Abstract
OGEE is an Online GEne Essentiality database. Gene essentiality is not a static and binary property, rather a context-dependent and evolvable property in all forms of life. In OGEE we collect not only experimentally tested essential and non-essential genes, but also associated gene properties that contributes to gene essentiality. We tagged conditionally essential genes that show variable essentiality statuses across datasets to highlight complex interplays between gene functions and environmental/experimental perturbations. OGEE v3 contains gene essentiality datasets for 91 species; almost doubled from 48 species in previous version. To accommodate recent advances on human cancer essential genes (as known as tumor dependency genes) that could serve as targets for cancer treatment and/or drug development, we expanded the collection of human essential genes from 16 cell lines in previous to 581. These human cancer cell lines were tested with high-throughput experiments such as CRISPR-Cas9 and RNAi; in total, 150 of which were tested by both techniques. We also included factors known to contribute to gene essentiality for these cell lines, such as genomic mutation, methylation and gene expression, along with extensive graphical visualizations for ease of understanding of these factors. OGEE v3 can be accessible freely at https://v3.ogee.info.
INTRODUCTION
Gene essentiality is a core concept of genetics, and has important relevance in relation to fundamental areas such as evolutionary, and systems and synthetic biology, and applicability in drug development. Essential genes (EGs) are often responsible for important biological processes and cellular fitness in an organism, and are critical for its survival. EGs are of particular importance in mechanistic biological studies and therapeutic applications, such as studying behavior of a biological system under perturbation (1), studying sequential gene deletions to define a minimal genome/organism (2,3) and identifying or prioritizing of therapeutic targets in pathogens (4–6) and human cancers (7–11). EGs complements have been identified in well-characterized small model organisms such as Saccharomyces cerevisiae and Caenorhabditis elegans (12,13) and some large and complex organisms, such as human (14), using various technologies.
The essentiality of a gene is highly dependent on various factors including the genetic context, genetic background of the host and environment (15). Hence, gene essentiality is not a static, but rather a context-dependent property of a gene. Since version 1, OGEE has been promoting the context-dependent concept and its importance for understanding on gene essentiality (16). In our last update (17), we focused on the importance of ‘conditionally essential genes’ (CEGs) or ‘differentially essential genes’ (DEGs) in cancer cell lines. The current update provides: (a) an increased coverage of essentiality tested genes and species, (b) up-to-date essentiality status of existing genes, (c) an increased coverage of human cell lines, (d) an extensive graphical visualization for various factors that influence gene essentiality and (e) a dedicated domain with secure HTTPS protocol, https://v3.ogee.info.
DATA GENERATION
Collection and organization of essentiality datasets
To find large-scale gene essentiality experiments, we examined publications from the NCBI PubMed database (18) identified using the key words ‘essential genes’, ‘gene essentiality’, ‘tumor dependency genes’, ‘cancer vulnerable genes’, and then downloaded all classified essential or non-essential genes. For model organisms such as Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae and Arabidopsis thaliana, we also obtained their tested genes from online public databases (19–30). The full list of reference databases and publication records are provided in Supplementary Table S1. The tested results (i.e. essential and non-essential statuses) were organized into datasets, according to their experimental conditions. In total, we collected 127 datasets for 91 non-human species, containing 213 608 tested genes. This collection represents a significant increase from our previous version, OGEE v2 (17), which contained 99 datasets from 48 non-human species, with in total 167 799 tested genes.
Essentiality datasets from human cancer cell lines
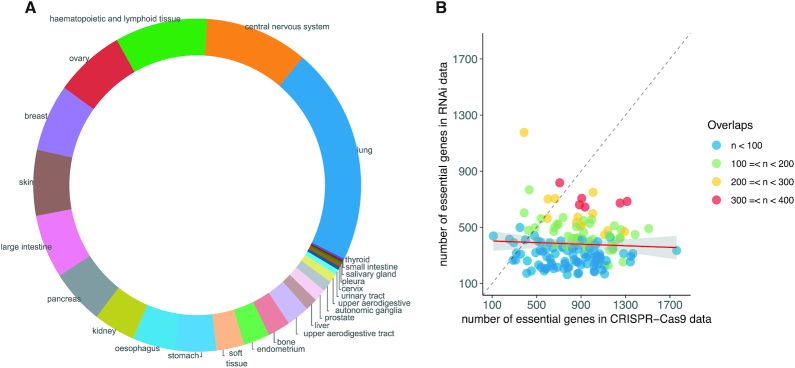
Tumor essential genes, also known as tumor dependency genes, are potential targets for treatment and/or drug development (31,32). During the last decade, cancer cell lines have been used extensively for tumor essential gene identification (32,33). We included results for 16 human cell lines in our last version. In OGEE v3, we expanded this collection to include a total of 581 cell lines which were tested by genome-wide CRISPR-Cas9 and RNAi screening. The 581 cell lines were related to 25 different tissues based on their histological origins, as shown in Figure 1A.
Figure 1.
Statistics on 581 human cell lines collected in OGEE v3. (A) The 581 cell lines were related to 25 different tissues by their histological origins. (B) CRISPR-based method often identified more essential genes than RNAi-based method. Each point indicates a cell line, and the x-axis and y-axis mean the number of identified essential genes of the cell line in CRISPR–Cas9 and RNAi datasets respectively. The colors (blue, green, red, yellow) illustrate the number of overlapping essential genes between CRISPR–Cas9 and RNAi data. The red line denotes the fitted regression line between numbers of essential genes in RNAi and CRISPR-Cas9 data. In total 150 cell lines were tested by both techniques.
In total, 150 cell lines were tested by both RNAi- and CRISPR-Cas9-based methods (Supplementary Table S2). As shown in Figure 1B, the CRISPR-Cas9-based method identified more essential genes than RNAi in most cell lines, likely because of higher efficiency of CRISPR-Cas9 in gene knockdown/out. Essential genes identified by RNAi are often not a subset of those identified by CRISPR–Cas9 in the same cell line, thus the combined data from two distinct methods could offer more robust results and coverage over human essential genes, as a previous study suggested (34).
In summary, the current version (v3) of OGEE include 16 datasets from 16 non-human eukaryotes and 111 datasets from 75 prokaryotes. Of the 213,608 collected genes, 31,177 were tested in multiple datasets, accounting up to 14.6% of all collected genes. Of the tested genes from multiple datasets, 15 440 genes were conditionally essential genes (CEGs) or context-dependent, representing around 49.52% of genes covered by multiple datasets. In addition, OGEE v3 also includes experimental essentiality results for 581 human cancer cell lines.
Collection of known factors influencing gene essentiality
The essentiality of a gene at any given time point depends on various factors. In OGEE v3, we included several factors known to influence gene essentiality, including paralogs and orthologs or duplication status (35), functional annotation of a gene (36), connectivity of protein-protein interaction (PPI) network (37) and early expression during embryonic development. OrthoFinder (38) was used to identify paralogs and orthologs for all species included here. The PPI data were downloaded from latest version of STRING v11.0 database (39); interactions with score ≥900 were used to compute the connectivity score for all the genes. As described in previous studies (40,41), the EGs had more interaction partners at the protein level. Protein sequences were obtained from NCBI protein database (18), while their functional annotations were obtained from the Gene Ontology (GO) database (42). For more information, please refer the ‘Help’ page of the OGEE database.
For human cell lines, factors known to influence gene essentiality included mutation, methylation and gene expression (43); in fact, factors including these have been used to predict gene essentiality in human (43,44). Thus, for each cell line, we collected data on gene mutations from cosmic (45), on methylation data from NCBI GEO database (18) and/or on gene expression data from the CCLE database (46).
Built-in tools for analysis
OGEE v3 also has integrated tools to analyze the impact of gene properties on the page ‘Analyze’. The available options include developmental vs. non-developmental and duplicates vs. singlets, calculated in terms of proportion of essentiality (PE). For more detail on built-in tools, please refer to our previous publication (17).
EXTENSIVE ANNOTATIONS OF HUMAN ESSENTIAL GENES
The identification and characterization of human EGs provides a better understanding of human diversity, has practical medical applications for disease genetics and clinical interpretation of disease-associated genetic variants; EGs are associated with the core developmental, metabolic and signaling pathways; and they are less likely to be tolerate missense variation, and more prone to pathogenicity (41). While effort has been put into identifying targets for cancer therapeutics, several research groups (32,33) have identified cancer dependencies genes and lineage-specific EGs in human cell lines using RNAi screening or CRISPR–Cas9 technology. Since both RNAi and CRISPR are promising complementary methods for identifying EGs, we annotated human EGs extensively by considering both of these techniques as well as factors influencing gene essentiality.
As shown in Figure 1A, the 581 human cell lines collected in OGEE v3 were assigned into 25 groups according to their tissue of origin. The largest number of cell lines were for lung (n = 123), followed by central nervous system (n = 59), haematopoietic and lymphoid tissue (n = 53), ovary (n = 40) and breast (n = 38).
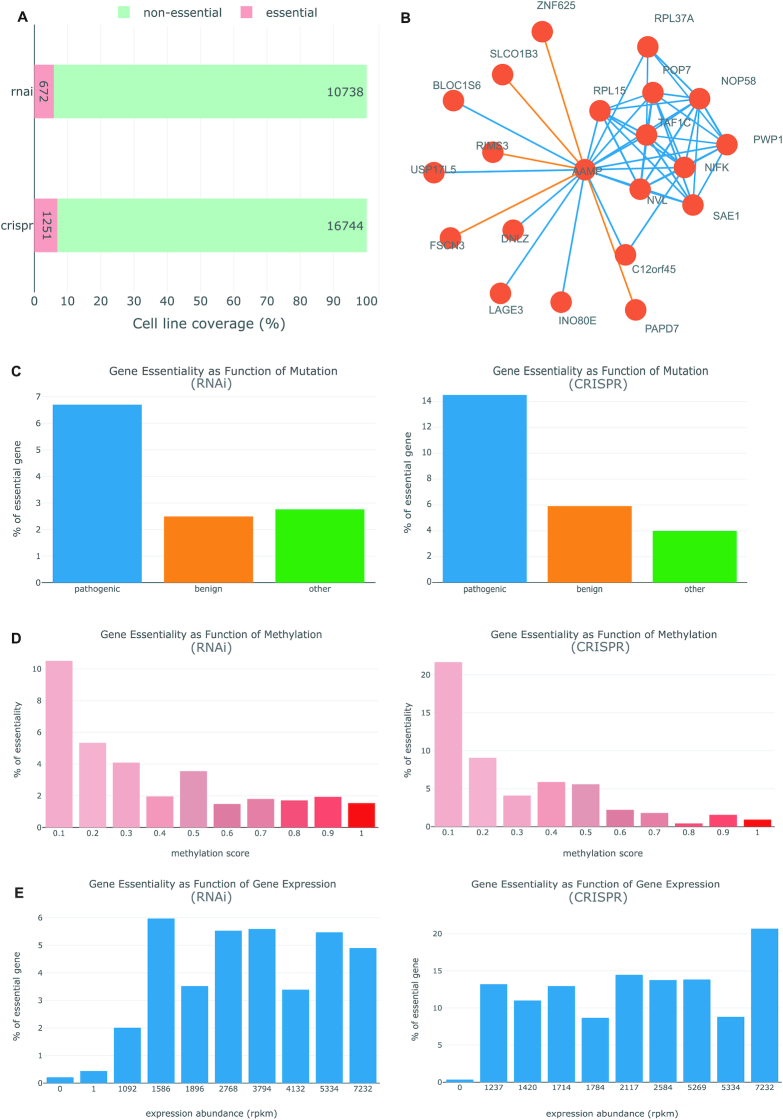
We selected the RKO cell line set to exemplify the annotation of essential gene in OGEE v3 (https://v3.ogee.info/#/cellline/large%20intestine/RKO/summary). The cell line RKO is inferred to have 672 essential genes based on RNAi screening, and 1251 essential genes based on CRISPR–Cas9 dataset (Figure 2A). To understand more about the characteristics of essential genes and the relationship with the influencing factors in human cell lines, we compared tested genes of RKO against the known influencing factors data that we had collected (Figure 2C–E). Mutations, methylation and gene expression all contributed significantly to gene essentiality in this cell line. For example, genes harboring pathogenic mutations are more likely to be essential regardless of the experimental methods used (Figure 2C); genes with less methylation at their transcription start site (TSS200) are also more likely to be essential (Figure 2D); in general, lowly-expressed genes are less likely to be essential (Figure 2E).
Figure 2.
Extensive annotations on human essential genes provided by OGEE v3. Shown here are graphical visualizations taken from the OGEE v3 (https://v3.ogee.info/#/cellline/large%20intestine/RKO/summary). (A) gene essentiality determined using different experimental methods. (B) Correlational co-essentiality chart of AAMP gene from central nervous system tissue tested by CRISPR-Cas9 technique. The orange edges denote negative correlation while the blue edges denote positive correlation among the connected genes. (C) Gene essentiality as function of mutation: mutation outcomes predicted using FATHMM were used to group all tested genes into three categories, the percentage of essential in each group was then calculated (please consult ‘Help’ page for more details); left panel: gene essentiality tested using RNAi, right panel: gene essentiality tested using CRISPR–Cas9. (D) Gene essentiality as function of methylation: genes are grouped based on methylation sites and calculate percentage of essential for each sites; red color gradient is used to denote the methylation score, increasing gradually from 0 to 1; users can select any methylations from the drop down menu. (E) Gene essentiality as function of gene expression: all 0 rpkm genes are in the first bin and rest of the genes are assigned into equal size nine bins; lastly, percentage of essential is calculated for each bin.
To better appraise the pattern of essentiality across human cell lines and identify potentially meaningful biological relationships between genes (47), we selected data of those tissues with at least ten cell lines from a conditioned experiment, and assessed gene-gene essentiality relationships among individual tissues by Spearman and Pearson correlation. Raw fitness scores instead of binary essentiality assignments (i.e. essential or non-essential genes) were used for such calculations. After filtering the genes with essentiality in at least 50% cell lines of a specific tissue, we then selected gene pairs with significant correlation results (P-value < 0.001). The results of this analysis are available via ‘Correlation co-essentiality’ in OGEE v3 for each human gene tested that met the cut-off (see Figure 2B for an example).
Orthology and comparisons with mouse essential genes
To date, mouse is the only mammalian species for which gene essentiality has been tested at organismal level. About 30% of tested mouse genes are essential for survival, which is markedly higher than that of human cell lines (21). Thus, unique essential genes in mouse experiments may be important during growth and development, while those uniquely essential in human cell lines may represent suitable treatment and/or drug targets with putatively less side-effects. To compare gene essentiality between mouse and human cells, we first selected tissues for which at least six cell lines were available, and defined 1607 core essential human genes in >50% of the cell lines (n ≥ 3). We downloaded the mouse knock-out and associated phenotype data from databases IMPC (21) and MGI (22) to deeply understand the functions of essential genes in mice; these databases contained information for 5799 and 9693 knockout genes, respectively. Both MGI and IMPC are generating a knockout mouse line for each protein-coding gene, collecting the corresponding phenotypes of mutants and controls, identifying the disease models, and building a complete functional catalogue for the mouse genome. From the orthogroup data generated by OrthoFinder (38) (see ‘Data generation’), we identified 3522 and 6214 knock-out genes for mice in the IMPC (21) and MGI (22) dataset, respectively. We also identified that 37.6% (4463/11 868) mouse genes were identified as lethal or sub-viable, and there existed 4192 genes corresponding to human orthologs, 13.5% (565/4192) of which are consistently essential in human cells (84.7%, 565/667) (shown in Figure 3). These results showed that the mice exhibit similar survival behavior with human cell line screened in their corresponding orthologous genes.
Figure 3.
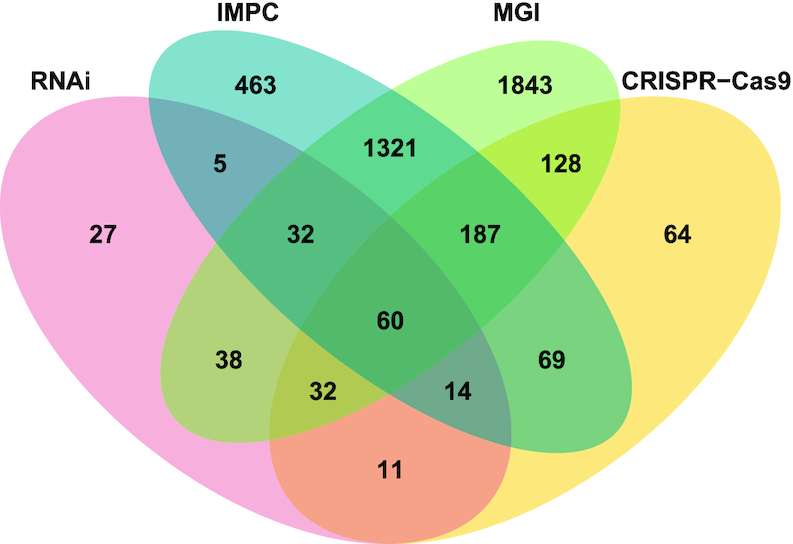
Comparing gene essentiality between human-mouse orthologs. Here only genes with one-to-one human-mouse orthologous relationships are included. Mouse essential genes were obtained from IMPC (blue) and MGI (green) databases. The two human essential gene dataset, RNAi and CRISPR–Cas9 were generated by selecting those that were essential in over 50% the cell lines of a tissue; tissues with less than six cell lines were excluded from this analysis.
CONCLUSION
Here, we introduce OGEE v3, an updated version of the Online GEne Essentiality database. This update almost doubles the number of species for which EGs have been inferred, and a marked increase in the number of genes that have been tested for essentiality. Furthermore, we thoroughly revisited information on conditionally essential genes in both existing and new datasets. We also included 581 datasets for human cell lines and genes tested using CRISPR–Cas9 and/or RNAi methods. In addition, we also included information on factors that influence gene essentiality to assist biological interpretation. Thus, OGEE v3 is expected to be a useful and an important database for biologists, bioinformatician and other colleagues from a range of scientific communities who are working on, or are interested in, exploring gene essentiality.
OUTLOOK
Over the coming years, the OGEE team aims to continue tracking all the available genes data sets and gene properties with the latest experimental records. Specific foci of the OGEE team will be to include a BLAST function (48) in the database and to improve the user-interface, visualizations and crosslinking of resources.
DATA AVAILABILITY
All data are freely accessible to all academic users. This work is licensed under a Creative Commons Attribution 3.0 Unported License (CC BY 3.0). Users can download datasets from the ‘Download’ page. Individual datasets or combined datasets of individual species can be downloaded via the ‘Browse’ page.
Supplementary Material
Contributor Information
Sanathoi Gurumayum, Key Laboratory of Molecular Biophysics of the Ministry of Education, Hubei Key Laboratory of Bioinformatics and Molecular-imaging, Center for Artificial Biology, Department of Bioinformatics and Systems Biology, College of Life Science and Technology, Huazhong University of Science and Technology (HUST), 430074 Wuhan, Hubei, China.
Puzi Jiang, Key Laboratory of Molecular Biophysics of the Ministry of Education, Hubei Key Laboratory of Bioinformatics and Molecular-imaging, Center for Artificial Biology, Department of Bioinformatics and Systems Biology, College of Life Science and Technology, Huazhong University of Science and Technology (HUST), 430074 Wuhan, Hubei, China.
Xiaowen Hao, Key Laboratory of Molecular Biophysics of the Ministry of Education, Hubei Key Laboratory of Bioinformatics and Molecular-imaging, Center for Artificial Biology, Department of Bioinformatics and Systems Biology, College of Life Science and Technology, Huazhong University of Science and Technology (HUST), 430074 Wuhan, Hubei, China.
Tulio L Campos, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia; Instituto Aggeu Magalhães, Fundação Oswaldo Cruz (IAM-Fiocruz), Recife, Pernambuco, Brazil.
Neil D Young, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
Pasi K Korhonen, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
Robin B Gasser, Department of Veterinary Biosciences, Melbourne Veterinary School, The University of Melbourne, Parkville, Victoria 3010, Australia.
Peer Bork, European molecular biology laboratory (EMBL), Meyerhof Strasse 1, 69117 Heidelberg, Germany; Molecular Medicine Partnership Unit, University of Heidelberg and European Molecular Biology Laboratory, 69120 Heidelberg, Germany; Max-Delbrück-Centre for Molecular Medicine, Robert-Rössle-Straße 10, 13125 Berlin, Germany; Department of Bioinformatics, Biocenter, University of Würzburg, 97074 Würzburg, Germany.
Xing-Ming Zhao, Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, 200433 Shanghai, China; Key Laboratory of Computational Neuroscience and Brain-Inspired Intelligence, Ministry of Education, China.
Li-jie He, Department of Medical Oncology, People's Hospital of Liaoning Province, 110016 Shenyang, China.
Wei-Hua Chen, Key Laboratory of Molecular Biophysics of the Ministry of Education, Hubei Key Laboratory of Bioinformatics and Molecular-imaging, Center for Artificial Biology, Department of Bioinformatics and Systems Biology, College of Life Science and Technology, Huazhong University of Science and Technology (HUST), 430074 Wuhan, Hubei, China; College of Life Science, Henan Normal University, 453007 Xinxiang, Henan, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFC0910502, 2019YFA0905601 to W.H.C.]; Natural Science Foundation of Shanghai [17ZR1445600]; Shanghai Municipal Science and Technology Major Project [2018SHZDZX01]; ZJLab; Research at the University of Melbourne was supported by the Australian Research Council (ARC); National Health and Medical Council (NHMRC) of Australia. Funding for open access charge: National Key Research and Development Program of China [2019YFA0905601].
Conflict of interest statement. None declared.
REFERENCES
- 1. Keller P.J., Knop M.. Evolution of mutational robustness in the yeast genome: a link to essential genes and meiotic recombination hotspots. PLoS Genet. 2009; 5:e1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glass J.I., Assad-Garcia N., Alperovich N., Yooseph S., Lewis M.R., Maruf M., Hutchison C.A., Smith H.O., Venter J.C.. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lluch‐Senar M., Delgado J., Chen W., Lloréns‐Rico V., O’Reilly F.J., Wodke J.A., Unal E.B., Yus E., Martínez S., Nichols R.J. et al.. Defining a minimal cell: essentiality of small ORFs and ncRNAs in a genome‐reduced bacterium. Mol. Syst. Biol. 2015; 11:780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hu W., Sillaots S., Lemieux S., Davison J., Kauffman S., Breton A., Linteau A., Xin C., Bowman J., Becker J. et al.. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 2007; 3:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu Y., Deng J., Rhodes J.C., Lu H., Lu L.J.. Predicting essential genes for identifying potential drug targets in Aspergillus fumigatus. Comput. Biol. Chem. 2014; 50:29–40. [DOI] [PubMed] [Google Scholar]
- 6. Paul M.L.S., Kaur A., Geete A., Sobhia M.E.. Essential gene identification and drug target prioritization in Leishmania species. Mol. Biosyst. 2014; 10:1184–1195. [DOI] [PubMed] [Google Scholar]
- 7. Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E. et al.. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data. 2014; 1:140035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo J., Emanuele M.J., Li D., Creighton C.J., Schlabach M.R., Westbrook T.F., Wong K.K., Elledge S.J.. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009; 137:785–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marcotte R., Brown K.R., Suarez F., Sayad A., Karamboulas K., Krzyzanowski P.M., Sircoulomb F., Medrano M., Fedyshyn Y., Koh J.L.Y. et al.. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012; 2:172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cheung H.W., Cowley G.S., Weir B.A., Boehm J.S., Rusin S., Scott J.A., East A., Ali L.D., Lizotte P.H., Wong T.C. et al.. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:12372–12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo B., Hiu W.C., Subramanian A., Sharifnia T., Okamoto M., Yang X., Hinkle G., Boehm J.S., Beroukhim R., Weir B.A. et al.. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B. et al.. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002; 418:387–391. [DOI] [PubMed] [Google Scholar]
- 13. Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M. et al.. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003; 421:231–237. [DOI] [PubMed] [Google Scholar]
- 14. Benjamin M., Davis, Glen F., Rall M.J.S.. Identification and characterization of essential genes in the human genome. Physiol. Behav. 2017; 176:139–148.28363838 [Google Scholar]
- 15. D’Elia M.A., Pereira M.P., Brown E.D.. Are essential genes really essential. Trends Microbiol. 2009; 17:433–438. [DOI] [PubMed] [Google Scholar]
- 16. Chen W.H., Minguez P., Lercher M.J., Bork P.. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012; 40:D901–D906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen W.H., Lu G., Chen X., Zhao X.M., Bork P.. OGEE v2: an update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res. 2017; 45:D940–D944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sayers E.W., Beck J., Brister J.R., Bolton E.E., Canese K., Comeau D.C., Funk K., Ketter A., Kim S., Kimchi A. et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2020; 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thurmond J., Goodman J.L., Strelets V.B., Attrill H., Gramates L.S., Marygold S.J., Matthews B.B., Millburn G., Antonazzo G., Trovisco V. et al.. FlyBase 2.0: The next generation. Nucleic Acids Res. 2019; 47:D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harris T.W., Arnaboldi V., Cain S., Chan J., Chen W.J., Cho J., Davis P., Gao S., Grove C.A., Kishore R. et al.. WormBase: a modern Model Organism Information Resource. Nucleic Acids Res. 2020; 48:D762–D767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muñoz-Fuentes V., Cacheiro P., Meehan T.F., Aguilar-Pimentel J.A., Brown S.D.M., Flenniken A.M., Flicek P., Galli A., Mashhadi H.H., Hrabě de Angelis M. et al.. The International Mouse Phenotyping Consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv. Genet. 2018; 19:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bult C.J., Blake J.A., Smith C.L., Kadin J.A., Richardson J.E., Anagnostopoulos A., Asabor R., Baldarelli R.M., Beal J.S., Bello S.M. et al.. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019; 47:D801–D806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R. et al.. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012; 40:D700–D705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berardini T.Z., Reiser L., Li D., Mezheritsky Y., Muller R., Strait E., Huala E.. The arabidopsis information resource: making and mining the ‘gold standard’ annotated reference plant genome. Genesis. 2015; 53:474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cerqueira G.C., Arnaud M.B., Inglis D.O., Skrzypek M.S., Binkley G., Simison M., Miyasato S.R., Binkley J., Orvis J., Shah P. et al.. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014; 42:D705–D710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruzicka L., Howe D.G., Ramachandran S., Toro S., Van Slyke C.E., Bradford Y.M., Eagle A., Fashena D., Frazer K., Kalita P. et al.. The zebrafish information network: new support for non-coding genes, richer gene ontology annotations and the alliance of genome resources. Nucleic Acids Res. 2019; 47:D867–D873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lock A., Rutherford K., Harris M.A., Hayles J., Oliver S.G., Bähler J., Wood V.. PomBase 2018: User-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 2019; 47:D821–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aurrecoechea C., Brestelli J., Brunk B.P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O.S. et al.. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009; 37:D539–D543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Harb O.S., Roos D.S.. ToxoDB: functional genomics resource for toxoplasma and related organisms. Methods in Molecular Biology. 2020; 2071:27–47. [DOI] [PubMed] [Google Scholar]
- 30. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X. et al.. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2009; 38:D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N. et al.. Identification of essential genes for cancer immunotherapy. Nature. 2017; 548:537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Behan F.M., Iorio F., Picco G., Gonçalves E., Beaver C.M., Migliardi G., Santos R., Rao Y., Sassi F., Pinnelli M. et al.. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature. 2019; 568:511–516. [DOI] [PubMed] [Google Scholar]
- 33. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M. et al.. Defining a cancer dependency map. Cell. 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morgens D.W., Deans R.M., Li A., Bassik M.C.. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016; 34:634–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen W.H., Trachana K., Lercher M.J., Bork P.. Younger genes are less likely to be essential than older genes, and duplicates are less likely to be essential than singletons of the same age. Mol. Biol. Evol. 2012; 29:1703–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Makino T., Hokamp K., McLysaght A.. The complex relationship of gene duplication and essentiality. Trends Genet. 2009; 25:152–155. [DOI] [PubMed] [Google Scholar]
- 37. Jeong H., Mason S.P., Barabási A.L., Oltvai Z.N.. Lethality and centrality in protein networks. Nature. 2001; 411:41–42. [DOI] [PubMed] [Google Scholar]
- 38. Emms D.M., Kelly S.. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P. et al.. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019; 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang H., Li W.H.. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007; 23:375–378. [DOI] [PubMed] [Google Scholar]
- 41. Bartha I., Di Iulio J., Venter J.C., Telenti A.. Human gene essentiality. Nat. Rev. Genet. 2018; 19:51–62. [DOI] [PubMed] [Google Scholar]
- 42. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al.. Gene ontology: tool for the unification of biology. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. et al.. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012; 483:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo F.B., Dong C., Hua H.L., Liu S., Luo H., Zhang H.W., Jin Y.T., Zhang K.Y.. Accurate prediction of human essential genes using only nucleotide composition and association information. Bioinformatics. 2017; 33:1758–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E. et al.. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., Barretina J., Gelfand E.T., Bielski C.M., Li H. et al.. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang T., Yu H., Hughes N.W., Liu B., Kendirli A., Klein K., Chen W.W., Lander E.S., Sabatini D.M.. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell. 2017; 168:890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L.. BLAST+: architecture and applications. BMC Bioinformatics. 2008; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are freely accessible to all academic users. This work is licensed under a Creative Commons Attribution 3.0 Unported License (CC BY 3.0). Users can download datasets from the ‘Download’ page. Individual datasets or combined datasets of individual species can be downloaded via the ‘Browse’ page.