Abstract
Purpose
CRISPR gene-editing technology has the potential to transform the diagnosis and treatment of infectious diseases, but most clinicians are unaware of its broad applicability. Derived from an ancient microbial defence system, these so-called “molecular scissors” enable precise gene editing with a low error rate. However, CRISPR systems can also be targeted against pathogenic DNA or RNA sequences. This potential is being combined with innovative delivery systems to develop new therapeutic approaches to infectious diseases.
Methods
We searched Pubmed and Google Scholar for CRISPR-based strategies in the diagnosis and treatment of infectious diseases. Reference lists were reviewed and synthesized for narrative review.
Results
CRISPR-based strategies represent a novel approach to many challenging infectious diseases. CRISPR technologies can be harnessed to create rapid, low-cost diagnostic systems, as well as to identify drug-resistance genes. Therapeutic strategies, such as CRISPR systems that cleave integrated viral genomes or that target resistant bacteria, are in development. CRISPR-based therapies for emerging viruses, such as SARS-CoV-2, have also been proposed. Finally, CRISPR systems can be used to reprogram human B cells to produce neutralizing antibodies. The risks of CRISPR-based therapies include off-target and on-target modifications. Strategies to control these risks are being developed and a phase 1 clinical trials of CRISPR-based therapies for cancer and monogenic diseases are already underway.
Conclusions
CRISPR systems have broad applicability in the field of infectious diseases and may offer solutions to many of the most challenging human infections.
Keywords: CRISPR gene editing, Infectious diseases, Viral infections, Resistant bacteria, Pandemic viruses
Introduction
The discovery of CRISPR
“Clustered regularly interspaced palindromic repeats” (CRISPR) were first discovered in the archaea species, Haloferax mediterranei, by a Spanish microbiologist, Dr. Francisco Mojica, They comprised palindromic sequences of 30 base pairs (bp) interspersed with spacer sequences of 36 bp [1]. Initially the discovery did not generate interest outside the field of microbiology, however, when similar palindromic repeats were identified in multiple bacterial species, including Escherichia coli, Mycobacterium tuberculosis, and Clostridium difficile [2], it became apparent that CRISPR repeats might serve an important conserved function. Protein-coding genes, known as “CRISPR-associated proteins” or Cas proteins, with nucleic acid unwinding and cleavage functions were also discovered in proximity to the CRISPR sequences [2], but their role remained a mystery.
The function of CRISPR repeats was finally elucidated in 2003, when Dr. Mojica made a key observation. While searching for homologies between CRISPR spacers and other known sequences, Mojica identifed matches between some CRISPR spacer sequences and bacteriophages, which are viruses that infect bacteria. Mojica postulated that CRISPR sequences and their associated Cas proteins might function as a microbial defense system, storing “memories” of bacteriophages, which could then be used to fight subsequent infections [1, 3].
The mechanism of CRISPR function in bacteria is remarkably simple (Fig. 1). When a bacterium is invaded by a bacteriophage or plasmid, portions of the invading DNA are cleaved by Cas proteins and inserted into the CRISPR locus, creating a “memory” of the sequence [4]. The bacterium constitutively transcribes the CRISPR locus to produce an RNA message that contains all of the CRISPR repeats and their spacers. This transcript is called the pre-crispr-ribonucleic acid (pre-crRNA). Cleavage of the pre-crRNA by a Cas protein produces the crispr-ribonucleic acids (crRNAs), each of which contains a single spacer and a single CRISPR repeat [5].
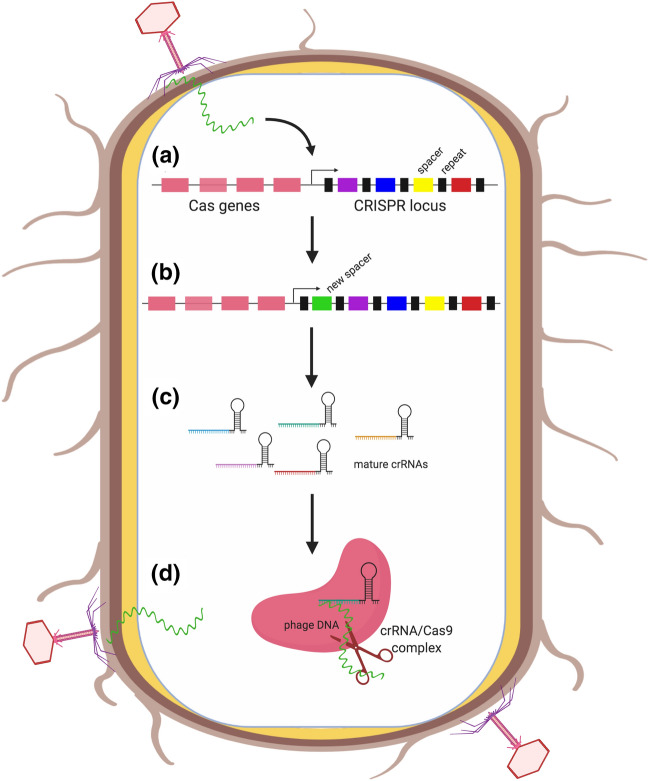
Fig. 1.
CRISPR/Cas9 function in bacteria. a Genetic material from a virus, phage, or plasmid enters the bacterium. b Short segments of genetic information from the invading agent are inserted into the CRISPR region interleaved with spacer segments. c crRNA segments consisting of CRISPR repeats and spacers are constitutively expressed in the bacterium. d Invading nucleic acid segments are recognized by crRNAs, leading to assembly of a cleavage complex containing the foreign DNA, crRNA, and Cas9 protein and resulting in cleavage of the invading DNA
To eliminate invading pathogens, the crRNA segments are released into the cytoplasm where they recognize and bind matching DNA or RNA sequences. When a match is made, a cleavage complex is assembled containing the crRNA and its target sequence, Cas nucleases, and a small RNA called the trans-activating CRISPR RNA (tracrRNA). This complex cleaves the target at a site adjacent to the crRNA target sequence, called the protospacer adjacent motif (PAM) (Fig. 1).
Cas proteins from different bacterial species mediate nucleic acid cleavage in different ways. Some Cas proteins, like Cas9, work individually, while others form cleavage complexes containing multiple Cas proteins. Irrespective of the mechanism of cleavage however, the CRISPR system acts like an microbial defense system, targeting the DNA or RNA of invading agents for degradation and elimination.
CRISPR and gene editing
In 2013, Dr. Feng Zhang at the Massachusetts Institute of Technology published the first report of CRISPR-based human gene editing [6]. His work was based on the ground-breaking studies of Emmanuelle Charpentier and Jennifer Doudna, who had previously identified the key elements required for CRISPR-mediated cleavage [7]. Zhang and his team recognized the potential of CRISPR systems to produce highly targeted modifications in human genes and designed a method to express guide RNAs (the equivalent of the crRNAs), tracrRNA, and the Cas9 cleavage protein in human cells [6].
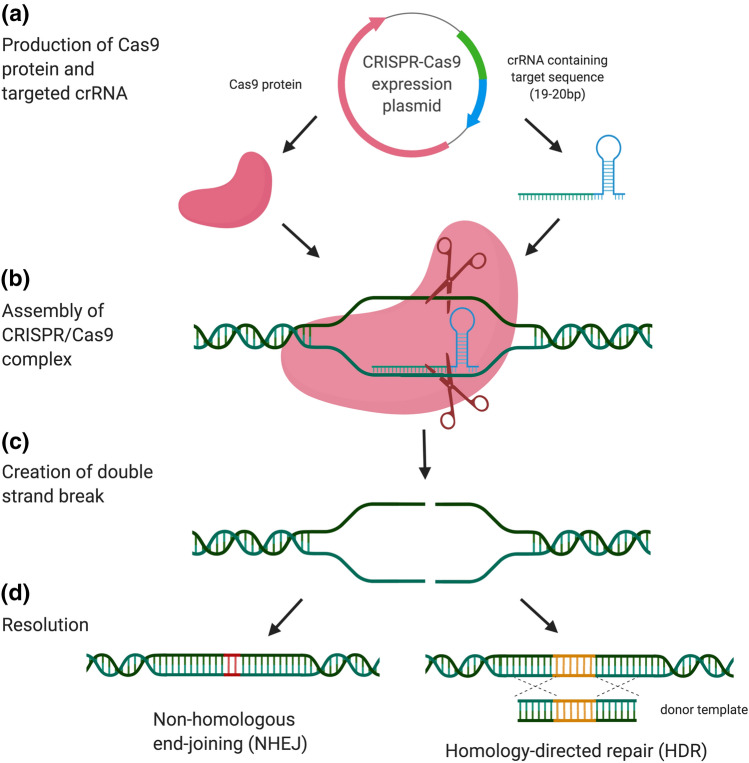
One of the key issues resolved by Zhang and his team was how to repair the cleaved DNA at the target site. In the absence of intervention, repair of the cleaved DNA ends occurs via non-homologous end joining (NHEJ) (Fig. 2). This is an error-prone process, often resulting in insertion or deletion of a small number of nucleotides (indels), and leading to unpredictable changes in the gene sequence. As a result, the target gene may or may not be inactivated after repair. Zhang and his team postulated that by introducing a homology repair template into the cells along with the CRISPR system, the cleaved DNA ends could be repaired via homology-directed repair (HDR), in a template-specific manner. Using this strategy, Zhang and his team were able to induce cleavage and repair via HDR at the EMX1 homeobox gene in human cells [6].
Fig. 2.
Mechanisms of CRISPR/Cas9 gene editing. a A construct that expresses a crRNA segment and the Cas9 protein is introduced into the target cell. b The target DNA is bound by the crRNA and Cas9 complex. c A double-strand break is introduced at the target site by the Cas9 nuclease. d The free ends of the DNA are subsequently repaired by the cell’s DNA repair mechanisms, either through error-prone non-homologous end joining (NHEJ) or, with the addition of a homology template, through homology-directed repair (HDR)
The principle advantages of CRISPR gene editing, relative to other gene-editing techniques, are its specificity and accuracy of editing, even for very small sequences. This is a product of the strong complementarity between the guide sequences (crRNAs) and their targets, as well as the precision of Cas cleavage at the target site [8, 9]. In addition, CRISPR has the advantage of simplicity, in that it requires, at a minimum, only a crRNA guide sequence, the Cas9 nuclease protein (from Streptococcus pyogenes), and a tracrRNA, to be functional [9, 10]. Additional functionality is available through the use of CRISPR-associated nucleases from different microorganisms, such as the Cas13 protein (from Leptotrichia shahii), which is an RNA-specific nuclease [8]. Cas proteins have also been engineered to perform different enzymatic functions, such as DNA methylation and base editing. These modifications have greatly expanded the nucleic-acid modifying potential of CRISPR-based systems [11, 12]
The possibilities offered by CRISPR technology are vast and apply to many areas of biology, from food production to environmental science. Within medicine, CRISPR-based treatments for genetic diseases and cancers are under development and some have already entered clinical trials [13].
One of the most intriguing applications for CRISPR technology is in the area of infectious diseases. Inexpensive and reliable CRISPR diagnostics for viruses and bacteria are under development and CRISPR-based therapeutics are in the pipeline to treat many challenging infections, including hepatitis B virus (HBV), human immunodeficiency virus-1 (HIV-1), and methicillin-resistant Staphylococcus aureus (MRSA). Unlike traditional diagnostic and therapeutic tools, CRISPR systems can be easily modified to target novel pathogens or resistance pathways. As such, they represent a powerful potential tool against emerging viruses, where speed of development is paramount. The risks of CRISPR-based therapeutics include both “off-target” and “on-target” effects, and these risks will need to be addressed as CRISPR technologies move from the laboratory to the bedside.
CRISPR-based diagnostics
CRISPR-based systems provide improved speed and accuracy in the detection of viral and bacterial nucleic acids [14, 15]. Two analogous diagnostic systems, known as DETECTR [16, 17] and SHERLOCK [18, 19], have been designed to detect RNA and DNA sequences respectively. In the first step, viral or bacterial sequences are amplified from clinical samples using recombinase polymerase amplification (RPA). The resulting DNA is then mixed with a CRISPR/Cas system that identifies and cleaves the sequence of interest. The Cas endonuclease used in this reaction has been engineered to display indiscriminate cleavage activity only after it is activated by binding to its target. For RNA (DETECTR), this is Cas12a ([16, 17] while for DNA (SHERLOCK) this is Cas13a ([18, 19]. After cleaving their target sequences, Cas12a and Cas13a begin indiscriminately cleaving other DNAs or RNAs in the solution. Thus, when a quenched reporter sequence is introduced, Cas12a or Cas13a will cleave the reporter sequence, resulting in a fluorescent signal. However, if the target sequence is not present in the solution, Cas12a and Cas13a will not be activated and the reporter sequence remains quenched.
SHERLOCK and DETECTR are so sensitive that they are capable of single copy viral detection. Moreover, the enzymatic reactions work at 37 ºC, and do not require expensive thermal cyclers. They are also rapid, taking only 1–2 hours from start to finish. For these reasons, there is tremendous interest in using SHERLOCK and DETECTR as field-deployable diagnostic tools for viruses such as Ebola, Zika, and Dengue [15, 20]. Recently, detection of SARS-CoV-2 using DETECTR was reported, with a turnaround time of only 45 min and a 95% positive and 100% negative predictive agreement with PCR techniques [21].
CRISPR strategies can also be used to facilitate detection of low-frequency sequences within clinical samples, such as antimicrobial-resistance genes, by improving the sensitivity of Next-Generation Sequencing (NGS). The Depletion of Abundant Sequences to Hybridization (DASH) strategy employs a pre-processing step in which high frequency targets that reduce the sensitivity of NGS, such as human mitochondrial ribosomal RNA genes, are cleaved with CRISPR/Cas9 [22]. After removal of these high-frequency targets, the pathogenic sequences of interest can then be easily amplified. This strategy was used to amplify fungal and parasitic sequences from cerebrospinal fluid samples with greatly increased sensitivity [22]. An alternative CRISPR-based strategy, called FLASH (Finding Low Abundance Sequences by Hybridization), uses alkaline phosphatase to block all sequences in the sample. A CRISPR/Cas system is then used to partially digest the sequence of interest, which is ligated to universal sequencing adapters and amplified, with very little amplification of other sequences in the sample [23]. FLASH-NGS was used to identify antibiotic-resistance genes in tracheal aspirates from patients with Staphylococcus aureus pneumonia [23]. The approach achieved 5000-fold enrichment of the target genes, relative to NGS alone. It was also used successfully to identify malaria-resistance genes from dried blood spot samples [23]. Once again, the sensitivity of detection with FLASH-NGS was much greater than with NGS alone.
These extraordinary techniques are likely to find quick application in clinical laboratories. Not only are they highly sensitive, they are also fast and inexpensive, an irresistible combination [21–24]. Moreover, unlike CRISPR therapeutics, there are no significant risks to patients that need to be addressed.
CRISPR-based therapies in the treatment of acute and chronic viral infections
Viruses are encapsulated pieces of viral DNA or RNA that invade living cells and hijack their intracellular processes to enable their own replication. Given their intracellular localization and their dependence on cellular proteins for their replication and survival, they are obvious targets for CRISPR-based gene editing therapies.
Viral infections exist in three distinct states: lytic infection, latent infection and chronic infection. In lytic infection, the virus is actively replicating and producing viral progeny, or virions. After the viral genomes are packaged into protein coats, or capsids, they are released through cell lysis, thereby destroying the host cell [25]. Some viruses can also cause latent infection, in which the virus enters a dormant state in which it can remain for years. Many important human pathogens, including Herpes Simplex 1 (HSV-1), Epstein–Barr virus (EBV), Cytomegalovirus (CMV), human herpesvirus 6 (HHV-6), Hepatitis B virus (HBV), polyomavirus, and parvovirus, cause latent infections [25, 26]. Finally, chronic infection represents a state of continuous low-level viral replication, leading to chronic organ damage. Chronic infection is typical of several important human pathogens, including HBV and HIV-1 [27]. These two infections cause enormous morbidity and mortality, with 250 million people worldwide currently suffering from chronic HBV infection [28], and almost 40 million from chronic HIV-1 infection [29].
In latent and chronic viral infection, the viral genome is maintained indefinitely inside the host cell, either integrated into the host genome or as free-floating viral minichromosomes. Once the virus has taken up residence inside the host cell, it is very challenging to treat. However, CRISPR-based systems can be used to target viral genomes inside the host cell, rendering them incapable for transcription and replication (see below). Thus, CRISPR gene editing offers the potential of a cure for these challenging chronic infections.
Although the concept makes intuitive sense, there are logistical challenges that need to be overcome. First, the CRISPR system has to be delivered to the target cells. Second, the system must accurately target viral genomes that are in a constant state of evolution; this is particularly challenging with HIV-1, due to its high rate of mutation and considerable genetic diversity even within a single host [30]. Third, there must be a strategy to prevent reinfection of the newly “cured” cells from other viral reservoirs within the body. Finally, the system must not induce unintended effects at “off-target” sites in the host genome.
In spite of these challenges, considerable progress has been made in developing CRISPR-based therapies for chronic viral infections. In one study, a CRISPR/Cas9 system was used to knock down HBV in a human hepatocyte cell line and in a mouse model of HBV [31]. Encouragingly, even after 4 weeks of continuous CRISPR/Cas9 expression in the mouse model, no off-target cleavage events were detected. Another study employed a modified “base editing” Cas9 protein to target HBV [32]. Instead of cleaving the DNA at the target site, the Cas9 base editing protein induced a nucleotide modification, resulting in a nonsense codon that prevented viral protein translation. By avoiding the creation of double-strand breaks, this base editing strategy reduces the risk of chromosomal rearrangements at the target site (with their potential to cause cancer), as well as limiting the danger associated with off-target effects.
HIV-1 is a complex and elusive virus and its ability to mutate allows it to develop resistance to most therapies over time [33]. However, several investigators have designed CRISPR systems that can target HIV-1 genes integrated into host DNA, thereby eliminating the virus in situ [33, 34]. In one study, investigators targeted the HIV-1 genome in a human T-cell line using a CRISPR system and either a single guide RNA or two guide RNAs simultaneously [33]. The use of a single-guide RNA lead to rapid viral resistance, but dual targeting with two guide RNAs caused complete suppression of viral replication, without evidence of viral escape [33]. Moreover, persistent co-expression of the CRISPR/Cas9 system in HIV-1-eradicated cells prevented reinfection [35, 36]. Recently, a CRISPR/Cas9 system was used to eradicate HIV-1 from a humanized mouse model of the disease [37]. No off-target effects were identified after sequencing more than 100 predicted off-target sites. Finally, in an ex vivo therapeutic approach, CRISPR/Cas9 technology was used to modify the CCR5 receptor in CD4 + germline cells, making them resistant to de novo HIV infection [38]. This raises the possibility of ex vivo treatment for HIV-1 patients, in which the patient’s own T cells are extracted, modified outside the body using CRISPR, and then reintroduced to provide the patient with a functioning HIV-1-resistant immune system [39, 40].
CRISPR/Cas9 systems have also been used to eradicate porcine endogenous retroviruses (PERVs) in pigs. PERVs are viruses that are integrated into the porcine genome and are a major barrier to xenotransplantation (i.e., transplant of pig organs into human recipients). In an effort to make pig organs PERV-free, CRISPR/Cas9 systems were used to eliminate PERVs from porcine cell lines by targeting the highly conserved PERV polymerase gene. This led to a 1000-fold reduction in PERV viral burden [16]. Subsequently, somatic cell nuclear transfer was used to generate PERV-free pigs that could potentially be used to grow organs for xenotransplantation [18].
A number of other human DNA viruses have been targeted using CRISPR/Cas9 systems. These include herpes simplex virus-1 (HSV-1) [41, 42], Epstein Barr Virus (EBV) [14, 43, 44], human cytomegalovirus (CMV) [41], human papilloma virus (HPV) [45–47], and JC polyomavirus [48]. However, many important human pathogens, including the flaviviruses (Zika virus, West Nile virus, Yellow fever, and dengue), rotaviruses, and coronaviruses (SARS-CoV, MERS-CoV, SARS-CoV-2) have RNA genomes. Recently, two CRISPR systems were developed that target RNA sequences. One such system uses the Cas9 protein, which has some intrinsic affinity for RNA, and a separate DNA oligomer containing the PAM sequence. The PAM oligomer binds to the target RNA sequence at the intrinsic PAM sequence thereby forming the double-stranded target necessary for Cas9 cleavage [4, 49]. A second RNA-modifying CRISPR system uses a family of RNA-targeted nucleases called Cas13 proteins [5, 8, 50]. This strategy has been proposed as a therapeutic approach for SARS-CoV-2, the virus causing Coronavirus 2019 (COVID-19) disease [51]. The concept entails introduction of a CRISPR/Cas13 system into airway epithelial cells, along with guide RNAs targeted against the replicase-transcriptase and spike protein genes of the SARS-CoV-2 virus, all under the control of a tissue-specific promoter. Delivery would be via aerosolization of a modified adeno-associated virus, a non-pathogenic human virus with affinity for the airway epithelium [51]. Recently, this strategy was tested in airway epithelial cell where it was successful in cleaving synthesized SARS-CoV-2 RNA fragments [52]. The authors noted that the strategy could be used to target 90% of known coronaviruses using only 6 guide RNAs targeted to conserved sequences. Further studies will be required to determine whether the strategy is effective against live SARS-CoV-2 virus and whether it is safe for use in patients.
Although CRISPR therapies for COVID-19 are unlikely to enter clinical practice during the current pandemic, they hold promise as a therapeutic approach that could be rapidly deployed for future viral epidemics.
CRISPR “VACCINES”
Traditional vaccination involves introducing viral or bacterial antigens into the host, resulting in the generation of a targeted antibody response and/or a targeted T cell response [53, 54]. The response is host-specific, however, with significant variability in the quantity and quality of the antibodies produced between individuals [53]. Moreover, some viruses, such as HIV-1, only generate neutralizing antibodies in a small minority of individuals, making vaccine design challenging.
An alternative strategy is to engineer patient B cells to produce neutralizing antibodies. Using CRISPR systems, human B cells have been modified to produce HIV-1-specific broadly neutralizing antibodies (bNAb), that can suppress HIV-1 viremia [55]. In a recent study, CRISPR-based cleavage and homology-directed repair were used to edit the heavy chain locus in activated B cells from human patients to produce bNAbs. These B cells were then transferred into mice, where they produced sufficient levels of antibody to confer immunity [55]. This strategy would potentially eliminate the inter-individual variability associated with traditional vaccination and might also be used to generate immunity against viruses, such as HIV-1, for which traditional vaccines have thus far proven ineffective.
CRISPR-based therapies for bacterial infections
Although CRISPR systems evolved in bacteria, they can also be used to create anti-bacterial therapeutics that can be delivered through the cell wall using bacteriophages, which are viruses that infect bacteria. Naturally occurring bacteriophages have previously been used to treat localized skin infections [56] as well as an intra-abdominal Acinetobacter baumanii infection [57] and a drug-resistant Staphylococcus aureus osteomyelitis [58]. In these cases, the bacteriophages were selected from phage libraries rather than engineered [59], however the use of bacteriophages to deliver engineered therapeutics is an area of growing interest [60].
Bacteriophages that can deliver CRISPR systems to pathogenic bacteria have been reported for both Staphylococcus aureus [61, 62] and Escherichia coli [63]. Cleavage targets for the CRISPR systems included: (1) bacterial virulence factors (resulting in decreased virulence) [25, 26, 61]; (2) antibiotic-resistance genes (resulting in improved susceptibility to antibiotics) [63]; and (3) genes unique to the bacterial species (resulting in highly specific bacterial killing) [62]. Unlike antibiotics, engineered bacteriophages should hypothetically have an extremely narrow spectrum of action, killing the bacteria of interest while sparing non-pathogenic commensal bacteria. Off-target effects on host cells should also be non-existent, as the CRISPR constructs are transcribed only inside the bacteria.
In vivo data indicate that CRISPR bacteriophages are highly effective against their target bacteria [61, 62, 64]. Results in vivo, however, have been more mixed. Treatment of skin and soft tissue infections in animal models was successful [63, 64], but treatment of Staphylococcus aureus osteomyelitis in a rat model was not [64]. This may be a result of poor bacteriophage penetration into the bone. Further in vivo research is required to optimize bacteriophage delivery and dosing, as well as to ensure safety and minimize side effects.
Impediments to CRISPR therapeutics
The principle risks of CRISPR-based therapeutics are “off-target” effects, namely DNA or RNA modifications at unanticipated sites, and “on-target” effects, namely unintended DNA or RNA modifications at the target site.
Off-target effects occur when partial homologies between the guide RNA(s) and off-target genomic sequences result in unintended modifications distant from the target site [65, 66]. These effects are not entirely predictable, as they depend not only on the degree and location of sequence homology but also on gRNA structure. To complicate matters further, the only way to identify all off-target effects is to perform whole-genome sequencing [67]. A number of strategies have been employed to reduce off-target effects. These include careful selection of gRNA sequences to minimize homologies as well as the use of truncated or 5′ guanine capped gRNAs that confer higher specificity [68, 69]. Engineered Cas nucleases with increased cleavage specificity can be also used to limit off-target effects [70, 71]. Finally, the use of paired Cas nucleases that only nick one DNA strand, thereby requiring the binding of two gRNA sequences to opposite DNA strands in close proximity, has also been shown to greatly improve specificity [72].
On-target effects comprise all unintended sequence modifications at the target site. These include base pair insertions and deletions, as well as inversions and recombination events [73, 74]. On-target effects are a significant concern in genetic engineering, where they reduce efficiency and potentially damage the target gene, but are less concerning when the target is a pathogenic sequence. However, they cannot be ignored given the risk of recombination events such as between an integrated viral sequence and the host genome. Strategies to reduce on-target effects include using a modified Cas9 nuclease that increases the frequency of homology repair and an optimized homology repair template, as well as inhibiting non-homologous end-joining through the use of NHEJ pathway inhibitors [75].
CRISPR Therapeutics in Human Trials
CRISPR systems are already being trialled in phase 1 studies in human patients. Most of these trials involve ex vivo CRISPR therapy, meaning that the patient’s cells (or donor cells) are harvested, modified in the laboratory, and then reintroduced into the patient. In 2019, a group from China reported the successful bone marrow transplantation of an HIV-1-positive patient with acute lymphocytic leukemia for whom the hematopoietic progenitor cells had been CCR5-ablated using CRISPR in order to prevent HIV infection [76]. CCR5 ablation was confirmed in a percentage of the patient’s T cells after engraftment and whole genome sequencing showed no evidence of off-target effects. Another small phase 1 trial included 3 patients with refractory cancer (two with advanced myeloma and one with liposarcoma) whose T cells were harvested, modified using CRISPR engineering to express a T cell receptor specific to the NY-ESO-1 tumor antigen, and then reintroduced into the patients [13]. Expression of the engineered T-cells was confirmed in all 3 patients for at least 100 days. At the end of the 9 month follow-up period, two patients were alive and one patient had died. Of note, chromosomal translocations were identified in all of the engineered T cell populations; however, these did not appear to confer a growth advantage to the modified T cells.
The first trial of in vivo CRISPR gene therapy in human patients began in early 2020. In the study, pediatric patients with a congenital retinal disease called Leber congenital aumaurosis will receive a CRISPR-based therapeutic system that will correct the single base pair mutation causing the disease and will be delivered to the retina via local injection of an engineered adenovirus vector [77]. Results of the trial are still pending.
Progress in the field of CRISPR therapeutics has been extremely fast, from the first CRISPR gene-editing system in human cells in 2013 to the first phase 1 clinical trial in 2020. Given the widespread interest in CRISPR-based therapies for many diseases, ranging from monogenic diseases such as sickle cell disease and hemophilia to complex diseases such as cancer, progress is likely to continue at breakneck speed. As safety issues are addressed, the feasibility of using CRISPR therapeutics in human patients with infectious diseases will only increase.
Conclusion
The discovery of CRISPR repeats in bacteria was initially considered of minor importance outside the field of microbiology. However, CRISPR technology has emerged as a landmark discovery in the field of genetic engineering and is poised to have a major impact in many areas of clinical medicine. In this review, we have outlined potential applications for CRISPR technology in infectious diseases, including the development of rapid, low-cost diagnostics, the treatment of acute and chronic viral infections, and the treatment of resistant bacterial infections. CRISPR-based diagnostics and therapeutics will likely appear in clinical practice in the near future, and clinicians should be aware of their extraordinary potential, as well as their potential risks.
Acknowledgements
We thank Dr Alexandra Gillespie for helpful comments on the text. Figures were created with Biorender.com.
Author contributions
AB, EF and PCB planned the article. AB, EF, and HAL performed the literature search and data analysis. All the authors critically reviewed the manuscript for publication.
Funding
This work was supported by the Fundação para a Ciência e Tecnologia (FCT) Portugal Research Center Grant UID/BIM/04773/2013 CBMR 1334.
Compliance with ethical standards
Conflict of interest
Prof. de Mello reports consultant/advisory board for Pfizer, Zodiac, MSD; speaker honoraria from AstraZeneca, Novartis; Educational Grants from Roche, Merck-Group; travel grant from BMS; and expert honoraria from National Science Center, Poland.
References
- 1.Lander ES. The Heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 2.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–4377. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60(2):174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Micro. 2014;12:317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 6.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown A, Woods WS, Perez-Pinera P. Targeted gene activation using RNA-guided nucleases. Methods Mol Biol. 2017;1468:235–250. doi: 10.1007/978-1-4939-4035-6_16. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Gaj T, Yang Y, Wang N, Shui S, Kim S, et al. Efficient delivery of nuclease proteins for genome editing in human stem cells and primary cells. Nat Protoc. 2015;10:1842–1859. doi: 10.1038/nprot.2015.117. [DOI] [PubMed] [Google Scholar]
- 11.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19:770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang JG, Park JS, Ko J-H, Kim Y-S. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Scientific Reports. 2019;9:11960. doi: 10.1038/s41598-019-48130-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtmauer EA, Fraietta JA, Davis MM, Cohen AD, Weber KL, Lancaster E. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuen K-S, Wang Z-M, Wong N-HM, Zhang Z-Q, Cheng T-F, Lui W-Y, et al. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9. Virus Res. 2018;244:296–303. doi: 10.1016/j.virusres.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 16.Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- 17.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niu D, Wei H-J, Lin L, George H, Wang T, Lee I-H, et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science. 2017;357:1303–1307. doi: 10.1126/science.aan4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu W, Crawford ED, O’Donovan BD, Wilson MR, Chow ED, Retallack H, et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016;17:41. doi: 10.1186/s13059-016-0904-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quan J, Langelier C, Kuchta A, Batson J, Teyssier N, Lyden A, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019;47:e83. doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ai J-W, Zhou X, Xu T, Yang M, Chen Y, He G-Q, et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg Microbes Infect. 2019;8:1361–1369. doi: 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traylen CM, Patel HR, Fondaw W, Mahatme S, Williams JF, Walker LR, et al. Virus reactivation: a panoramic view in human infections. Future Virol. 2011;6:451–463. doi: 10.2217/fvl.11.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman PM. Epigenetics and genetics of viral latency. Cell Host Microbe. 2016;19:619–628. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Virgin HW, Wherry EJ, Ahmed R. Redefining chronic viral infection. Cell. 2009;138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 28.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 29.Joint United Nations Programme on HIV AIDS. Fact sheet–latest global and regional statistics on the status of the AIDS epidemic. 2019. Geneva: Joint United Nations Programme on HIV.
- 30.Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR. Targeted DNA mutagenesis for the cure of chronic viral infections. J Virol. 2012;86:8920–8936. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific Reports. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y-C, Chen Y-H, Kao J-H, Ching C, Liu I-J, Wang C-C, et al. Permanent inactivation of HBV genomes by CRISPR/Cas9-mediated non-cleavage base editing. Mol Ther Nucleic Acids. 2020;20:1–47. doi: 10.1016/j.omtn.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebbink RJ, de Jong DCM, Wolters F, Kruse EM, van Ham PM, Wiertz EJHJ, et al. A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Scientific Reports. 2017;7:41968. doi: 10.1038/srep41968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 35.Kaminski R, Chen Y, Fischer T, Tedaldi E, Napoli A, Zhang Y, et al. Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Scientific Reports. 2016;6:22555. doi: 10.1038/srep22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bella R, Kaminski R, Mancuso P, Young W-B, Chen C, Sariyer R, et al. Removal of HIV DNA by CRISPR from patient blood engrafts in humanized mice. Mol Ther Nucleic Acids. 2018;12:275–282. doi: 10.1016/j.omtn.2018.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dash PK, Kaminski R, Bella R, Su H, Mathews S, Ahooyi TM, et al. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nature Communications. 2019;10:2753. doi: 10.1038/s41467-019-10366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Guan X, Du T, Jin W, Wu B, Liu Y, et al. Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol. 2015;96:2381–2393. doi: 10.1099/vir.0.000139. [DOI] [PubMed] [Google Scholar]
- 39.Savić N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15–21. doi: 10.1016/j.trsl.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Hirakawa MP, Krishnakumar R, Timlin JA, Carney JP, Butler KS. Gene editing and CRISPR in the clinic: current and future perspectives. Biosci Rep. 2020;40:BSR20200127. doi: 10.1042/BSR20200127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Diemen FR, Kruse EM, Hooykaas MJG, Bruggeling CE, Schürch AC, van Ham PM, et al. CRISPR/Cas9-Mediated genome editing of herpesviruses limits productive and latent infections. PLOS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin C, Li H, Hao M, Xiong D, Luo Y, Huang C, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Scientific Reports. 2016;6:34531. doi: 10.1038/srep34531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu JL, Glaser SL. Epstein-barr virus-associated malignancies: epidemiologic patterns and etiologic implications. Crit Rev Oncol Hematol. 2000;34:27–53. doi: 10.1016/S1040-8428(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 44.Kanda T, Furuse Y, Oshitani H, Kiyono T. Highly efficient CRISPR/Cas9-Mediated cloning and functional characterization of gastric cancer-derived epstein-barr virus strains. J Virol. 2016;90:4383–4393. doi: 10.1128/JVI.00060-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy EM, Kornepati AVR, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang C, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhen S, Hua L, Takahashi Y, Narita S, Liu Y-H, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 48.Chou Y-Y, Krupp A, Kaynor C, Gaudin R, Ma M, Cahir-McFarland E, et al. Inhibition of JCPyV infection mediated by targeted viral genome editing using CRISPR/Cas9. Scientific Reports. 2016;6:36921. doi: 10.1038/srep36921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(665–676):e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nguyen TM, Zhang Y, Pandolfi PP. Virus against virus: a potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Res. 2020;30:189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181:865–76. [DOI] [PMC free article] [PubMed]
- 53.Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist C-A, Marchant A, et al. Genetic regulation of immune responses to vaccines in early life. Genes Immun. 2004;5:122–129. doi: 10.1038/sj.gene.6364051. [DOI] [PubMed] [Google Scholar]
- 54.Gilbert SC. T-cell-inducing vaccines - what’s the future. Immunology. 2012;135:19–26. doi: 10.1111/j.1365-2567.2011.03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartweger H, McGuire AT, Horning M, Taylor JJ, Dosenovic P, Yost D, et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J Exp Med. 2019;216:1301–1310. doi: 10.1084/jem.20190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morozova VV, Vlassov VV, Tikunova NV. Applications of bacteriophages in the treatment of localized infections in humans. Front Microbiol. 2018;9:1696. doi: 10.3389/fmicb.2018.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61:e00954. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fish R, Kutter E, Bryan D, Wheat G, Kuhl S. Resolving digital staphylococcal osteomyelitis using bacteriophage—a case report. Antibiotics (Basel) 2018;7:87. doi: 10.3390/antibiotics7040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson SB, Green SI, Liu CG, Salazar KC, Clark JR, Terwilliger AL, et al. Constructing and characterizing bacteriophage libraries for phage therapy of human infections. Front Microbiol. 2019;10:2537. doi: 10.3389/fmicb.2019.02537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romero-Calle D, Guimarães Benevides R, Góes-Neto A, Billington C. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics. 2019;8:138. doi: 10.3390/antibiotics8030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Scientific Reports. 2017;7:44929. doi: 10.1038/srep44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cobb LH, Park J, Swanson EA, Beard MC, McCabe EM, Rourke AS, et al. CRISPR-Cas9 modified bacteriophage for treatment of Staphylococcus aureus induced osteomyelitis and soft tissue infection. PLoS ONE. 2019;14:e0220421. doi: 10.1371/journal.pone.0220421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimberland ML, Hou W, Alfonso-Pecchio A, Wilson S, Rao Y, Zhang S, et al. Strategies for controlling CRISPR/Cas9 off-target effects and biological variations in mammalian genome editing experiments. J Biotechnol. 2018;284:91–101. doi: 10.1016/j.jbiotec.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 67.Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 68.Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR–Cas9 nucleaseswith no detectable genome-wideoff-target effects. Nat Nat Publ Group. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee H, Kim J-S. Unexpected CRISPR on-target effects. Nat Biotechnol. 2018;36:703–704. doi: 10.1038/nbt.4207. [DOI] [PubMed] [Google Scholar]
- 74.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jamal M, Khan FA. Da, L, Habib Z, Dai J, Cao G. Keeping CRISPR/Cas on-target. Current Issues Mol Biol. 2016;20:1–12. [PubMed] [Google Scholar]
- 76.Xu L, Wang J, Liu Y, Xie L, Su B, Mou D, et al. CRISPR-Edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 77.First CRISPR therapy dosed. Nat Biotechnol. 2020;38:382. [DOI] [PubMed]