Virus-carrying animal hosts
The coronavirus disease 2019 (COVID-19) pandemic is becoming the greatest public health crisis since the influenza pandemic in 1918 (Sanders et al. 2020). Indeed, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread to 219 countries and regions, causing over 46.5 million human infections including more than 1.2 million deaths as of November 3, 2020 (WHO 2020). So far, research has focused on its transmission from animals to humans—‘zoonosis’—to explain its spillover to humans, yet its further transmission from humans to animals—‘anthroponosis’ or ‘reverse zoonosis’—has been much less considered. Knowledge concerning the relationships between its human and animal hosts is still limited, and a consensus has yet to be reached on the animal origin of COVID-19. It is known that SARS-CoV-2 shares 79.6% of its genome sequences with SARS-CoV-1, a strain that has infected humans, bats and palm civets in 2003. Also, SARS-CoV-2 shares 96.2% of its sequences with a bat coronavirus isolated from Rhinolophus affinis, suggesting that bats may be the natural hosts of SARS-CoV-2 (Lu et al. 2020; Paraskevis et al. 2020; Zhou et al. 2020). Cats, ferrets, minks, pangolins, snakes and turtles have been postulated as intermediate hosts of the novel coronavirus (Ji et al. 2020; Li et al. 2020; Nabi et al. 2020; Wu et al. 2020). Here we show that COVID-19 transmission from humans to animals is likely to amplify mutations and, in turn, to re-infect humans with deadlier mutants.
Infectable vertebrates
The infection of SARS-CoV-2 is induced by interactions between the SARS-CoV-2 receptor-binding domain and angiotensin-converting enzyme 2 (ACE2) receptor proteins (Santini and Edwards 2020; Wu et al. 2020; Zhou et al. 2020). As a consequence, a diverse range of vertebrates can be potentially infected by SARS-CoV-2 via their ACE2 receptors. This assumption is supported by the findings of ACE2 receptors in over 400 vertebrate species including both domestic and wild animals (Damas et al. 2020; Wu et al. 2020). SARS-CoV-2 infections have been recently confirmed in mammals including cats, monkeys, ferrets and hamsters (Santini and Edwards 2020). Studies have also shown that infected cats, ferrets and hamsters were capable of spreading SARS-CoV-2 to other animals in laboratory settings (Kim et al. 2020; Shi et al. 2020; Sia et al. 2020). Overall, contamination and cross-infection of vertebrate species are likely.
Transmission via human-animal contact
About 70% of emerging infectious diseases and almost all recent pandemics caused by zoonoses are correlated with aggressive land reclamation, intensified livestock production, increased wildlife hunting and trading activities by humans (Di Marco et al. 2020). Conversely, regular or inadvertent interactions between infected persons and wild or domestic animals allow COVID-19 to be transmitted in the reverse direction. Anthroponosis is more likely to occur in areas with high prevalence of human infection and co-existing wild habitants, e.g., in city suburbs. This hypothesis is supported by recent incidents and studies. For instance, domestic cats and dogs owned by infected individuals have been tested positive for SARS-CoV-2, some showing no specific symptoms (Santini and Edwards 2020; Sit et al. 2020). Following an incident at the Bronx Zoo in New York City, direct human-to-animal transmission has been proposed from the epidemiological data and genetic similarities of SARS-CoV-2 strains isolated from tigers and their keepers (McAloose et al. 2020). Minks infected with SARS-CoV-2 have also been identified on farms in the Netherlands, Denmark, Spain and the USA, where infected farm workers were speculated as sources of infection (CDC 2020; Oreshkova et al. 2020). Human infection acquired from infected minks, i.e., reverse anthroponosis, was also suspected on two mink farms in the Netherlands reporting human-to-mink transmission (Oreshkova et al. 2020). Converging evidence thus points to possible underestimated transmission from humans to animals and further spillover back to humans.
Anthropogenic wastes as secondary routes
Apart from direct human-to-animal transmission, e.g., by physical contact with domestic and wild animals, transmission to animals via human wastes is also likely. Potentially, animals may come into contact with wastes from infected households that contain infectious human biological matter, e.g., nasal discharge, phlegm, saliva, semen, vaginal fluids, blood, urine and feces (Franklin and Bevins 2020; Han and He 2020; Nabi et al. 2020). The main route is probably water contamination due to the enteric propagation of the virus. Also, improper disposal of contaminated personal protective equipment constitutes a source of COVID-19 infection for wild and stray animals (Fadare and Okoffo 2020; Kalina and Tilley 2020). Particularly, masks, face coverings, gloves, tissues and wipes are routinely used in response to the current pandemic, creating enormous amounts of single-use items that could be potentially infected with the virus (UNEP 2020). When not managed timely and soundly, these are subject to open dumping, which pose risks of infection for domestic and wild animals in surrounding environments. This is supported by the fact that SARS-CoV-2 could survive for several days on face masks, tissues, and other household materials under common conditions (Chin et al. 2020; Han et al. 2020; van Doremalen et al. 2020). In addition, water contamination is possible due to the enteric propagation of the virus and substantial viral loadings found in human excreta, especially in communities with unimproved sanitary facilities or recurring combined sewage overflows (Han and He 2020; Sun and Han 2020).
Mutations and gene mayonnaise of SARS-CoV-2
The genetic diversity of coronaviruses is caused by accumulation of mutations and high-frequency homologous recombination, which favor tissue tropism, breaking of interspecies barriers and adapting to ecological variations (Amer 2018). There is also a form of template switching similar to homologous recombination, although the mechanism is different. The in-host genetic diversity and host-switching events could incite the virus to adapt to a wider array of selective pressures, ultimately inducing different diseases and symptoms in hosts (Borucki et al. 2013).
Members of the coronaviridae family, e.g., SARS and the Middle East respiratory syndrome (MERS)-related coronavirus, have been shown to cross the interspecies barrier (Amer 2018). Graham and Baric (2010) found that the recombination of SARS in the spike glycoprotein genes might have mediated the initial cross-species transmission event from bats to other mammals. Moreover, Ji et al. (2020) pointed out that a homologous recombination of SARS-CoV-2 may occur between a bat coronavirus and a coronavirus of an unknown origin within the viral spike glycoprotein gene. This study found that SARS-CoV-2 has highly similar genetic information with bat coronavirus. Lau et al. (2020) further showed that the entire genome of SARS-CoV-2 is most closely related to the SARSr-Ra-BatCoV RaTG13 from an intermediate horseshoe bat, except for its receptor-binding domain which is closest to pangolin-SARSr-CoVs, suggesting that SARS-CoV-2 is a recombinant virus. Overall, these findings indicate the complex origin of SARS-CoV-2, where evolutionary recombination and strong purifying selection occurred between strains from distinct host species before it spilled over to humans (Li et al. 2020; Wu et al. 2020; Sallard et al. 2020). The back-and-forth transmission between human and wild species, when it occurs, may further enhance the genetic diversity of the virus.
Lessons from flu vaccines
Reverse zoonosis of SARS-CoV-2 may seed unrestrained spread and mutations in infectable wild species and, perhaps at some stage, transmission of novel strains back to humans. Without human intervention, the dense roosting behavior and long foraging range of some wild species may facilitate exchange of viruses and recombination. Lau et al. (2010) found that recombination events occurred between strains of Rousettus bat coronavirus Ro-BatCoV HKU9 from different bat individuals, which might have allowed the same bat to be infected by at least two distinct genotypes. Likewise, different species coinfected with influenza A genotypes potentially facilitated genetic reassortment of the virus, for instance, between human and avian virus strains. These activities can create new genotypes with substantial antigenic changes that can result in an influenza pandemic and produce a virulent strain (Carrat et al. 2007). It should be noted that genetic recombination is a known contributor to major shifts in influenza antigenicity. As a consequence, the influenza season vaccine becomes less effective due to genetic shift by mutations of the virus with time (Carrat et al. 2007).
A meta-analysis of data in 30 selected publications showed that, between 2010–2011 and 2014–2015, the pooled seasonal influenza vaccine effectiveness was 51% for any influenza among people aged 18–64 and 37% among those 65 and older (Rondy et al. 2020). Notably, the study found that vaccine effectiveness against influenza A (H3N2) was 43% among persons aged ≥ 65 in seasons when circulating and vaccine strains were antigenically similar, but only 14% in seasons when variant viruses of influenza A (H3N2) predominated (Rondy et al. 2020). In contrast, the effectiveness of human vaccines against bacterial pathogens is generally much higher, given the low mutation rates of the latter. In a study enrolling 782 cases and 2,512 controls, Whitney et al (2006) found that the effectiveness of seven-valent pneumococcal conjugate vaccine (PCV-7) for bacteraemic pneumonia was 98% in healthy children. We hypothesize that similar risks may exist in the current and future development of vaccines against the novel coronavirus. Once SARS-CoV-2 spills from human to wildlife, the spontaneous behaviors of wild species could facilitate not only the establishment of the virus within the animal group and the transmission to other species, but the mutation of the virus itself (Keesing et al. 2010). Foraging, predation, mating and open defecation enable the spread of pathogens carried by infected animals to others in new terrestrial and aquatic environments, causing clustered and cross-species infections, while the virus continuously evolves in different wildlife reservoirs.
Deadly reinfection
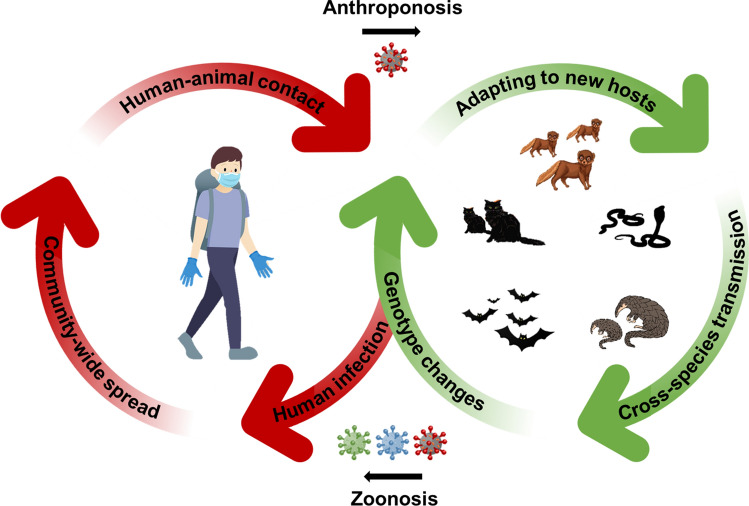
It is commonly accepted that, once already infected then cured, an individual is better protected against the infectious agent, yet this might not be true in some cases of rapidly mutating viruses such as COVID-19. At least seven cases of SARS-CoV-2 reinfections have been reported since the first case was reported in Hong Kong on August 24, 2020 (Larson et al. 2020; Mulder et al. 2020; Prado-Viviat et al. 2020; Tillett et al. 2020; To et al. 2020; Torres et al. 2020; Van Elslande et al. 2020). Remarkably, in four cases, patients showed more severe symptoms than previous symptoms from their first infection, including one severe case where the individual died shortly after the reinfection. In all seven cases, viral strains isolated from the reinfected individuals showed substantial genetic differences from the initial strains causing their first infection. Less severe secondary infections have also been reported. The emerging cases of SARS-CoV-2 reinfections are likely to be caused by mutations and continuing evolution of the novel coronavirus. There are risks that novel strains of SARS-CoV-2 may emerge in wildlife reservoirs by accumulating point mutations and homologous recombination as it spreads in an uncontrolled manner between different hosts and species. Reverse anthroponosis of novel strains, once it occurs, may render current COVID-19 vaccines less effective (Dai et al. 2020). The virus may spill back and forth between humans and animals, thus forming a vicious loop with prolonged spread, recurrent infections and outbreaks, and continuing fights with evolved viral strains (Fig. 1).
Fig. 1.
Virus transmission from wildlife to human (zoonosis), human to wildlife (reverse zoonosis or anthroponosis) and then back to human (reverse anthroponosis). Back-and-forth transmission of SARS-CoV-2 between humans and animals is likely to induce more mutations and adaptation of the virus, leading to novel strains that could be deadlier than original strains. Spontaneous behaviors of wild species such as foraging, predation, mating and defecating enable both in-species and cross-species transmission, facilitating genotype changes and evolution of SARS-CoV-2 in various wildlife reservoirs. Mutated viruses may then spillover to humans again through different natural or intermediate hosts, forming a dangerous loop of zoonosis, anthroponosis and reverse anthroponosis of an evolving virus between human and animal hosts. Once transmitted to humans, those novel strains will pose significant challenges for infection control by causing reinfections, re-emergent outbreaks and rendering current vaccines less effective
Early intervention could be implemented to minimize the risks of forming a loop of zoonosis, anthroponosis and reverse anthroponosis during the current pandemic. Since mid-October 2020, Denmark, the world’s largest producer of mink pelts, has opted to cull 2.5 million minks in the coming months as a precautionary measure to control the spread of COVID-19 in minks and farm workers (Olsen and Scislowska 2020). A series of control measures may be put in place, such as: (1) quarantining and safely disposing of domesticated animals or livestock with suspected SARS-CoV-2 infection; (2) advising the public to keep safe distances from wildlife, particularly species that are known to be susceptible to SARS-CoV-2 infections, for instance, by issuing emergency orders and temporary bans on hunting, trading and other non-essential contact with wildlife; and (3) monitoring novel strains of SARS-CoV-2, including those emerged in animal hosts, and continually developing vaccines for mutated virus strains.
Acknowledgement
This work was funded by the 'Young Talent Support Plan' of Xi’an Jiaotong University. We thank Prof. Etienne Decroly, Aix-Marseille University, for advices on the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Amer HM. Bovine-like coronaviruses in domestic and wild ruminants. Anim Health Res Rev. 2018;9:113–124. doi: 10.1017/S1466252318000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki MK, Allen JE, Chen-Harris H, Zemla A, Vanier G, Mabery S, Torres C, Hullinger P, Slezak T. The role of viral population diversity in adaptation of Bovine Coronavirus to new host environments. PLoS ONE. 2013;8(1):e52752. doi: 10.1371/journal.pone.0052752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25(39–40):6852–6862. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- CDC (Centers for Diseases Control and Prevention) (2020) Animals & COVID-19. https://www.cdc.gov/coronavirus/2019-ncov/daily-life-coping/animals.html. Accessed 3 Nov 2020
- Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen H, Chan MCW, Peiris M, Poon LLM. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1(1):e10. doi: 10.1016/s2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H, Han J, Lichtfouse E. Who is running faster, the virus or the vaccine? Environ Chem Lett. 2020;18:1761–1766. doi: 10.1007/s10311-020-01110-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA. 2020;117(36):22311–22322. doi: 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco M, Baker ML, Daszak P, De Barro P, Eskew EA, Godde CM, Harwood TD, Herrero M, Hoskins AJ, Johnson E, Karesh WB, Machalaba C, Garcia JN, Paini D, Pirzl R, Smith MS, Zambrana-Torrelio C, Ferrier S. Opinion: sustainable development must account for pandemic risk. Proc Natl Acad Sci USA. 2020;117(8):3888–3892. doi: 10.1073/pnas.2001655117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadare OO, Okoffo ED. Covid-19 face masks: A potential source of microplastic fibers in the environment. Sci Total Environ. 2020;737:140279. doi: 10.1016/j.scitotenv.2020.140279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AB, Bevins SN. Spillover of SARS-CoV-2 into novel wild hosts in North America: a conceptual model for perpetuation of the pathogen. Sci Total Environ. 2020;733:139358. doi: 10.1016/j.scitotenv.2020.139358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84(7):3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, He S. Urban flooding events pose risks of virus spread during the novel coronavirus (COVID-19) pandemic. Sci Total Environ. 2020;755:142491. doi: 10.1016/j.scitotenv.2020.142491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang X, He S, Jia P. Can the coronavirus disease be transmitted from food? A review of evidence, risks, policies and knowledge gaps. Environ Chem Lett. 2020 doi: 10.1007/s10311-020-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalina M, Tilley E. "This is our next problem": Cleaning up from the COVID-19 response. Waste Manage. 2020;108:202–205. doi: 10.1016/j.wasman.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, Hudson P, Jolles A, Jones KE, Mitchell CE, Myers SS, Bogich T, Ostfeld RS. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Kim S, Kim S, Kim E, Park S, Yu K, Chang J, Kim EJ, Lee S, Casel MAB, Um J, Song M, Jeong HW, Lai VD, Kim Y, Chin BS, Park J, Chung K, Foo S, Poo H, Mo I, Lee O, Webby RJ, Jung JU, Choi YK. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27(5):704–709.e2. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson D, Brodniak SL, Voegtly LJ, Cer RZ, Glang LA, Malagon FJ, Long KA, Potocki R, Smith DR, Lanteri C, Burgess T, Bishop-Lilly KA. A case of early re-infection with SARS-CoV-2. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SKP, Poon RWS, Wong BHL, Wang M, Huang Y, Xu H, Guo R, Li KSM, Gao K, Chan K, Zheng B, Woo PCY, Yuen K. Coexistence of different genotypes in the same bat and serological characterization of Rousettus Bat Coronavirus HKU9 belonging to a novel betacoronavirus subgroup. J Virol. 2010;84(21):11385–11394. doi: 10.1128/JVI.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lau SKP, Luk HKH, Wong ACP, Li KSM, Zhu L, He Z, Fung J, Chan TTY, Fung KSC, Woo PCY. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):1542–1547. doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Giorgi EE, Marichannegowda MH, Foley B, Xiao C, Kong X, Chen Y, Gnanakaran S, Korber B, Gao F. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci Adv. 2020 doi: 10.1126/sciadv.abb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAloose D, Laverack M, Wang L, Killian ML, Caserta LC, Yuan F, Mitchell PK, Queen K, Mauldin MR, Cronk BD, Bartlett SL, Sykes JM, Zec S, Stokol T, Ingerman K, Delaney MA, Fredrickson R, Ivančić M, Jenkins-Moore M, Mozingo K, Franzen K, Bergeson NH, Goodman L, Wang H, Fang Y, Olmstead C, McCann C, Thomas P, Goodrich E, Elvinger F, Smith DC, Tong S, Slavinski S, Calle PP, Terio K, Torchetti MK, Diel DG. From people to Panthera: Natural SARS-CoV-2 infection in tigers and lions at the Bronx Zoo. mBio. 2020 doi: 10.1128/mBio.02220-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M, van der Vegt DSJM, Munnink BBO, GeurtsvanKessel CH, van de Bovenkamp J, Sikkema RS, Jacobs EMG, Koopmans MPG, Wegdam-Blans MCA. Reinfection of SARS-CoV-2 in an immunocompromised patient: a case report. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi G, Khan S. Risk of COVID-19 pneumonia in aquatic mammals. Environ Res. 2020;188:109732. doi: 10.1016/j.envres.2020.109732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JM, Scislowska M (2020) Danes start culling 2.5 million minks after virus hits farms. https://www.newsnationnow.com/world/danes-start-culling-2-5-million-minks-after-virus-hits-farms/. Accessed 3 Nov 2020
- Oreshkova N, Molenaar RJ, Vreman S, Harders F, Munnink BBO, der Honing RWH, Gerhards N, Tolsma P, Bouwstra R, Sikkema RS, Tacken MGJ, de Rooij MMT, Weesendorp E, Engelsma MY, Bruschke CJM, Smit LAM, Koopmans M, van der Poel WHM, Stegeman A. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. 2020;25(23):2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Viviat B, Becerra-Wong M, Guadalupe JJ, Marquez S, Gutierrez B, Rojas-Silva P, Grunauer M, Trueba G, Barragan V, Cardenas P. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN. 2020 doi: 10.2139/ssrn.3686174. [DOI] [Google Scholar]
- Rondy M, Omeiri NE, Thompson MG, Levêque A, Moren A, Sullivan SG. Effectiveness of influenza vaccines in preventing severe influenza illness among adults: a systematic review and meta-analysis of test-negative design case-control studies. J Infect. 2017;75(5):381–394. doi: 10.1016/j.jinf.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard E, Halloy J, Casane D, Decroly E, Helden J. Tracing the origins of SARS-CoV-2 in coronavirus phylogenies. Med Sci (Paris) 2020;36(8–9):783–796. doi: 10.1051/medsci/2020123. [DOI] [PubMed] [Google Scholar]
- Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19) A Review. JAMA. 2020;323(18):1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Santini JM, Edwards SJL. Host range of SARS-CoV-2 and implications for public health. Lancet. 2020;1(4):e141–e142. doi: 10.1016/S2666-5247(20)30069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y, Liu P, Liang L, Cui P, Wang J, Zhang X, Guan Y, Tan W, Wu G, Chen H, Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia SF, Yan L, Chin AWH, Fung K, Choy K, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, Peiris M, Yen H. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, Sims LD, Tsang DNC, Chu DKW, Perera RAPM, Poon LLM, Peiris M. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Han J (2020) Open defecation and squat toilets pose elevated risks of fecal transmission of COVID-19 and other pathogens in developing communities. Environ Chem Lett (in press) [DOI] [PMC free article] [PubMed]
- Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, Laverdure C, Verma SC, Rossetto CC, Jackson D, Farrell MJ, Van Hooser S, Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KK, Hung IF, Ip JD, Chu AW, Chan W, Tam AR, Fong CH, Yuan S, Tsoi H, Ng AC, Lee LL, Wan P, Tso EY, To W, Tsang DN, Chan K, Huang J, Kok K, Cheng VC, Yuen K. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres DA, Ribeiro LCB, Riello APFL, Horovitz DDG, Pinto LFP, Croda J. Reinfection of COVID-19 after 3 months with a distinct and more aggressive clinical presentation: case report. J Med Virol. 2020 doi: 10.1002/jmv.26637. [DOI] [PubMed] [Google Scholar]
- UNEP (UN Environment Programme) (2020) COVID-19 waste management factsheets. https://www.unep.org/resources/factsheet/covid-19-waste-management-factsheets. Accessed 3 Nov 2020
- van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elslande J, Vermeersch P, Vandervoort K, Wawina-Bokalanga T, Vanmechelen B, Wollants E, Laenen L, André E, Van Ranst M, Lagrou K, Maes P. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney CG, Pilishvili T, Farley MM, Schaffner W, Craig AS, Lynfield R, Nyquist A, Gershman KA, Vazquez M, Bennett NM, Reingold A, Thomas A, Glode MP, Zell ER, Jorgensen JH, Beall B, Schuchat A. Effectiveness of seven-valent pneumococcal conjugate vaccine against invasive pneumococcal disease: a matched case-control study. Lancet. 2006;368(9546):1495–1502. doi: 10.1016/S0140-6736(06)69637-2. [DOI] [PubMed] [Google Scholar]
- WHO (World Health Organization) (2020) WHO coronavirus disease (COVID-19) dashboard. https://covid19.who.int/. Accessed 3 Nov 2020
- Wu L, Chen Q, Liu K, Wang J, Han P, Zhang Y, Hu Y, Meng Y, Pan X, Qiao C, Tian S, Du P, Song H, Shi W, Qi J, Wang H, Yan J, Gao GF, Wang Q. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang X, Wang X, Hu B, Zhang L, Zhang W, Si H, Zhu Y, Li B, Huang C, Chen H, Chen J, Luo Y, Guo H, Jiang R, Liu M, Chen Y, Shen X, Wang X, Zheng X, Zhao K, Chen Q, Deng F, Liu L, Yan B, Zhan F, Wang Y, Xiao G, Shi Z. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s415+86-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]