol significantly inhibits epidermal growth factor (EGF)-induced proliferation and inhibits the recruitment of tumor-associated macrophages (47). In HERB-REF, cannabidiol was supported by 6/17/4 references from only target-based search, disease-based search, and both searches, respectively.
Aside from reference mining, we merged 157 722 ingredient-target pairs related to 7781 ingredients and 12 838 gene targets from previous TCM databases, including SymMap, HIT, TCMSP and TCMID. We statistically inferred the indirect relationships between ingredient-disease, herb-target, and herb-disease (Supplementary Table S3). The number of each component and their relationships, based on different cutoffs, are shown in Supplementary Figure S6. As a result, we ultimately linked 12 933 targets and 28 212 diseases to 7263 herbs and 49 258 ingredients to obtain HERB, the most comprehensive database for TCM available to date.
Using the HERB database
HERB is freely accessible at http://herb.ac.cn. It provides browse and search pages for users to navigate herbs, ingredients, targets, and diseases. Examples and the types of searchable keywords are shown in or below the search menu for each component (Figure 3A). For the two novel features added in HERB, experiments and references, we provide additional browsing and searching pages. In the experiment search page (Figure 3B), users can click on an experiment ID to view the details page for each EXP. Descriptive information about each EXP includes experimental subjects, experimental conditions, administration methods, sample information for treatment groups and control groups, etc. These descriptions can be found in both the search page and detailed page of each EXP. The analyzed results of EXP-level, high-throughput data are shown in the details page of experiments in the same format as herbs/ingredients, which are described in the following paragraph. In the reference search page (Figure 3C), users can click on the reference ID to view the detail page for each REF. Details of references shown in the browse and details page include the journal name, publication date, experimental subjects, experimental type, and PubMed ID. In the detailed page, we show the pairing relationships between herbs/ingredients and targets/diseases that were manually curated from each corresponding reference. Furthermore, we visualized the abstract for each reference by using a word cloud.
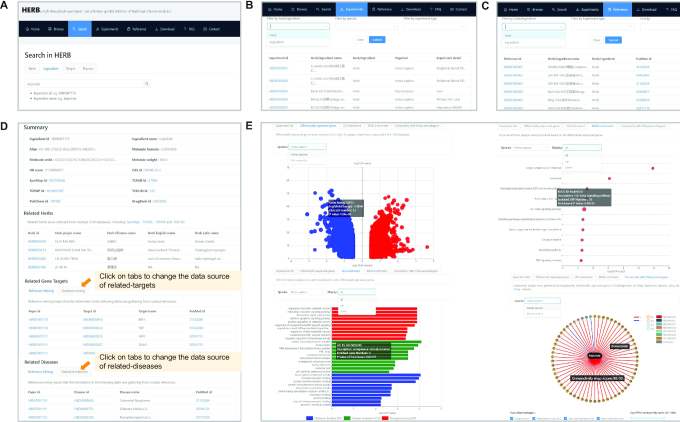
Figure 3.
An illustration of an HERB search. (A) The search page of HERB shows the main 4 components of HERB, including herbs, ingredients, targets, and diseases. (B) The experiment page of HERB shows all high-throughput data related to herbs/ingredients and analyzed by HERB. (C) The reference page of HERB shows all published references with curated targets/diseases information for herbs/ingredients. (D) The details page for an example ingredient, triptolide. The summary panel comes first, and shows relevant descriptive information for this ingredient. The following panels show the herbs, gene targets, and diseases related to this ingredient, respectively. Users can click on the tabs in each panel to navigate information from different data sources. (E) The subsequent detailed page for triptolide. Four figures were used to visualize the high-throughput data related to triptolide. For example, the DEGs are shown in a volcano plot (upper-left). The enriched GO terms and KEGG pathways are shown in bar plot and dot plot (lower-left and upper-right). The connectivity between triptolide and its related compounds in CMap are shown in a network. Note that all figures are implemented in an interactive way where users can see the details of each dot/bar/node/edge upon mousing over them.
The detail page for an example ingredient, triptolide, is shown in Figure 3D and E. The first section includes descriptive information about triptolide and its related herbs, gene targets, and diseases (Figure 3D). The summary information includes the ingredient name, alias, formula, molecule SMILES, molecular weight, and external IDs from other databases. Related herbs for triptolide were integrated from multiple TCM databases. The related targets and diseases for triptolide were derived from reference mining, database mining, or statistical inference, which are each displayed separately under different subpages. And help information about the data source is shown for each table. Among these, the records from reference mining, i.e. manual curation, give the highest-confidence data and can be cross-referenced into the reference page of HERB. The records from database mining are also of high quality, with the original database name and links provided. The significance of the records from statistical inference, i.e. Fisher's exact, is provided, in both P-values and FDRs followed by BH adjustment.
The high-throughput data analyzed by HERB is visualized in both tables and plots. Users can rank and filter records according to each column in all tables. We introduced four figures in the website to better visualize the data in these tables, with each representing an important aspect of the high-throughput data (Figure 3E). Note that human and mouse data are displayed separately, and users can navigate between the two datasets using drop-down menus. For example, the first volcano plot, shown in the upper left, visualizes the DEGs related to triptolide, which has been merged from multiple EXPs of triptolide. One dot in this figure represents a gene, with the color indicating the direction (up or down) of its dysregulation, and the size is proportional to the number of EXPs that support the gene as a DEG. The bar plot shown in the lower left and the dot plot shown in the upper right show the enriched GO terms and KEGG pathways. Each term in the underlying tables can be linked back to the GO and KEGG pages (48–50). Moreover, drop-down menus were provided for users to navigate GO/KEGG results for all, up, and down-regulated genes. The three colors in the GO figure represents three types of GO terms, molecular function (MF), cellular component (CC), and biological process (BP). The size of dots in the KEGG figures indicates the number of EXPs that support that particular item. Detailed information for each bar or dot are visible when the mouse is moved over the element. The last figure in the lower right is a network showing the modern compounds (in CMap) that are mappable to triptolide. The node in the center is triptolide. Other nodes around it are related compounds in four different colors that stand for the data type of the CMap perturbation, including compound (CP), perturbational Class (PCL), gene over-expression (OE), gene knock-down (KD). Furthermore, we embedded a pie chart in each surrounding node to show the specific EXPs that support its mapping to triptolide. Edges between triptolide and its mapped compounds are shown in two colors, with red for a positive correlation and blue for a negative correlation between gene expression profiles. The widths of the edges are proportional to the absolute value of connectivity score. By default, we visualized mapped results with an absolute connectivity score >95. Users are free to select different cutoffs and subsets of the data types of CMap to focus on specific data of interest. And users can use the zoom in/out buttons at the upper-left to control the connectivity figure.
DISCUSSION
Transcriptome analysis allows quantitative measurements of the transcriptional responses and regulatory changes resulting from the perturbations due to compounds, making it a powerful approach for discovering drug targets, evaluating the therapeutic efficacy of drugs, and revealing possible side effects. In recent years, high-throughput data from laboratory or clinical studies of TCM herbs and ingredients have quickly accumulated, but to date there has been no good system of organization. Besides, new TCM-related references published within the last decade have not been curated. As a result, in this work, we reanalyzed all available microarray and RNA-seq data for TCM herbs/ingredients, and curated high-confidence targets and diseases information from recently published TCM references. Together with database mining and statistical inference, we ultimately built HERB, the most comprehensive database for TCM available to date.
The novelty of the HERB database includes: (i) HERB provides a comprehensive and unified pharmacotranscriptomics database of TCM, by reanalyzing all available high-throughput experiments for TCM. Using HERB, researchers and drug developers can view primary data as well as the data-driven mapping results between TCM herbs/ingredients and modern compounds, allowing easy exploration of potential mechanism of actions for herbs/ingredients as well as the identification of new potentially effective therapeutics. (ii) HERB gives high-confidence targets and diseases information related TCM herbs/ingredients based on manual curation of novel references published within the last decade, which bridging the large gap since the creation of HIT (2011) and TCMID (2012). Using these newly curated references in HERB, users can easily search and rank promising herbs or ingredients for a disease with a set of candidate targets. We believe this intuitive workflow will help researchers make use of the significant volume of published data related to TCM. (iii) HERB provides the most comprehensive list of herbs, ingredients, targets, and diseases by integrating multiple data resources. Further, HERB gives comprehensive pairwise relationships among them by combing diverse strategies including database mining, reference mining, and statistical inference.
HERB provides a convenient web interface for users to browse, search, visualize and download key information, and we believed that HERB will intensively support the modernization of TCM to guide drug discovery efforts. However, the currently available data set for TCM pharmacotranscriptomics is remain insufficient, not only in the number of data sets, but also in the diversity of sequencing technologies. In the future, we plan to continuously add as much data as possible. In particular, we would like to add new types of data, such as proteomic, metabolomic, and meta-genomic datasets, to HERB for further improvements. With more and diverse data becoming available, further studies based on these data will be required for analyzing the gene regulation networks under the herb/ingredient perturbation (51,52), and identifying new types of disease-relevant genes (53,54). In a word, we plan to continuously improve the HERB database to provide a high-quality resource for the field of TCM big data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Lihong Liu for her help using the National Database for Chemical Composition in TCM. We thank Dr Jingyu Wang for her help curating the experimental details in HERB.
Contributor Information
ShuangSang Fang, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
Lei Dong, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
Liu Liu, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
JinCheng Guo, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
LianHe Zhao, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
JiaYuan Zhang, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
DeChao Bu, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
XinKui Liu, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
PeiPei Huo, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
WanChen Cao, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
QiongYe Dong, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
JiaRui Wu, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
Xiaoxi Zeng, West China Biomedical Big Data Center, Division of Nephrology, Kidney Research Institute, West China Hospital of Sichuan University, Chengdu 610041, China.
Yang Wu, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Yi Zhao, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China; Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFC1704100, 2019YFC1709801, 2018YFC1313000, 2018YFC1313001, 2018YFD1000604, 2018YFC1704106]; National Natural Science Foundation for Young Scholars of China [31701141, 31701149]; National Natural Science Foundation of China [91740113, 32070670]; Zhejiang Provincial Natural Science Foundation of China [LY21C060003]; BMICC of National Population Health Data Center; Innovation Project for Institute of Computing Technology, CAS [20186060]; China Postdoctoral Science Foundation [2019M660033]; China Postdoctoral Innovative Talent Foundation [BX20200068]. Funding for open access charge: China Postdoctoral Innovative Talent Foundation [BX20200068].
Conflict of interest statement. None declared.
REFERENCES
- 1. Duran-Frigola M., Pauls E., Guitart-Pla O., Bertoni M., Alcalde V., Amat D., Juan-Blanco T., Aloy P.. Extending the small-molecule similarity principle to all levels of biology with the Chemical Checker. Nat. Biotechnol. 2020; 38:1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kwon O.S., Kim W., Cha H.J., Lee H.. In silico drug repositioning: from large-scale transcriptome data to therapeutics. Arch. Pharm. Res. 2019; 42:879–889. [DOI] [PubMed] [Google Scholar]
- 3. Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. et al.. A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell. 2017; 171:1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clough E., Barrett T.. The gene expression omnibus database. Methods Mol. Biol. 2016; 1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Musa A., Ghoraie L.S., Zhang S.D., Glazko G., Yli-Harja O., Dehmer M., Haibe-Kains B., Emmert-Streib F.. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2018; 19:506–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu J., Lee J., Salazar Hernandez M.A., Mazitschek R., Ozcan U.. Treatment of obesity with celastrol. Cell. 2015; 161:999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee J., Liu J., Feng X., Salazar Hernández M.A., Mucka P., Ibi D., Choi J.W., Ozcan U.. Withaferin A is a leptin sensitizer with strong antidiabetic properties in mice. Nat. Med. 2016; 22:1023–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen K.K. A pharmacognostic and chemical study of ma huang (Ephedra vulgaris var. helvetica). 1925. J. Am. Pharm. Assoc. 2012; 52:406–412. [DOI] [PubMed] [Google Scholar]
- 9. Tu Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011; 17:1217–1220. [DOI] [PubMed] [Google Scholar]
- 10. Harvey A.L., Edrada-Ebel R., Quinn R.J.. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015; 14:111–129. [DOI] [PubMed] [Google Scholar]
- 11. Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.-Y., Tang S.-H., Zhang X.-B., Zhang W., Li Z.-Y., Zhou R.-R. et al.. ETCM: an encyclopaedia oftraditional Chinese medicine. Nucleic Acids Res. 2019; 47:D976–D982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lv C., Wu X., Wang X., Su J., Zeng H., Zhao J., Lin S., Liu R., Li H., Li X. et al.. The gene expression profiles in response to 102 traditional Chinese medicine (TCM) components: a general template for research on TCMs. Sci. Rep. 2017; 7:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marquardt J.U., Gomez-Quiroz L., Arreguin Camacho L.O., Pinna F., Lee Y.H., Kitade M., Domínguez M.P., Castven D., Breuhahn K., Conner E.A. et al.. Curcumin effectively inhibits oncogenic NF-κB signaling and restrains stemness features in liver cancer. J. Hepatol. 2015; 63:661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoo M., Shin J., Kim H., Kim J., Kang J., Tan A.C.. Exploring the molecular mechanisms of Traditional Chinese Medicine components using gene expression signatures and connectivity map. Comput. Methods Programs Biomed. 2019; 174:33–40. [DOI] [PubMed] [Google Scholar]
- 15. Ye H., Ye L., Kang H., Zhang D., Tao L., Tang K., Liu X., Zhu R., Liu Q., Chen Y.Z. et al.. HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 2011; 39:D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xue R., Fang Z., Zhang M., Yi Z., Wen C., Shi T.. TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 2013; 41:D1089–D1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y. et al.. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020; 48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinero J., Ramirez-Anguita J.M., Sauch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I.. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020; 48:D845–D855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohler S., Carmody L., Vasilevsky N., Jacobsen J.O.B., Danis D., Gourdine J.P., Gargano M., Harris N.L., Matentzoglu N., McMurry J.A. et al.. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019; 47:D1018–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schriml L.M., Arze C., Nadendla S., Chang Y.W., Mazaitis M., Felix V., Feng G., Kibbe W.A.. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012; 40:D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Y., Zhang F., Yang K., Fang S., Bu D., Li H., Sun L., Hu H., Gao K., Wang W. et al.. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019; 47:D1110–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C.. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018; 46:D1117–D1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y. et al.. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X., Zhou H., Liu Y.B., Wang J.F., Li H., Ung C.Y., Han L.Y., Cao Z.W., Chen Y.Z.. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 2006; 149:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haldeman M., Vieira B., Winer F., Knutsen L.J.. Exploration tools for drug discovery and beyond: applying SciFinder to interdisciplinary research. Curr Drug Discov Technol. 2005; 2:69–74. [DOI] [PubMed] [Google Scholar]
- 26. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al.. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2018; 47:D1102–D1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherry S., Xiao C., Durbrow K., Kimelman M., Rodarmer K., Shumway M., Yaschenko E.. Ncbi sra toolkit technology for next generation sequence data. Plant and Animal Genome XX Conference (January 14–18, 2012). 2012; Plant and Animal Genome. [Google Scholar]
- 29. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 31. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carvalho B.S., Irizarry R.A.. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mosteller F., Fisher R.A.. Questions and answers. The American Statistician. 1948; 2:30–31. [Google Scholar]
- 35. Moulos P., Hatzis P.. Systematic integration of RNA-Seq statistical algorithms for accurate detection of differential gene expression patterns. Nucleic Acids Res. 2014; 43:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statal Soc. 1995; 57:289–300. [Google Scholar]
- 37. Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y. et al.. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics. 2016; 54:1.30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- 38. Jin H., Lian N., Zhang F., Chen L., Chen Q., Lu C., Bian M., Shao J., Wu L., Zheng S.. Activation of PPARgamma/P53 signaling is required for curcumin to induce hepatic stellate cell senescence. Cell Death. Dis. 2016; 7:e2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Di Pascoli M., Divi M., Rodriguez-Vilarrupla A., Rosado E., Gracia-Sancho J., Vilaseca M., Bosch J., Garcia-Pagan J.C.. Resveratrol improves intrahepatic endothelial dysfunction and reduces hepatic fibrosis and portal pressure in cirrhotic rats. J. Hepatol. 2013; 58:904–910. [DOI] [PubMed] [Google Scholar]
- 40. Shi Y.L., Feng S., Chen W., Hua Z.C., Bian J.J., Yin W.. Mitochondrial inhibitor sensitizes non-small-cell lung carcinoma cells to TRAIL-induced apoptosis by reactive oxygen species and Bcl-X(L)/p53-mediated amplification mechanisms. Cell Death. Dis. 2014; 5:e1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Min K.J., Kim H.S., Park E.J., Kwon T.K.. Melatonin enhances thapsigargin-induced apoptosis through reactive oxygen species-mediated upregulation of CCAAT-enhancer-binding protein homologous protein in human renal cancer cells. J. Pineal Res. 2012; 53:99–106. [DOI] [PubMed] [Google Scholar]
- 42. Zhang S., Pang G., Chen C., Qin J., Yu H., Liu Y., Zhang X., Song Z., Zhao J., Wang F. et al.. Effective cancer immunotherapy by Ganoderma lucidum polysaccharide-gold nanocomposites through dendritic cell activation and memory T cell response. Carbohydr. Polym. 2019; 205:192–202. [DOI] [PubMed] [Google Scholar]
- 43. Liu Z., Cai C., Du J., Liu B., Cui L., Fan X., Wu Q., Fang J., Xie L.. TCMIO: A comprehensive database of traditional chinese medicine on immuno-oncology. Front. Pharmacol. 2020; 11:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tang J., Pearce L., O’Donnell-Tormey J., Hubbard-Lucey V.M.. Trends in the global immuno-oncology landscape. Nat. Rev. Drug Discov. 2018; 17:783–784. [DOI] [PubMed] [Google Scholar]
- 45. Tang J., Shalabi A., Hubbard-Lucey V.M.. Comprehensive analysis of the clinical immuno-oncology landscape. Ann. Oncol. 2018; 29:84–91. [DOI] [PubMed] [Google Scholar]
- 46. Lim S.O., Li C.W., Xia W., Cha J.H., Chan L.C., Wu Y., Chang S.S., Lin W.C., Hsu J.M., Hsu Y.H. et al.. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016; 30:925–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elbaz M., Nasser M.W., Ravi J., Wani N.A., Ahirwar D.K., Zhao H., Oghumu S., Satoskar Abhay R., Shilo K., Carson W.E. III et al.. Modulation of the tumor microenvironment and inhibition of EGF/EGFR pathway: novel anti-tumor mechanisms of Cannabidiol in breast cancer. Mol. Oncol. 2015; 9:906–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li B., Ma C., Zhao X., Hu Z., Du T., Xu X., Wang Z., Lin J.. YaTCM: Yet another traditional chinese medicine database for drug discovery. Comput. Struct. Biotechnol. J. 2018; 16:600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2018; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2018; 47:D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liao Q., Liu C., Yuan X., Kang S., Miao R., Xiao H., Zhao G., Luo H., Bu D., Zhao H. et al.. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011; 39:3864–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao Q., Xiao H., Bu D., Xie C., Miao R., Luo H., Zhao G., Yu K., Zhao H., Skogerbø G. et al.. ncFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res. 2011; 39:W118–W124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X. et al.. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018; 46:D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo J.C., Fang S.S., Wu Y., Zhang J.H., Chen Y., Liu J., Wu B., Wu J.-R., Li E.-M., Xu L.-Y. et al.. CNIT: a fast and accurate webtool for identifying protein-coding and long non-coding transcripts based onintrinsic sequence composition. Nucleic Acids Res. 2019; 47:W516–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.