Abstract
NONCODE (http://www.noncode.org/) is a comprehensive database of collection and annotation of noncoding RNAs, especially long non-coding RNAs (lncRNAs) in animals. NONCODEV6 is dedicated to providing the full scope of lncRNAs across plants and animals. The number of lncRNAs in NONCODEV6 has increased from 548 640 to 644 510 since the last update in 2017. The number of human lncRNAs has increased from 172 216 to 173 112. The number of mouse lncRNAs increased from 131 697 to 131 974. The number of plant lncRNAs is 94 697. The relationship between lncRNAs in human and cancer were updated with transcriptome sequencing profiles. Three important new features were also introduced in NONCODEV6: (i) updated human lncRNA-disease relationships, especially cancer; (ii) lncRNA annotations with tissue expression profiles and predicted function in five common plants; iii) lncRNAs conservation annotation at transcript level for 23 plant species. NONCODEV6 is accessible through http://www.noncode.org/.
INTRODUCTION
There are vast majority of transcribed sequences that do not encode proteins called non-coding RNAs (ncRNAs). Long non-coding RNAs (lncRNAs) are ncRNAs that are >200 nt in length (1–3). Many types of research showed that lncRNAs play key roles in different kinds of biological processes in animals, such as circuitry controlling pluripotency and differentiation, imprinting control, immune responses and chromosome dynamics (4–7). LncRNAs are also implicated in human diseases, with much attention focused on their involvement in cancer progression and development (8–9).
Of the articles retrieved from PubMed (10) about lncRNAs, we found that a large number of studies focused on lncRNA functions, especially the relationships between lncRNAs and cancers (11). It is increasingly evident that many genomic variations related to cancer reside inside regions that do not encode proteins. However, these regions are often transcribed into lncRNAs (12). The last version of NONCODE collected relationships between lncRNAs and various diseases through literature mining and mutation analyses from public genome-wide association study (GWAS) data (13). NONCODEV6 obtains relationships between lncRNAs and cancers from known databases like Lnc2Cancer (14) and literature mining. Recently, studies on transcriptome sequencing profiles revealed that lncRNAs may involve in cancer by interaction with cancer-related genes, such as RAS family (15). The recent application of next-generation sequencing to a growing number of cancer transcriptomes has indeed revealed thousands of lncRNAs whose aberrant expression is associated with different cancer types (16).
The numbers of lncRNAs found in plants are usually an order of magnitude lower than in animals, but they still constitute an important component of their transcriptomes (17). Since great importance has been attached to lncRNAs, large-scale identification and deep analysis on plant lncRNAs are in need (18). To this end, we took advantage of publicly available RNA-seq datasets for calculating their expression values and identifying homologs of lncRNAs. We then aimed to explore their potential functions (19).
In this update of NONCODE database, we collected high-confidence datasets of plant lncRNA and developed a pipeline to annotate lncRNAs in plants. The pipeline was applied to 23 plant species. NONCODEV6 provides information including sequence, genome position, CNIT score, conservation, tissue expression profiles and functional prediction for five common plant species. The goal of NONCODEV6 is to be a meeting point for lncRNA research in both plants and animals. NONCODEV6 is freely available at http://www.noncode.org/.
Data collection and processing
Similar to the former update, the current release incorporates data sources of the previous versions of NONCODE (13,20–24), public literature and other lncRNA databases. We used ‘ncrna’, ‘noncoding’, ‘non-coding’, ‘non-code’, ‘lncrna’ and ‘lincrna’ as keywords to retrieve literature related to lncRNAs in PubMed. We found 57 872 articles in the last 10 years about lncRNAs in plants and 51 771 new articles since 22 June 2017 (the last update of NONCODE) about lncRNAs in human and mice. The newly identified lncRNAs and their annotations were retrieved from the supplementary material or corresponding websites. The related lncRNA databases include Ensembl (25), RefSeq (26), lncRNAdb (27), LNCipedia (28), CANTATAdb (29), GREENC (30) and the old versions of NONCODE.
All of the collected data were processed through a standard pipeline, which includes the following steps:
Format normalization. All input data were processed into bed or gtf formats based on one assembly version. For example, TAIR 10 and TAIR 9 are two different assembly versions of Arabidopsis thaliana. All of the related data were converted into the TAIR 10 version.
Multi-source data combination. All of the normalized data files were combined using the Cuffcompare program in the Cufflinks suite (31).
Protein-coding RNA filtration. We filtered out protein-coding RNA using two methods. First, all RNAs were compared with the coding RNAs in RefSeq and Ensembl. Second, CNIT (Coding-NonCoding Identifying Tool) (32) was used to filter the RNAs and only the RNAs considered noncoding by CNIT were kept.
General information presentation. Location, exons, length, assembly sequence, source are listed in each transcript.
Expression profiles and functional prediction in plants. Corresponding information in five common plants out of 23 is shown. Their expression profiles were curated from multiple tissues. Detailed data sources were listed in Supplementary Table S1. Functions for lncRNAs were predicted based on their co-expressed coding genes (34).
Conservation analysis at transcript level. The conservation of plant lncRNAs analysis was conducted with BLAST (35,36). The E-value cut-off was 1e-10. Each transcript in a plant species was blasted against each transcript in other 22 plant species.
Web presentation. New web pages, especially for plants, were constructed in NONCODEV6. More annotation information has been updated (52).
Statistical analysis of NONCODEV6
NONCODEV6 contains 644 510 lncRNA transcripts from 39 species including 16 animals and 23 plants. A total of 96 411 and 87 890 genes were generated from 173 112 and 131 974 transcripts in human and mouse transcripts, respectively. Corresponding expression profiles and predicted functions were provided.
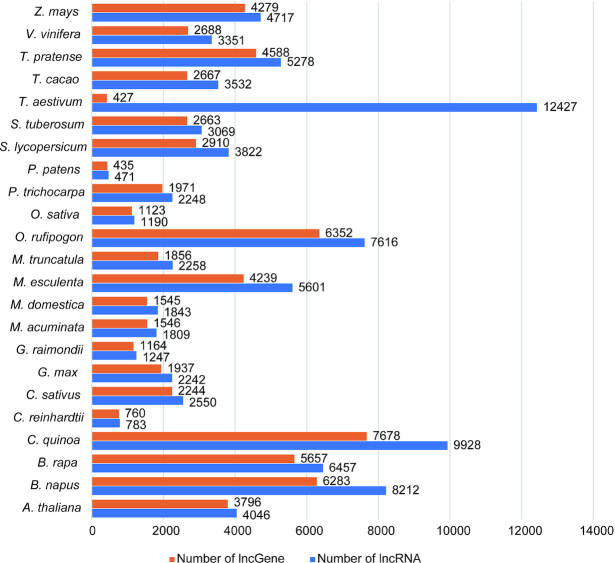
A total of 94 697 lncRNAs from 23 plant species (A. thaliana, Cucumis sativus, Brassica napus, Brassica rapa, Chenopodium quinoa, Chlamydomonas reinhardtii, Glycine max, Gossypium raimondii, Malus domestica, Manihot esculenta, Medicago truncatula, Musa acuminata, Oryza rufipogon, Oryza sativa Japonica Group, Physcomitrella patens, Populus trichocarpa, Solanum lycopersicum, Solanum tuberosum, Triticum aestivum, Theobroma cacao, Trifolium pratense, Vitis vinifera, Zea mays) are included. The number of lncRNAs and long non-coding RNA genes (lncGenes) in plants are shown in Figure 1. The total number of plant lncGenes was 68 808. LncRNAs in plants were annotated using strict standards for assurance of high confidence. NONCODEV6 follows the nomenclature of the last version (13). Both lncRNA transcripts and genes are designated systematically: NON + three characters (representing a species) + T (transcript) or G (gene) + six sequential numbers.
Figure 1.
Summary of long non-coding RNAs collected in 23 plant species from literature and databases.
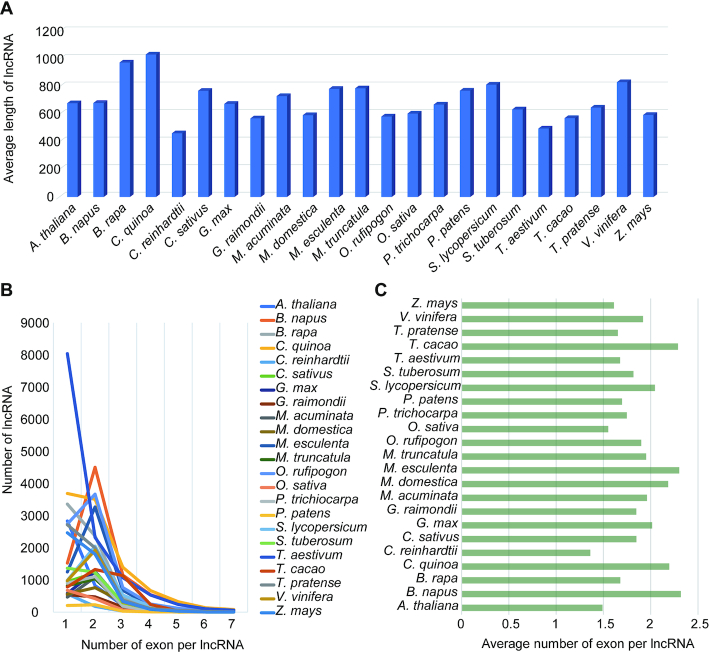
The addition of lncRNAs in plants is a very important update in NONCODEV6. Figure 2 shows the distribution of lncRNAs in 23 plant species. In Figure 2A, the average length of lncRNAs in plants ranged from 462 bp of C. reinhardtii to 1 033 bp of C. quinoa. Most lncRNAs in plants have two or three exons and the distribution of exon number was shown in Figure 2B. The average number of exons per lncRNA in plants ranged from 1.3 of C. reinhardtii to 2.3 of B. napus in Figure 2C.
Figure 2.
Distribution of lncRNAs in 23 plant species
LncRNAs in human and mouse
We used a set of keywords to search in PubMed and proceeded data collection from other databases. After data collection and filtration, the number of lncRNAs in human increased from 172 216 to 173 112. The number of lncRNAs in mouse increased from 131 697 to 131 974. LncRNAs in human and mouse were updated using the last version of NONCODE pipeline.
Recently, researches have been focusing on the relationships between lncRNAs and cancers in human. There is a strong need to collect high-quality lncRNA profiles in cancers, which will help to explore the function and mechanism of lncRNAs in cancer. We summarized lncRNA-cancer associated information from related databases and literature mining. Information from six databases (LncSpA (37), LncTarD (38), Lnc2Cancer (14), LncRNADisease (39), LncRNAWiki (40) and MNDR (41)) was collected. Lnc2Cancer is a manually curated database that provides comprehensive experimentally supported associations between lncRNA and human cancer. LncSpA is a lncRNA spatial atlas of expression across normal and cancer tissues. LncTarD provides key lncRNA-target regulations, their functions and lncRNA-mediated regulatory mechanisms in human diseases.
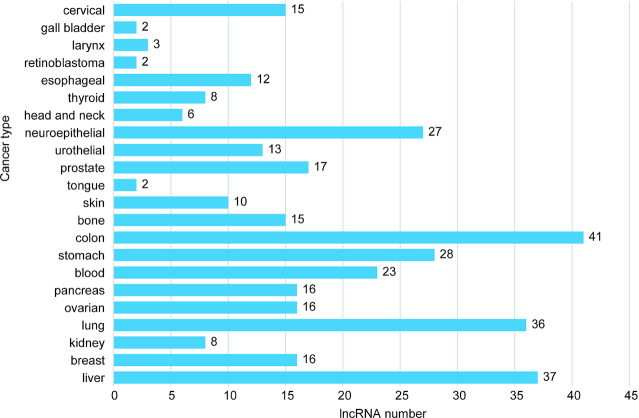
LncRNA-disease associations predicted by computational methods are not included in NONCODEV6 due to the uncertainty; only experimentally supported relationships are integrated into NONCODEV6. In summary, we obtained 13 749 records of lncRNA and disease related information. The basic statistics about lncRNAs related to different cancer types were shown in Figure 3.
Figure 3.
The number of functional lncRNAs in top 20 cancers.
Plant lncRNAs expression profiles in tissues
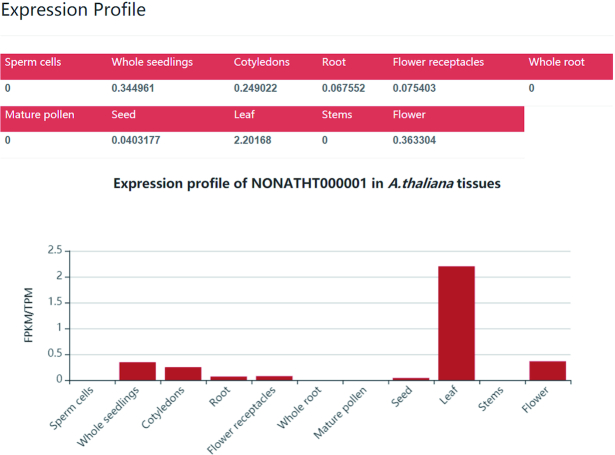
RNA-seq data of different tissues in five common plants were collected, including A. thaliana, Z. mays, S. lycopersicum, C. sativus and O. sativa (Table 1). Tissue expression profiles were analyzed using STAR (42) and StringTie (43). Clean reads were mapped to the corresponding genome by STAR with the parameters ‘--outSAMtype BAM SortedByCoordinate --outSAMattributes All’. Then, the expression levels of all lncRNAs and mRNAs were quantified as transcripts per kilobase million (TPM) using StringTie. The detailed expression information can be queried by searching for specific lncRNAs in NONCODEV6 (Figure 4).
Table 1.
The tissue RNA-seq data used in NONCODEV6
| Species | Tissue | No. of tissuesa |
|---|---|---|
| Arabidopsis thaliana | flower receptacles, flower, root, whole seedlings, cotyledons, leaf, stems, sperm cells, mature pollen, seed | 10 |
| Cucumis sativus | ovary, root, stem, leaf, male flower, tendril, tendril base, female flower | 8 |
| Solanum lycopersicum | stem, floral, leaf, root, vegetative meristem, seedling | 6 |
| Zea mays | seedling, root, shoot, ovule, pollen, embryo sac, leaf, immature tassel, endosperm | 9 |
| Oryza sativa | shoot, caryopsis, crown root, egg, embryonic root, endosperm, lateral root, leaves, pistil, seedling root, unicellular zygote | 11 |
a Detailed data sources were listed in Supplementary Table S1.
Figure 4.
An example of tissue expression profiles for one lncRNA of A.thaliana with id NONATHT000001.1.
Plant lncRNAs and function annotation
Functional annotation analysis was conducted on five plants, including A. thaliana, Z. mays, S. lycopersicum, C. sativus and O. sativa. To investigate the potential functions of lncRNAs, the RNA-seq data collected from different tissues were used to analyze the co-expression between coding genes and lncRNAs.
Genes with the expression of variance ranked in the top 75% were retained, then the expression was used to calculated Pearson's correlation coefficient (Pcc) and P-value of Pcc for each pair of retained genes using WGCNA (34) package of R. Gene pairs with P-value <0.05 and with Pcc >0.999 or <−0.999 were considered to be co-expressed (33). To predict the potential functions of lncRNAs in five plants, GO (49) annotation was performed by the co-expressed coding genes of lncRNAs using online tools in PANTHER classification system (45). Users can obtain the co-expressed coding genes and related GO (50) terms by searching lncRNAs.
Plant lncRNAs conservation at transcript level
NONCODE provided the human lncRNA conservation analysis in previous versions. Therefore, when we added lncRNA annotation for plant species in NONCODEV6, plant lncRNA conservation analysis was provided, as well. With sequence homology and conservation accepted as indicators of biological function, more attention has been paid to understanding the evolutionary dynamics of lncRNAs. The evolutionary history of lncRNAs can provide insights into their functionality (44), but the absence of lncRNA annotations in plants has precluded comparative analyses. To understand the biological significance and evolution of lncRNAs in plants, we conducted transcript level assessments of the conservation of lncRNAs across the 23 plant species (46). To assess the primary sequence conservation of the plant lncRNAs, we performed a multi-species comparison. The lncRNA sequences of each plant species were aligned against the other 22 plant species by BLASTn (35) with parameter –E-value 1e−10 (47). The genomes of all plant species were downloaded from plant Ensembl and GenBank database (48). Our study provides the first step to understand the evolutionary conservation of lncRNAs in plants for further functional studies.
We calculated the lncRNA sequence similarity at the transcript level to facilitate lncRNAs study across species. Pair-wise conservation comparisons of lncRNAs in 23 plant species were conducted by BLAST. Same as previous plant conservation study (35), reciprocal best hits for each pair were extracted with a matched query proportion ≥50% and E-value ≤1e−10. Only 122 orthologous lncRNAs were found to meet the thresholds. Most of the plant species show limited conserved orthologs at transcript level filtered by E-value and coverage. Three plant pairs have more orthologous lncRNAs at the transcript level, including B. napus and B. rapa; O. rufipogon and O. sativa; S. lycopersicum and S. tuberosum. Generally speaking, there is low sequence conservation among the majority of lncRNAs at the transcript level. The cut-off values can be customized according to users’ specific needs, and the output results could be further investigated at genome or other level analysis.
DISCUSSION
Determining the protein-coding ability of a transcript is a critical part of the identification of lncRNAs, yet it represents quite a challenging task. CNIT, as a newly updated version of CNCI (51), shows 99.3% accuracy when tested on plant protein-coding and long non-coding transcripts (29). CNIT can provide the confidence of lncRNAs noncoding status. The standard for lncRNAs in NONCODEV6 is very high compared with other methods.
The focus has shifted from novel lncRNAs detection to more in-depth research on lncRNAs including function and multiple annotations, along with the progress of related studies. Therefore, in the update of human lncRNA, we focused on the relationship between lncRNAs and cancer. NONCODEV6 is a comprehensive database dedicated to lncRNA annotation in both animals and plants. We provide complete annotations of lncRNAs and detailed information, including function prediction and expression profiles.
For plants, there are not comprehensive tissue RNA-seq data due to difficulties in genome alignment and scarce annotation data for lncRNAs. In consideration of the limited resources, tissue RNA-seq data were collected for only five plant species. Data from other species still needs to be collected. We will follow up with the latest released datasets to enrich the annotation of lncRNAs in plants continuously. The lncRNA conservation analysis in plants was difficult without comprehensive construction tools and datasets. In the future, we will add more levels of analysis for plant lncRNAs conservation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Heng Zhou for help with the server adjustment of NONCODEV6.
Contributor Information
Lianhe Zhao, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jiajia Wang, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yanyan Li, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Tingrui Song, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Yang Wu, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Shuangsang Fang, Beijing University of Chinese Medicine, Chaoyang District, Beijing 100029, China.
Dechao Bu, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Hui Li, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Liang Sun, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Dong Pei, State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry, Beijing 100091, China.
Yu Zheng, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jianqin Huang, Nurturing Station for the State Key Laboratory of Subtropical Silviculture, Zhejiang A & F University, Lin’an, Hangzhou 311300, China.
Mingqing Xu, Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Shanghai Jiao Tong University, Shanghai 518102, China.
Runsheng Chen, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; Guangdong Geneway Decoding Bio-Tech Co. Ltd, Foshan 528316, China; National Genomics Data Center, Chinese Academy of Sciences, Beijing 100101, China.
Yi Zhao, Key Laboratory of Intelligent Information Processing, Advanced Computer Research Center, Institute of Computing Technology, Chinese Academy of Sciences, Beijing 100190, China.
Shunmin He, Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China; National Genomics Data Center, Chinese Academy of Sciences, Beijing 100101, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2018YFD1000604, 2016YFC0901702, 2019YFC1709801, 2018YFC1313000, 2018YFC1313001]; National Natural Science Foundation of China [31672126, 91940306, 31871294, 91740113, 32070670, 31871294]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDA12030100 and XDB38040300]; Natural Science Foundation for Young Scholars of China [31701141]; Innovation Project for Institute of Computing Technology, CAS [20186060]; China Postdoctoral Science Foundation [2019M660033]; China Postdoctoral Innovative Talent Foundation [BX20200068]; 13th Five-year Informatization Plan of Chinese Academy of Sciences [XXH13505-05-02]; Science and Technology Service Program of the Chinese Academy of Sciences [KFJ-STS-ZDTP-060]. Funding for open access charge: National Key Research and Development Program [2016YFC0901702].
Conflict of interest statement. None declared.
REFERENCES
- 1. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponting C.P., Oliver P.L., Reik W.. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–641. [DOI] [PubMed] [Google Scholar]
- 3. Pennisi E. Shining a light on the genome's ‘dark matter’. Science (New York, N.Y.). 2010; 330:1614. [DOI] [PubMed] [Google Scholar]
- 4. Chen J., Lan J., Ye Z., Duan S., Hu Y., Zou Y., Zhou J.. Long noncoding RNA LRRC75A-AS1 inhibits cell proliferation and migration in colorectal carcinoma. Exp. Biol. Med. (Maywood). 2019; 244:1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin Y., Xu L., Wei W., Zhang X., Ying R.. Long noncoding RNA H19 in digestive system cancers: a meta-analysis of its association with pathological features. Biomed. Res. Int. 2016; 2016:4863609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sallam T., Sandhu J., Tontonoz P.. Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ. Res. 2018; 122:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Y., Zhu P., Wang J., Zhu X., Luo J., Meng S., Wu J., Ye B., He L., Du Y. et al.. Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J. Hepatol. 2018; 69:861–872. [DOI] [PubMed] [Google Scholar]
- 8. Romero-Barrios N., Legascue M.F., Benhamed M., Ariel F., Crespi M.. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018; 46:2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorenzi L., Avila Cobos F., Decock A., Everaert C., Helsmoortel H., Lefever S., Verboom K., Volders P.J., Speleman F., Vandesompele J. et al.. Long noncoding RNA expression profiling in cancer: challenges and opportunities. Genes Chromosomes Cancer. 2019; 58:191–199. [DOI] [PubMed] [Google Scholar]
- 10. NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 2018; 46:D8–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang J., Wei X., Liu Z., Cao D., Tang Y., Zou Z., Zhou C., Lu Y.. Long noncoding RNA CYTOR in cancer: A TCGA data review. Clin. Chim. Acta. 2018; 483:227–233. [DOI] [PubMed] [Google Scholar]
- 12. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 13. Fang S., Zhang L., Guo J., Niu Y., Wu Y., Li H., Zhao L., Li X., Teng X., Sun X. et al.. NONCODEV5: a comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018; 46:D308–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao Y., Wang P., Wang Y., Ma X., Zhi H., Zhou D., Li X., Fang Y., Shen W., Xu Y. et al.. Lnc2Cancer v2.0: updated database of experimentally supported long non-coding RNAs in human cancers. Nucleic Acids Res. 2019; 47:D1028–D1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jia Y., Shi L., Yun F., Liu X., Chen Y., Wang M., Chen C., Ren Y., Bao Y., Wang L.. Transcriptome sequencing profiles reveal lncRNAs may involve in breast cancer (ER/PR positive type) by interaction with RAS associated genes. Pathol. Res. Pract. 2019; 215:152405. [DOI] [PubMed] [Google Scholar]
- 16. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 17. Wang H.V., Chekanova J.A.. Long noncoding RNAs in Plants. Adv. Exp. Med. Biol. 2017; 1008:133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun X., Zheng H., Sui N.. Regulation mechanism of long non-coding RNA in plant response to stress. Biochem. Biophys. Res. Commun. 2018; 503:402–407. [DOI] [PubMed] [Google Scholar]
- 19. Guo X., Gao L., Liao Q., Xiao H., Ma X., Yang X., Luo H., Zhao G., Bu D., Jiao F., Shao Q., Chen R., Zhao Y.. Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucleic Acids Res. 2013; 41:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q. et al.. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016; 44:D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y.. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014; 42:D98–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bu D., Yu K., Sun S., Xie C., Skogerbø G., Miao R., Xiao H., Liao Q., Luo H., Zhao G. et al.. NONCODE v3.0: integrative annotation of long noncoding RNAs. Nucleic Acids Res. 2012; 40:D210–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He S., Liu C., Skogerbø G., Zhao H., Wang J., Liu T., Bai B., Zhao Y., Chen R.. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008; 36:D170–D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu C., Bai B., Skogerbø G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R.. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005; 33:D112–D115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Howe K.L., Contreras-Moreira B., De Silva N., Maslen G., Akanni W., Allen J., Alvarez-Jarreta J., Barba M., Bolser D.M., Cambell L. et al.. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020; 48:D689–D695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R. et al.. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quek X.C., Thomson D.W., Maag J.L., Bartonicek N., Signal B., Clark M.B., Gloss B.S., Dinger M.E.. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015; 43:D168–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Volders P.J., Anckaert J., Verheggen K., Nuytens J., Martens L., Mestdagh P., Vandesompele J.. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019; 47:D135–D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szcześniak M.W., Bryzghalov O., Ciomborowska-Basheer J., Makałowska I.. CANTATAdb 2.0: expanding the collection of plant long noncoding RNAs. Methods Mol. Biol. 2019; 1933:415–429. [DOI] [PubMed] [Google Scholar]
- 30. Paytuví Gallart A., Hermoso Pulido A., Anzar Martínez de Lagrán I., Sanseverino W., Aiese Cigliano R.. GREENC: a Wiki-based database of plant lncRNAs. Nucleic Acids Res. 2016; 44:D1161–D1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghosh S., Chan C.K.. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016; 1374:339–361. [DOI] [PubMed] [Google Scholar]
- 32. Guo J.C., Fang S.S., Wu Y., Zhang J.H., Chen Y., Liu J., Wu B., Wu J.R., Li E.M., Xu L.Y. et al.. CNIT: a fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019; 47:W516–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liao Q., Liu C., Yuan X., Kang S., Miao R., Xiao H., Zhao G., Luo H., Bu D., Zhao H. et al.. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011; 39:3864–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deng P., Liu S., Nie X., Weining S., Wu L.. Conservation analysis of long non-coding RNAs in plants. Science China. Life Sciences. 2018; 61:190–198. [DOI] [PubMed] [Google Scholar]
- 36. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 37. Lv D., Xu K., Jin X., Li J., Shi Y., Zhang M., Jin X., Li Y., Xu J., Li X.. LncSpA: LncRNA spatial atlas of expression across normal and cancer tissues. Cancer Res. 2020; 80:2067–2071. [DOI] [PubMed] [Google Scholar]
- 38. Zhao H., Shi J., Zhang Y., Xie A., Yu L., Zhang C., Lei J., Xu H., Leng Z., Li T. et al.. LncTarD: a manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2020; 48:D118–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D.. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019; 47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cui T., Zhang L., Huang Y., Yi Y., Tan P., Zhao Y., Hu Y., Xu L., Li E., Wang D.. MNDR v2.0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018; 46:D371–D374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015; 33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Necsulea A., Soumillon M., Warnefors M., Liechti A., Daish T., Zeller U., Baker J.C., Grützner F., Kaessmann H.. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014; 505:635–640. [DOI] [PubMed] [Google Scholar]
- 45. Mi H., Muruganujan A., Huang X., Ebert D., Mills C., Guo X., Thomas P.D.. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 2019; 14:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ponjavic J., Ponting C.P., Lunter G.. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007; 17:556–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng P., Liu S., Nie X., Weining S., Wu L.. Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2018; 61:190–198. [DOI] [PubMed] [Google Scholar]
- 48. Benson D.A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Ostell J., Pruitt K.D., Sayers E.W.. GenBank. Nucleic Acids Res. 2018; 46:D41–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. (2019); 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. et al.. Gene ontology: tool for the unification of biology. Nat Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun L., Luo H., Bu D., Zhao G., Yu K., Zhang C., Liu Y., Chen R., Zhao Y.. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013; 41:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liao Q., Xiao H., Bu D., Xie C., Miao R., Luo H., Zhao G., Yu K., Zhao H., Skogerbø G. et al.. ncFANs: a web server for functional annotation of long non-coding RNAs. Nucleic Acids Res. 2011; 39:W118–W124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.