Abstract
Malignant pleural mesothelioma (MPM) is a type of cancer originating from the pleura with high aggressiveness and poor prognosis. A timely diagnosis is crucial to improve its prognosis. Laboratory biomarkers have significant advantages of reduced invasiveness, low cost, and are observer‐independent, and therefore represent a promising diagnostic tool for MPM. MicroRNA is a family of non‐coding RNA that regulates gene expression at the post‐transcriptional level. Accumulated studies showed that microRNA, either in tissue, circulating, and body fluid, has potential diagnostic value for various disorders. Here, we reviewed the diagnostic value of microRNA for MPM.
Keywords: Diagnosis, malignant pleural mesothelioma, microRNA, review

Malignant pleural mesothelioma (MPM) is a type of cancer originating from the pleura with high aggressiveness and poor prognosis, Laboratory biomarkers have significant advantages of reduced invasiveness, low cost, and are observer‐independent, and therefore represent a promising diagnostic tool for MPM. Here, we reviewed the diagnostic value of microRNA for MPM.
Introduction
Malignant pleural mesothelioma (MPM) is a type of cancer originating from the pleura with high aggressiveness. 1 Due to a lack of specific symptoms and signs, it usually diagnosed at an advanced stage, which is associated with a poor prognosis. A study using the Surveillance, Epidemiology and End Results (SEER) database showed that the median survival time of MPM is only one year. 2 Therefore, timely and accurate diagnosis is crucial to improve its prognosis.
Diagnostic pleural aspiration and thoracoscopy are the gold standards for diagnosing MPM 3 ; however, they have limitations, including sampling error, invasiveness, 4 being observer‐dependent, with special training required. Cytology is an alternative diagnostic tool with high specificity, but its sensitivity is lower than 30%. 5 , 6 Imaging is another choice for the diagnosis of MPM, but the equipment is large and special training in its operation is required which can be prohibitive in resource‐poor areas. By contrast, laboratory tests, with the advantages of observer‐independent, low cost, and reduced invasiveness, are of considerable value for diagnosing MPM. 7
Serum or pleural effusion (PE) soluble mesothelin‐related peptides (SMRP), osteopontin, and fibulin‐3 are three well recognized diagnostic markers for MPM. However, some evidence from the systematic review showed that the areas under summary receiver operating characteristics (sROC) curve (AUCs) of these biomarkers are between 0.80 and 0.90, 8 , 9 , 10 , 11 indicating that their diagnostic accuracy is insufficient. Accordingly, the current guidelines do not support using biomarkers alone to confirm or exclude MPM. 12 , 13 , 14 Therefore, further studies are needed to identify more biomarkers for the diagnosis of MPM. 3
MicroRNA is a family of non‐coding RNAs with a length of 18–25 bp regulating target gene expression at the post‐transcriptional level. 15 It is estimated that microRNA regulates nearly one‐third of human gene expression, and it, therefore, represents a potential therapeutic target for various types of diseases. 15 Previous studies have shown that microRNA can be determined in tissue, circulating, body fluid, and their levels are of diagnostic value for various disorders, such as cancer, tissue injury, and infectious diseases. 16 , 17 , 18 This review aims to summarize the research progress of microRNA as a diagnostic tool for MPM.
Diagnostic value of tissue microRNA for MPM
The first step in the identification of differentially expressed microRNAs is to identify the tissue microRNAs with potential diagnostic value for MPM. Three approaches are usually used to identify differentially expressed microRNAs. The first approach employed microRNA array to compare the microRNA profile of mesothelioma tissue with normal pleural tissue, 19 , 20 pleura of asbestos‐exposed patients, 21 non‐neoplastic pleura, 22 and other types of cancer. 23 , 24 The second approach compares the microRNA profile of mesothelioma and human mesothelial cell lines. 25 , 26 The third approach is data mining with a publicly accessible database. 27 With these approaches, some differentiated expressed microRNA were identified, such as miR‐126, 26 miR‐200c, 24 and miR‐130a, 27 and their relative expressions were further validated by polymerase chain reaction (PCR). The diagnostic accuracy of these microRNAs was assessed using the receiver operating characteristics (ROC) curve. Generally, the diagnostic accuracy of these microRNAs was unsatisfactory, with areas under the curve (AUCs) below 0.80. Notably, a study reported that the diagnostic accuracy of microRNAs could be improved by building a mathematic model (AUC = 0.96), 22 indicating that the microRNA panel represents a promising diagnostic tool for MPM.
In summary, some tissue microRNAs have been identified as a diagnostic tool for MPM, and their diagnostic accuracy has been evaluated in some studies. Generally, their diagnostic accuracy is unsatisfactory. In addition, the invasive nature of tissue microRNA determination may impair its feasibility as a diagnostic tool.
Diagnostic value of PE microRNA for MPM
PE is a common sign of MPM. In patients with undiagnosed PE, approximately 3% are MPM, 6 and in patients with MPM, 53% complain of dyspnea, which is caused by the presence of PE. 13 Therefore, detecting microRNAs in PE represents a promising diagnostic tool for MPM.
To date, only one study has investigated the diagnostic value of PE microRNA for MPM. 28 In 2019, Birnie et al. investigated the microRNA profile of PE and cell culture medium with microRNA array and compared it with that of non‐MPM induced PE. 28 They found that four PE microRNAs (miR‐944, miR‐139‐5p, miR‐210 and miR‐320) were upregulated and seven (miR‐200b, miR‐200c, miR‐143, miR‐200a, miR‐203, miR‐31 and miR‐874) were downregulated. PCR further validated the levels of four microRNA (miR‐210, miR‐143, miR‐139‐5p, and miR‐200c), and the ROC curve was used to evaluate their diagnostic accuracy. The AUCs of these four microRNAs were below 0.80, indicating that their diagnostic accuracy is inadequate. However, a logistic regression model containing miR‐200c, miR‐143, and miR‐210, has an AUC of 0.94, indicating that their combination can improve the diagnostic accuracy.
Diagnostic value of circulating microRNA for MPM
To date, three studies have investigated the circulating microRNA profile of MPM patients with microRNA array and compared it with the circulating microRNA profile of healthy individuals, and workers ex‐exposed to asbestos.29–31 Some differentially expressed microRNA were identified, including miR‐197‐3p, 29 miR‐625‐3p, 31 miR‐29c, 31 miR‐9231 and miR‐132. 30 Further studies with clinical specimens indicated that the diagnostic accuracy of miR‐197‐3p, miR‐625‐3p, miR‐132 was moderate, with AUCs of 0.76, 0.82, and 0.75.
In addition to the microRNA array, the public database was also used to identify microRNAs with potential diagnostic value. Matboli et al. analyzed the serum microRNA profile of MPM with three public microRNA databases (miR2Disease, miRWalk, Human MiR, and Disease Database). They found that miR‐548a‐3p and miR‐20a were potential diagnostic markers for MPM. 32 In a clinical study with 60 MPM patients (76% of them had a history of asbestos exposure) and 20 workers formerly exposed to asbestos, a high diagnostic accuracy of miR‐548a‐3p and miR‐20a was observed, with AUCs of 0.92 and 0.98. These results indicate that these two circulating microRNAs are excellent for identifying MPM in patients with a history of asbestos exposure.
Because a previous study indicated that miR‐126 was downregulated in tumor tissue of MPM, some studies also investigated the diagnostic accuracy of circulating miR‐126 for MPM, but the results were varied. In a study with non‐small cell lung cancer (NSCLC) as the control, the AUC of miR‐126 was only 0.75. 20 , 33 Healthy individuals were used as the control in two studies, and their results were inconsistent. The AUC in one study was as high as 0.95, 34 but in another study, the AUC was only 0.71. 35 Notably, in a head‐to‐head comparison study, the AUC of miR‐126 was lower than SMRP (0.71 vs. 0.72), indicating that the diagnostic accuracy of miR‐126 was inferior to that of SMRP. However, the combination of miR‐126 and SMRP can improve diagnostic accuracy. 35
Exosomes are a subtype of extracellular vesicles with a diameter between 30 and 100 nm, containing miRNA, mRNA, protein, and DNA, protected by a lipid bilayer. Cancer cells can secrete exosomes into the circulation and the signature of proteins and nucleic acids contained in exosomes are significantly correlated with those in primary tumor cells. 36 Therefore, serum exosomal microRNA has been proposed to be a potential diagnostic marker for MPM. 37 , 38 To date, only one study has investigated the diagnostic accuracy of exosomal microRNA for MPM. Cavalleri et al. compared the serum exosomal microRNA profile of MPM patients and patients with a history of asbestos exposure and found several dysregulated microRNAs. 39 Among these microRNAs, miR‐103, miR‐98, miR‐148b, miR‐744, and miR‐30e‐3p had high diagnostic accuracy for MPM, with AUCs larger than 0.83. These results indicate that exosomal microRNA represents a good diagnostic accuracy for MPM.
In addition to circulating cell‐free microRNA and exosomal microRNA, microRNA in blood cells has been proposed to be a diagnostic marker for MPM. By comparing the blood cell microRNA profile of MPM patients and healthy individuals, a study revealed that miR‐103 was significantly decreased in MPM patients. In a clinical study with small sample size, the AUC of miR‐103 was 0.76. 40 A further study with a relatively large sample size showed a similar AUC, and the AUC of miR‐103 was lower than that of SMRP. However, it has been previously reported that a logistic regression model contained miR‐103 and SMRP had an AUC of 0.90, indicating that miR‐103 can improve the diagnostic accuracy of SMRP. 41
In conclusion, several studies have investigated the diagnostic accuracy of tissue, PE, and circulating microRNA for MPM. By using microRNA array or public databases, some microRNAs with potential diagnostic values have been identified. PCR was used to detect the levels of these candidate microRNAs and their diagnostic accuracy was assessed with the ROC curve. Based on the available evidence, we concluded that many microRNAs can assist in MPM diagnosis (Fig 1), the diagnostic accuracy of the majority of candidate microRNAs is moderate, but the microRNA panel represents a promising diagnostic tool for MPM.
Figure 1.
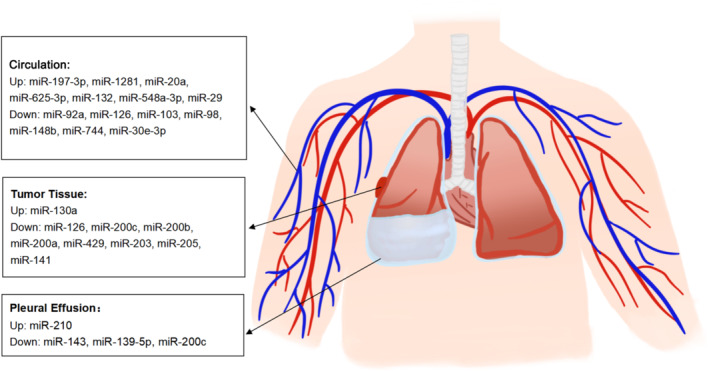
Diagnostic microRNAs in tumor tissue, pleural effusion and circulation.
Notably, the sample sizes of all of the available studies are small, and no clear and uniform criteria were used for enrollment of patients. In addition, whether the subjects were consecutively enrolled was not reported by the investigators. Therefore, patient selection bias cannot be excluded. 42 , 43 Further studies are required to identify more candidate microRNAs and validate the findings of the available studies.
Disclosure
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by the Natural and Science Foundation of Inner Mongolia Autonomous Region for Distinguished Young Scholars (2020JQ07).
Contributor Information
Wen‐Qi Zheng, Email: zhengwenqi2011@163.com.
Zhi‐De Hu, Email: hzdlj81@163.com.
References
- 1. Tsao AS, Wistuba I, Roth JA, Kindler HL. Malignant pleural mesothelioma. J Clin Oncol 2009; 27 (12): 2081–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meyerhoff RR, Yang CF, Speicher PJ et al Impact of mesothelioma histologic subtype on outcomes in the surveillance, epidemiology, and end results database. J Surg Res 2015; 196 (1): 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bianco A, Valente T, De Rimini ML et al Clinical diagnosis of malignant pleural mesothelioma. J Thorac Dis 2018; 10 (Suppl 2): S253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang XJ, Yang Y, Wang Z et al Efficacy and safety of diagnostic thoracoscopy in undiagnosed pleural effusions. Respiration 2015; 90 (3): 251–5. [DOI] [PubMed] [Google Scholar]
- 5. Arnold DT, De Fonseka D, Perry S et al Investigating unilateral pleural effusions: The role of cytology. Eur Respir J 2018; 52 (5): 1801254. [DOI] [PubMed] [Google Scholar]
- 6. Porcel JM, Esquerda A, Vives M, Bielsa S. Etiology of pleural effusions: Analysis of more than 3,000 consecutive thoracenteses. Arch Bronconeumol 2014; 50 (5): 161–5. [DOI] [PubMed] [Google Scholar]
- 7. Hollevoet K, Reitsma JB, Creaney J et al Serum mesothelin for diagnosing malignant pleural mesothelioma: An individual patient data meta‐analysis. J Clin Oncol 2012; 30 (13): 1541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cui A, Jin XG, Zhai K, Tong ZH, Shi HZ. Diagnostic values of soluble mesothelin‐related peptides for malignant pleural mesothelioma: Updated meta‐analysis. BMJ Open 2014; 4 (2): e004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu ZD, Liu XF, Liu XC et al Diagnostic accuracy of osteopontin for malignant pleural mesothelioma: A systematic review and meta‐analysis. Clin Chim Acta 2014; 433: 44–8. [DOI] [PubMed] [Google Scholar]
- 10. Ren R, Yin P, Zhang Y et al Diagnostic value of fibulin‐3 for malignant pleural mesothelioma: A systematic review and meta‐analysis. Oncotarget 2016; 7 (51): 84851–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gillezeau CN, van Gerwen M, Ramos J, Liu B, Flores R, Taioli E. Biomarkers for malignant pleural mesothelioma: A meta‐analysis. Carcinogenesis 2019; 40 (11): 1320–31. [DOI] [PubMed] [Google Scholar]
- 12. Scherpereel A, Opitz I, Berghmans T et al ERS/ESTS/EACTS/ESTRO guidelines for the management of malignant pleural mesothelioma. Eur Respir J 2020; 55 (6): 1900953. [DOI] [PubMed] [Google Scholar]
- 13. Woolhouse I, Bishop L, Darlison L et al British Thoracic Society guideline for the investigation and management of malignant pleural mesothelioma. Thorax 2018; 73 (Suppl 1): i1–i30. [DOI] [PubMed] [Google Scholar]
- 14. Kindler HL, Ismaila N, Armato SG et al Treatment of malignant pleural mesothelioma: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol 2018; 36 (13): 1343–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bartel DP. MicroRNAs. Target recognition and regulatory functions. Cell 2009; 136 (2): 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartels CL, Tsongalis GJ. MicroRNAs. Novel biomarkers for human cancer. Clin Chem 2009; 55 (4): 623–31. [DOI] [PubMed] [Google Scholar]
- 17. Adachi T, Nakanishi M, Otsuka Y et al Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem 2010; 56 (7): 1183–5. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y, Jia Y, Zheng R et al Plasma microRNA‐122 as a biomarker for viral‐, alcohol‐, and chemical‐related hepatic diseases. Clin Chem 2010; 56 (12): 1830–8. [DOI] [PubMed] [Google Scholar]
- 19. Guled M, Lahti L, Lindholm PM et al CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma ‐a miRNA microarray analysis. Genes Chromosomes Cancer 2009; 48 (7): 615–23. [DOI] [PubMed] [Google Scholar]
- 20. Santarelli L, Strafella E, Staffolani S et al Association of MiR‐126 with soluble mesothelin‐related peptides, a marker for malignant mesothelioma. PLOS One 2011; 6 (4): e18232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ak G, Tomaszek SC, Kosari F et al MicroRNA and mRNA features of malignant pleural mesothelioma and benign asbestos‐related pleural effusion. Biomed Res Int 2015; 2015: 635748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersen M, Grauslund M, Ravn J, Sørensen JB, Andersen CB, Santoni‐Rugiu E. Diagnostic potential of miR‐126, miR‐143, miR‐145, and miR‐652 in malignant pleural mesothelioma. J Mol Diagn 2014; 16 (4): 418–30. [DOI] [PubMed] [Google Scholar]
- 23. Benjamin H, Lebanony D, Rosenwald S et al A diagnostic assay based on microRNA expression accurately identifies malignant pleural mesothelioma. J Mol Diagn 2010; 12 (6): 771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gee GV, Koestler DC, Christensen BC et al Downregulated microRNAs in the differential diagnosis of malignant pleural mesothelioma. Int J Cancer 2010; 127 (12): 2859–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Busacca S, Germano S, De Cecco L et al MicroRNA signature of malignant mesothelioma with potential diagnostic and prognostic implications. Am J Respir Cell Mol Biol 2010; 42 (3): 312–9. [DOI] [PubMed] [Google Scholar]
- 26. Cappellesso R, Nicole L, Caroccia B et al Young investigator challenge: MicroRNA‐21/MicroRNA‐126 profiling as a novel tool for the diagnosis of malignant mesothelioma in pleural effusion cytology. Cancer Cytopathol 2016; 124 (1): 28–37. [DOI] [PubMed] [Google Scholar]
- 27. Cappellesso R, Galasso M, Nicolè L, Dabrilli P, Volinia S, Fassina A. miR‐130A as a diagnostic marker to differentiate malignant mesothelioma from lung adenocarcinoma in pleural effusion cytology. Cancer Cytopathol 2017; 125 (8): 635–43. [DOI] [PubMed] [Google Scholar]
- 28. Birnie KA, Prêle CM, Musk AWB et al MicroRNA signatures in malignant pleural mesothelioma effusions. Dis Markers 2019; 2019: 8628612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bononi I, Comar M, Puozzo A et al Circulating microRNAs found dysregulated in ex‐exposed asbestos workers and pleural mesothelioma patients as potential new biomarkers. Oncotarget 2016; 7 (50): 82700–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weber DG, Gawrych K, Casjens S et al Circulating miR‐132‐3p as a candidate diagnostic biomarker for malignant mesothelioma. Dis Markers 2017; 2017: 9280170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirschner MB, Cheng YY, Badrian B et al Increased circulating miR‐625‐3p: A potential biomarker for patients with malignant pleural mesothelioma. J Thorac Oncol 2012; 7 (7): 1184–91. [DOI] [PubMed] [Google Scholar]
- 32. Matboli M, Shafei AE, Azazy AE et al Clinical evaluation of circulating miR‐548a‐3p and ‐20a expression in malignant pleural mesothelioma patients. Biomark Med 2018; 12 (2): 129–39. [DOI] [PubMed] [Google Scholar]
- 33. Tomasetti M, Staffolani S, Nocchi L et al Clinical significance of circulating miR‐126 quantification in malignant mesothelioma patients. Clin Biochem 2012; 45 (7–8): 575–81. [DOI] [PubMed] [Google Scholar]
- 34. Mozzoni P, Ampollini L, Goldoni M et al MicroRNA expression in malignant pleural mesothelioma and asbestosis: A pilot study. Dis Markers 2017; 2017: 9645940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Santarelli L, Staffolani S, Strafella E et al Combined circulating epigenetic markers to improve mesothelin performance in the diagnosis of malignant mesothelioma. Lung Cancer 2015; 90 (3): 457–64. [DOI] [PubMed] [Google Scholar]
- 36. Rabinowits G, Gercel‐Taylor C, Day JM et al Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer 2009; 10 (1): 42–6. [DOI] [PubMed] [Google Scholar]
- 37. Cortez MA, Bueso‐Ramos C, Ferdin J, Lopez‐Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids–the mix of hormones and biomarkers. Nat Rev Clin Oncol 2011; 8 (8): 467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou L, Lv T, Zhang Q et al The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett 2017; 407: 84–92. [DOI] [PubMed] [Google Scholar]
- 39. Cavalleri T, Angelici L, Favero C et al Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos‐exposed subjects suggest a 2‐miRNA signature as potential biomarker of disease. PLOS One 2017; 12 (5): e0176680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weber DG, Johnen G, Bryk O, Jöckel KH, Brüning T. Identification of miRNA‐103 in the cellular fraction of human peripheral blood as a potential biomarker for malignant mesothelioma–A pilot study. PLOS One 2012; 7 (1): e30221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weber DG, Casjens S, Johnen G et al Combination of MiR‐103a‐3p and mesothelin improves the biomarker performance of malignant mesothelioma diagnosis. PLOS One 2014; 9 (12): e114483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang M, Hu ZD. Suggestions for designing studies investigating diagnostic accuracy of biomarkers. Ann Transl Med 2019; 7 (23): 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu ZD. Circulating biomarker for malignant pleural mesothelioma diagnosis: Pay attention to study design. J Thorac Dis 2016; 8 (10): 2674–6. [DOI] [PMC free article] [PubMed] [Google Scholar]


