Abstract
Natural plant populations encounter strong pathogen pressure and defence-associated genes are known to be under selection dependent on the pressure by the pathogens. Here, we use populations of the wild tomato Solanum chilense to investigate natural resistance against Cladosporium fulvum, a well-known ascomycete pathogen of domesticated tomatoes. Host populations used are from distinct geographical origins and share a defined evolutionary history. We show that distinct populations of S. chilense differ in resistance against the pathogen. Screening for major resistance gene-mediated pathogen recognition throughout the whole species showed clear geographical differences between populations and complete loss of pathogen recognition in the south of the species range. In addition, we observed high complexity in a homologues of Cladosporium resistance (Hcr) locus, underlying the recognition of C. fulvum, in central and northern populations. Our findings show that major gene-mediated recognition specificity is diverse in a natural plant-pathosystem. We place major gene resistance in a geographical context that also defined the evolutionary history of that species. Data suggest that the underlying loci are more complex than previously anticipated, with small-scale gene recombination being possibly responsible for maintaining balanced polymorphisms in the populations that experience pathogen pressure.
Keywords: major gene resistance, Solanum chilense, Cladosporium fulvum, tomato, loss of resistance
1. Introduction
Plant–pathogen interactions are often studied in crops. How plants and their pathogens adapt to one another and interact molecularly in their natural habitat is still poorly understood. Technical difficulties with obtaining the right samples, high manpower required to maintain the species and perform experiments in controlled conditions and most importantly, difficulties in developing methods to evaluate the resistance hamper studies on plant–pathogen interactions in a natural context. In spite of these difficulties, a few studies have highlighted such interactions very nicely: in the wild flax–flax rust pathosystem more resistant wild flax harboured more virulent strains of the rust [1,2]. Similarly, the co-occurrence of virulent strains of the powdery mildew Podosphaera plantaginis and more resistant plants of Plantago lanceolata have also been documented [3,4]. Furthermore, complex multi-host and multi-pathogen systems, with clear differences at a regional scale, have been observed in anther smut-fungi-infecting Dianthus hosts in the southern European Alps [5].
One molecular mechanism that is thought to drive mutual adaptation is a gene-for-gene interaction [6], where single-gene-encoded pathogen molecules (avirulence factors) are recognized by specific receptors (major resistance genes) present in resistant host plants.
With an aim to understand the major gene-mediated resistance diversity in a geographical context, we use Solanum chilense, 1 of the 17 wild tomato species known. The species range spreads from southern Peru, where it inhabits both arid lowland and relatively wet, high-altitude regions (up to 3500 m above sea level), down to the edges of the Atacama Desert in Chile [7,8]. This varied distribution results in sub-populations that encounter different environmental challenges. Ultimately, these different habitats lead to genetic differentiation within the species. The S. chilense populations can be clustered in four groups: north, central, southern highlands, and southern lowlands. The southern highlands and southern lowlands populations are derived from the central group [9]. This divergence was confirmed by whole-genome sequencing. Coalescent simulations revealed that migrations happened from the central group southward 50 000 to 200 000 years ago [10]. Adaptations to abiotic factors have been highlighted in these populations [11]. In addition, these habitats are expected to be home to different biotic stressors, which are anticipated to result in genetic variation in pathogen response. Differences were observed in resistance properties of the various S. chilense populations against three pathogens, Alternaria solani, Phytophthora infestans, and a Fusarium sp. [12]. Moreover, large genetic variation has been observed within S. chilense populations in major pathogen resistance genes of the nucleotide-binding leucine-rich repeat (NLR) family [13]. A study covering the whole species range of S. chilense shows evidence that the selection pressure imposed on the individual NLRs differs for each of the populations [10]. This phenotypic and genotypic diversity observed in former studies in natural populations of S. chilense makes it a system of choice for this study.
To evaluate defence response in S. chilense, we use the model system of tomato and the fungus Cladosporium fulvum syn. Passalora fulva. C. fulvum causes tomato leaf mould and has been a major problem as a greenhouse pathogen. C. fulvum is a globally occurring pathogen with high genetic diversity [14]. It is thought to be a natural pathogen of tomato and originated in the Andean region [15]. The fungus secretes effectors as avirulence factors into the apoplast, following the gene-for-gene model of interaction for successful colonization, which can be recognized by a resistant host. As a result of selection pressure imposed by virulent strains of C. fulvum, several of the wild tomato species have evolved resistance genes whose products mediate recognition of the Avrs secreted by C. fulvum [16]. Resistance is achieved when Avr-activated defence leads to a hypersensitive response (HR). C. fulvum (Cf) resistance genes belong to the receptor-like proteins (RLP) and were first identified in wild tomato species [17]. Cf-9 first found from S. pimpinellifolium [18] and Cf-4 from S. habrochaites [19] are two very well-studied Cf genes. Their gene products recognize Avr9 and Avr4, respectively. Cf-variants resulting from single nucleotide polymorphisms (SNPs) or intragenic recombination have been reported [20,21]. Cf-9 and Cf-4 and their homologues are reported to be located on the same locus, called Homologues of Cladosporium fulvum resistance gene Cf-9 (Hcr9s). For a related gene family, Hcr2, in S. pimpinellifolium, several putative homologues show geographic diversity, but their function remains unknown [22,23].
Species-wide studies showing the interaction of a pathogen and a wild plant species in a well-resolved geographical context with a focus on major gene resistance are limited. We investigated the interaction of C. fulvum with S. chilense throughout populations covering the complete geographical range of the host. We show that S. chilense plants from different locations show differences in their resistance to the fungus, including the complete loss of resistance in some populations. By investigating the well-characterized genes of the Hcr9 locus, we furthermore place major gene-mediated immunity in a geographical context.
2. Materials and methods
(a). Plants and fungal material
Seeds of 15 accessions (LA1958, LA1963, LA2746, LA2747, LA2931, LA2932, LA2750, LA2959, LA3111, LA3784, LA3786, LA4107, LA4117A, LA4118, and LA4330) of S. chilense were obtained from the C. M. Rick Tomato Genetics Resource Center of the University of California, Davis (TGRC UC-Davis) (http://tgrc.ucdavis.edu/). The geographical locations of the accessions and the number of plants tested are described in electronic supplementary material, table S1. Each accession represents a collection of seeds from a wild population and is propagated in a glasshouse by the TGRC. It has been shown that such propagation does not lead to the loss of genetic diversity [9]. Hence, each accession can be considered to represent the diversity in the wild plant populations. Introgression lines S. lycopersicum cv Moneymaker (MM) with Cf-9, Cf-4, and Cf-5 were available [24], and S. pimpinellifolium (LP12) was provided by the Center for Genomic Resources of the Netherlands.
Per accession, 8–17 plants were grown in a glasshouse (minimum daytime temperature: 24°C, 16 h of light). Adult plants were cut at bi-weekly intervals to assure the presence of mature, fully developed leaves during each repetition of the experiment.
A strain of C. fulvum race 5 was used in this study and is known to be virulent on various S. lycopersicum varieties. The strain was maintained on 1/2 potato dextrose agar (PDA) medium and incubated at 16°C in the dark.
(b). Visualization of infection phenotype and quantification of Cladosporium fulvum biomass from tomato leaves
To test the interaction of S. chilense and C. fulvum, plants of population LA3111, LA4330 and MM-Cf-9, MM-Cf-5 were propagated using stem cutting and were maintained for three weeks in a glasshouse (minimum daytime temperature: 24°C, 16 h of light). Spray inoculation of C. fulvum (20 000 conidia/ml) was performed on the plants and water-inoculated plants served as a negative control. Plants after inoculation were maintained at 24°C and 16 h of light, 100% humidity for the first 2 days and 80% humidity afterwards.
Photographs of inoculated plants were taken 14 days post-inoculation (dpi). Microscopy (Zeiss Imager Z1 m) was performed from 7–19 dpi on bleached leaves following staining in acetic acid (25%) 1 : 9 + ink (Königsblau, Pelikan, 4001) and washing in water.
Leaflets were collected at 14 dpi. DNA was isolated using cetyl trimethyl ammonium bromide (CTAB) [25]. Infection loads in inoculated leaves were quantified by qPCR using the Maxima SYBR Green Master Mix (2X) with ROX solution (Thermo Scientific). qPCR reactions were performed on an AriaMx Real-Time PCR system (Agilent Technologies, Waldbronn, Germany) with the primer pairs RS158 (5′-GTCTCCGGCTGAGCAGTT-3′)/ ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and RS001 (5′-GCCTACCATGAGCAGCTTTC-3′)/ RS002 (5′-CAATGCGTGAGAAGACCTCA-3′), annealing to the internal transcribed spacer (ITS) region of C. fulvum and α-tubulin of the plant, respectively. The experiment was performed on three plants of MM-Cf-9 and MM-Cf-5 each and five plants of LA3111 and LA4330 each. All samples were evaluated in three technical replicates and Cq differences higher than one among technical replicates were not considered. The Cq values and the linear equations from the sensitivity graphs were used to calculate the quantities of pathogen and plant DNA in each sample (electronic supplementary material, figure S1). ANOVA, with post hoc Tukey honest significant difference test, was performed using R command TukeyHSD.
(c). Apoplastic washing fluid and Avr infiltration assays
To phenotype the populations of S. chilense, apoplastic washing fluid (AF) containing the complete set of C. fulvum Avrs, except for Avr5, was obtained by isolating AF from leaves of MM-Cf-5 plants colonized by race 5 of C. fulvum, at 10 to 14 days after inoculation. A preparation of Avr9 concentrated from AF was obtained by acetone precipitation, and Avr4 was produced in the yeast Pichia pastoris [26]. The AF, Avr9, and Avr4 preparations were infiltrated from the lower side of fully expanded leaves of the different plants. As controls, infiltrations were performed in the introgression lines Cf-0, Cf-4, and Cf-9 (electronic supplementary material, figure S2). Experiments were performed in adult plants in three independent biological replicates, with two technical replicates per plant. Scoring was done 2 to 7 days post infiltration.
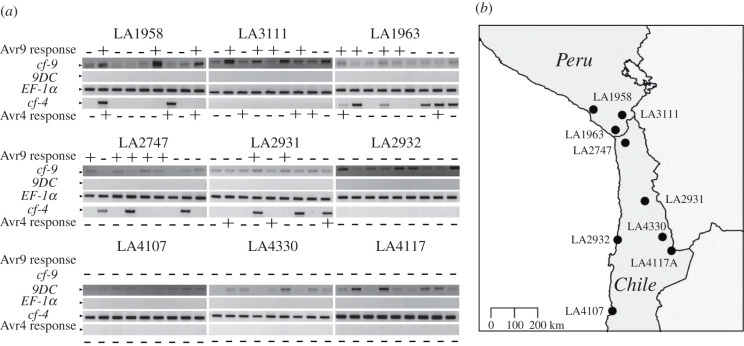
(d). Presence of Cf-9-, 9DC-, and Cf-4-specific regions in different Solanum chilense populations
We screened nine S. chilense populations (nine plants each) to test the presence of unique regions of Cf-9, 9DC, and Cf-4 with PCR amplification using gene-specific primers CS5-CS1 and DS1-CS1 for Cf-9 and 9DC, respectively [21]. Primer pair PSK047 (5′-ACGACAGAAGAACTC-3′)/ PSK050 (5′-GATGGAATTGGTCCTT-3′) was designed to amplify the canonical Cf-4 domain (electronic supplementary material, figure S3). DNA samples were obtained as in a previous study [10]. PCR on gDNA was performed using Promega Green GoTaq® Polymerase. Products were analysed in 1% agarose gel electrophoresis. As PCR control, we used elongation factor 1 alpha, amplified with the primer pair 5′-GTCCCCATCTCTGGTTTTGA-3′/ 5′-GGGTCATCTTTGGAGTTGGA-3′. MM-Cf-9 and MM-Cf-4, and LP12 served as positive and negative controls.
(e). Semi-quantitative evaluation of the expression of a Cf-9 homologue
Leaf material was flash frozen 8 h after Avr9 or water infiltration. RNA was extracted using the RNeasy plant mini kit (Qiagen) and cDNA was synthesized using the QuantiTect reverse transcription kit (Qiagen). Amplification of the Cf-9 homologue was performed with primer pair binding on the start and end of the open reading frame (ORF), PSK009 (5′-ATGGATTGTGTAAAACTTGTATTCCT -3′) /PSK010 (5′-CTAATATCTTTTCTTGTGCTTTTTCA -3′). The product was visualized in 1% agarose gel.
(f). Whole-genome sequence analyses for Cf co-receptors in Solanum chilense
We extracted the sequences for four components of the Cf protein signalling complex: SUPPRESSOR OF BIR1 (SORBIR1), SOMATIC EMBRYOGENESIS RECEPTOR-KINASE 3A (SERK3a), and AVR9/CF-9-INDUCED KINASE 1 (ACIK1) [27], and for an additional potential co-receptor, BIR2 [28] from National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/. To identify the genomic sequences in S. chilense, we performed a BLAST search against the S. chilense reference genome [10]. SOBIR1, SEKR3a, and ACIK1 yielded one unequivocal target sequence. BIR2 yielded two hits, of which we dubbed the second one BIR2b.
To look for polymorphisms in one responsive plant from LA3111, and one each from non-responsive southern populations LA2932 and LA4330 [13], we obtained the genomic coordinates of the genes of interest from our reference genome and called the SNPs in the region of interest (samtools mpileup and bcftools call (-mv -Oz)) [29,30] and removed low-quality indels. The resulting vcf files were tabulated (tabix) and consensus sequences extracted using bcftools consensus. We used gffread from gffutils [31] to obtain the correct coding sequence for each of the plants. Multiple sequence alignments were made and inspected using aliview [32].
3. Results
(a). Solanum chilense populations show differences in resistance against Cladosporium fulvum
We found that distinct populations of S. chilense showed different outcomes of leaf mould pathogenesis after spray inoculation of the plants with a conidial suspension of C. fulvum race 5. Plants from LA3111 showed similar phenotypes to our resistant control plant MM-Cf-9, a recombinant inbred line of S. lycopersicum cv. MoneyMaker with introgressed Cf-9 (resistant to C. fulvum race 5 as this race produces Avr9). Plants from LA4330 showed a phenotype similar to our susceptible control MM-Cf-5, a Cf-5 introgression line of S. lycopersicum (resistance is circumvented due to the loss of the Avr5 gene by race 5) [33] (figure 1a,b). We further quantified C. fulvum DNA load and found significant differences in pathogen DNA contents between the tested plants (p < 0.001, ANOVA). Differences can be seen between the resistant and susceptible control plants (p = 0.05, TukeyHSD) and LA4330 showed a significantly higher (p < 0.001, TukeyHSD) presence of C. fulvum DNA than LA3111 (figure 1c). These findings show that C. fulvum can infect S. chilense and that the populations of this host differ in resistance.
Figure 1.
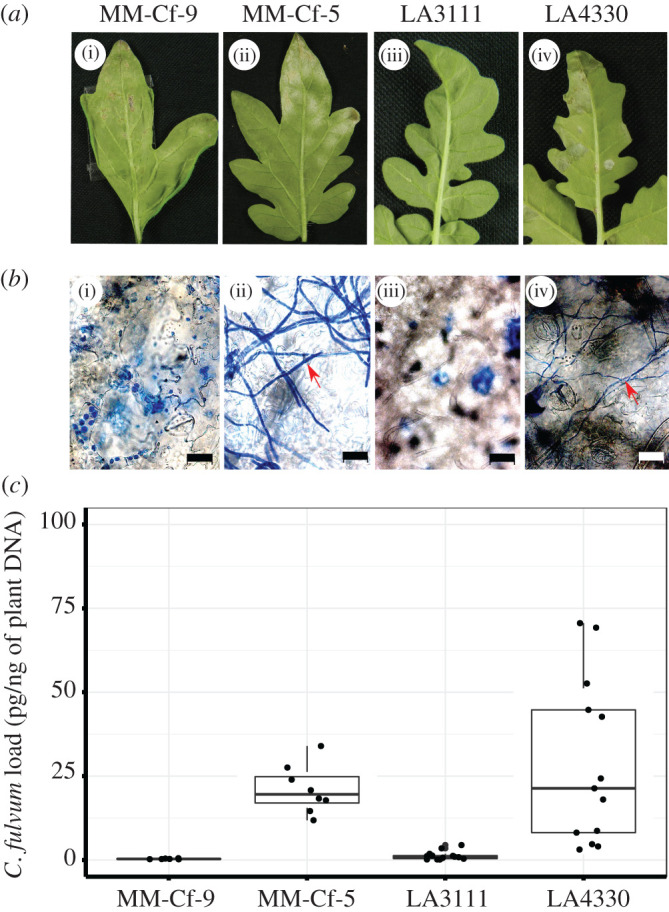
Tomato leaves inoculated with 2 × 104 conidia ml−1. (a) i–iv inoculated leaves harvested at 14 dpi of MM-Cf-9, MM-Cf-5, and S. chilense population LA3111 and LA4330. (b) Microscopic pictures of bleached leaves (scale bar = 20 µm), after staining with ink (mycelium of pathogen indicated with red arrows). (c) Quantification of C. fulvum DNA load in pg/ng of plant DNA after 14 dpi of inoculation in three plants per MM control and five plants per S. chilense population. All the plants were evaluated in three technical replicates and each data point shows pathogen load per technical replicate. (Online version in version.)
(b). Southern S. chilense populations do not recognize Cladosporium fulvum effectors
The appearance of visible HR upon effector infiltration is an efficient and reliable way to test for the resistance properties of tomato plants. We performed an infiltration with AF of infected tomato plants, which is sufficient to trigger an HR in plants carrying matching Cf genes. C. fulvum race 5 was used for the inoculation, thus the AF contains all known secreted C. fulvum effectors, except Avr5. We infiltrated the AF in leaves of 155 individuals, representing 15 different populations. We did not observe any HR-associated recognition of an effector present in the AF by any of the populations belonging to the southern highlands and southern coastal groups (figure 2, electronic supplementary material, table S1). Populations from the northern and central regions recognized at least one of the effectors present in the AF. However, recognition did not happen in all tested plants of these populations. Rather, we observed differences in recognition capacities within the population. Populations from central regions showed recognition, in 10–100% of the plants tested in the different populations. Two northern populations showed AF recognition in 75% and 80% of the plants (figure 2, electronic supplementary material, table S1).
Figure 2.
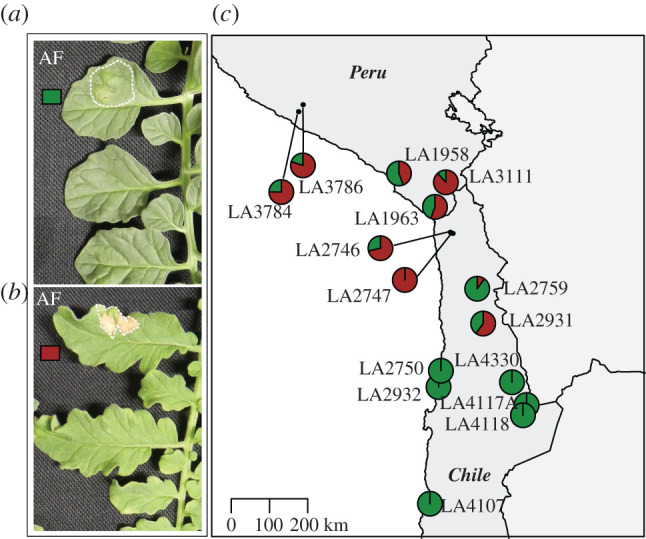
Infiltration of AF in populations of S. chilense. Leaf of an individual plant with no recognition of components present in the infiltrated AF (a) and leaf of an individual plant recognizing at least one component present in the AF (b). The infiltrated areas are indicated with white dotted lines. The map on the right shows the geographical distribution of AF component perception in S. chilense. The map shows the geographically distinct populations of S. chilense and their response to infiltration with AF. Each pie chart indicates one population with 8 to 17 individuals (c). The brick red fraction represents plants that respond to C. fulvum AF, whereas green indicates the fraction of non-responding plants. (Online version in colour.)
(c). Differential Avr9 and Avr4 recognition patterns are present in Solanum chilense
Recognition of Avr9 and Avr4 has been postulated to be an important conserved feature in wild tomato species in order to maintain resistance to C. fulvum [20]. To understand the recognition of these Avrs in detail, we performed additional infiltration assays with individual effectors.
To test whether S. chilense plants are able to specifically recognize Avr9, we infiltrated the plants with a preparation enriched for Avr9. Populations from the central region showed a large variation in the ability to recognize Avr9, ranging from 10 to 80% of the plants that were tested in individual populations (electronic supplementary material, table S2). The two northern populations that were tested showed Avr9 recognition for 20% and 37.5% (figure 3).
Figure 3.
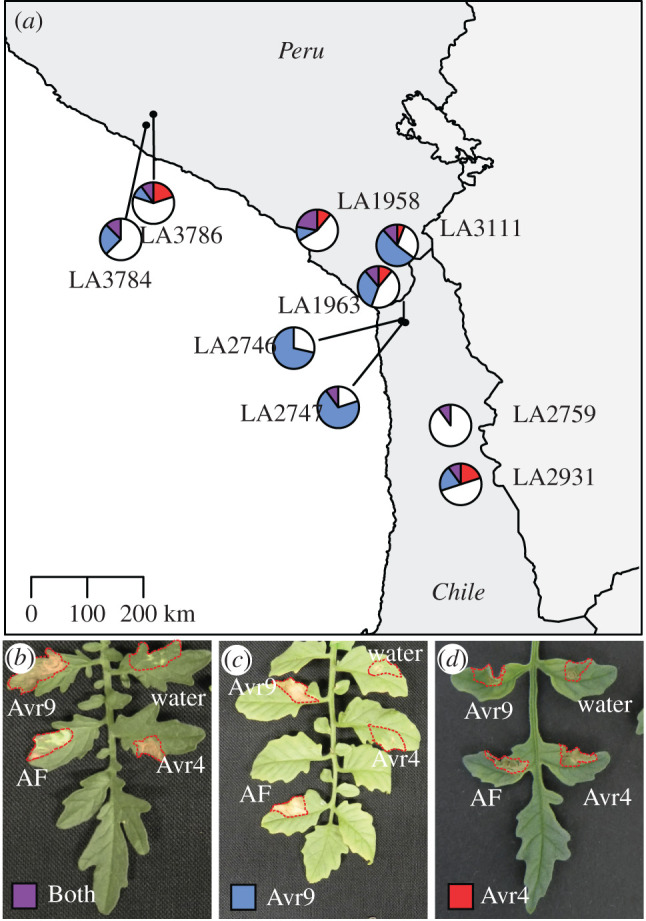
(a–d) Geographical distribution of Avr9 and Avr4 perception in S. chilense. The map shows populations of S. chilense and their response to infiltration with Avr9 and Avr4 (purple when there is an HR-associated response to both), Avr9 (blue when there is an HR-associated response), and Avr4 (red when there is an HR-associated response). White sectors indicate plants showing no recognition of either Avr9 or Avr4. The infiltrated areas are indicated with red dotted lines. Each pie chart indicates one population with 8 to 17 individuals. (Online version in colour.)
We tested the ability of S. chilense to recognize Avr4 in a similar fashion, using Avr4 produced in Pichia pastoris. In the northern populations, we observed 12.5% and 30% Avr4 recognition and in the central populations 0–33%. Contrary to previous reports that Cf-4 and Cf-9 are mutually exclusive, we also found plants that were able to recognize both Avr4 and Avr9 (figure 3 and electronic supplementary material, table S2).
(d). The presence of canonical Cf-9-, 9DC-, and Cf-4-specific regions in different Solanum chilense populations does not correlate with their recognition properties
In S. pimpinellifolium, either Cf-9 or closely related 9DC are responsible for Avr9 recognition [21]. Cf-9 or 9DC gene sequences in S. chilense have not been reported to date and putative full-length Cf-9 or 9DC genes cannot be found in the currently available reference genome sequence, likely due to misassemblies of the repetitive leucine-rich repeat regions [13]. To investigate, which gene is responsible for Avr9 recognition in S. chilense, we looked into the presence of Cf-9 in genomic DNA using gene-specific primer sequences that were used before to identify these genes [21]. MM-Cf-9, LP12, and MM-Cf-4 served as positive and negative controls. The Cf-9-specific sequence was amplified from all plants, although with different efficiency. Strikingly, there was no association between the presence or abundance of the amplicon and the response to Avr9 (figure 4a). We found a complete absence of the 9DC canonical region in our populations (figure 4a).
Figure 4.
Amplification of the canonical Cf-9, 9DC, and Cf-4 region in geographically distinct populations of S. chilense. (a) The CS5-CS1 primer pair was used to amplify a fragment of Cf-9 (378 bp), the DS1-CS1 primer pair for 9DC (507 bp), and the PSK047/PSK050 primer pair for Cf-4 (728 bp). As a PCR control, part of the coding region of elongation factor 1 alpha (EF-1α) was amplified using the primer pair RS003/RS004 (400 bp). MM-Cf-9 and MM-CF-4, and LP12 (9DC) were used as controls (electronic supplementary material, figure S2). + or – are indicative of Avr9 and Avr4 responsiveness upon infiltration of these effectors (figure 2). (b) Map shows the populations used in the analysis. (Online version in colour.)
We also evaluated the presence of Cf-4, using newly designed primers that bind to the intron that defines the difference between Cf-4 and Cf-9 (electronic supplementary material, figure S3) and amplify a 786 bp product that is only present in Cf-4. This Cf-4-specific region was evident in few plants belonging to central populations, suggesting that unlike reports about other Solanum. spp., Cf-4 and Cf-9 are not mutually exclusive in S. chilense (figure 4, electronic supplementary material, table S3). The presence of this gene fragment corresponding to this canonical Cf-4 region in plants non-responsive to Avr4 and the absence in responders suggests that other or new recombinant gene products with differences in their function exist in the different populations.
(e). Cf-9 is expressed in a southern population
To test whether failed Cf-9(-like) gene expression is responsible for the complete loss of Avr9-triggered HR in the southern populations, we extracted RNA from 10 individuals from the LA4330 population. We used a primer pair that should allow us to amplify cDNA originating from Cf-9 orthologues and performed semi-quantitative RT-PCR on plants infiltrated with water and Avr9.
Without treatment, we evidenced transcripts in all tested S. chilense plants, as well as in our MM-Cf-9 control plant (electronic supplementary material, figure S4), indicating that steady-state Cf-9 gene expression is not affected in the southern S. chilense populations. Using templates from the MM-Cf-9 plant, we saw a stronger band at 8 h after treatment, indicative of the upregulation and positive feedback induced by Cf-9. This was not visible in the southern S. chilense population (electronic supplementary material, figure S4).
(f). Mutations in Cf-co-receptors in the southern populations
Seeing that steady-state expression levels of Cf-9 are not affected in the southern populations and complete loss of resistance is unlikely to result from detrimental mutations on all individual Cf genes. We hypothesize that general loss of Cf responses likely results from changes in Cf regulatory genes, rather than mutations in the individual Cf genes themselves. Several co-receptors are known to regulate the Cf protein function. We examined genome sequence data that are available for three plants, representative for the central (LA3111), southern coastal (LA2932), and southern highlands (LA4330) groups. [13]. We extracted and aligned the genomic sequences of the co-receptors SERK3a (also known as BRI1-ASSOCIATED KINASE 1, BAK1) and SOBIR1, the adaptor ACIK1, required for regulation of the complex, as well as BIR2 and its homologue BIR2b (electronic supplementary material, table S4). In all cases, the extracted sequence data show polymorphisms between the reference genome of S. lycopersicum Heinz1706 and the Avr4 and Avr9 responsive plant from LA3111. In-frame indels were found for BIR2b between its sequence in S. lycopersicum and those in the three S. chilense populations, yet, the three S. chilense populations show similar, complete sequences. Several unique polymorphisms exist in the southern populations, e.g. occurring in the plants from LA2932 and LA4330, but not in the plant from population LA3111 from the central region or in the S. lycopersicum reference genome sequence, resulting in up to 18 non-synonymous amino acid changes for SERK3a in the plant from LA4330 (table 1). In ACIK1, both the LA2932 and the LA4330 plant have unique indels neither present in the S. lycopersicum reference genome sequence nor in the LA3111 plant (table 1). These results suggest that there are various amino acid changes in these receptors, co-receptors, and adaptors that could be responsible for the observed loss of resistance.
Table 1.
Number of non-synonymous SNPs or indels observed in the open reading frame of genes encoding co-receptors or adaptors of Cf genes in the two southern populations LA2932 and LA4330.
| gene name | SNPs/indels in LA2932 | SNPs/indels in LA4330 | SNPs/indels in both LA2932 and LA4330 |
|---|---|---|---|
| SOBIR1 | 4/0 | 1/0 | 0/0 |
| SERK3a | 4/0 | 18/1 | 1/0 |
| ACIK1 | 1/0 | 3/0 | 2/0 |
| BIR2 | 2/0 | 3/0 | 0/0 |
| BIR2b | 2/0 | 1/0 | 0/1 |
4. Discussion
C. fulvum is a pathogen of tomato and known functional Cf resistance genes have been identified in several wild tomato species. Yet, the physiological interaction between C. fulvum and wild tomato species had not been documented. We show that C. fulvum is able to infect S. chilense. Plants from a S. chilense population from the southern edge of the species range (LA4330) show higher susceptibility when compared to plants from a population from the central part of the range (LA3111). Thus, we show a compatible interaction between C. fulvum and wild tomato species, but also clear differences in resistance between the host populations.
To test further if these differences follow a geographical pattern in a species-wide context, we exploited the well-defined interaction between C. fulvum and tomato, which is governed by gene-for-gene interactions, in which secreted Avrs from C. fulvum are recognized by corresponding Cf genes from tomato [16]. We tested recognition of such Avrs in 15 populations of S. chilense from different locations, to understand whether the observed resistance and susceptibility within S. chilense populations to C. fulvum follows a specific geographical pattern and reached two important conclusions. First of all, Avr recognition in the northern and central regions is not as conserved as previously hypothesized [20], and second, Avr recognition appears to have been completely lost at the southern edge of the species range.
Previous reports suggest that Avr9 and Avr4 are the main factors in the recognition of C. fulvum and this recognition is conserved throughout all wild tomato species [20]. In previous work, S. chilense Avr4 recognition is documented in 16 out of 20 tested populations [20]. In line with our results, four of the tested populations in the previous study belonged to the southern genotype groups. We observe Avr9 recognition to be present more often than Avr4 recognition. This is somewhat contrasting with the previous study where they did not report Avr9 recognition, although similar Avr9 has been used. The differences in results can be explained by the fact that all previous studies used young seedlings to allow quick screening of the plants. We used adult plants to better reflect natural situations, and we performed repeated infiltration of the same plant. Indeed, we observed differences in the strength of the HR upon Avr9 infiltration in young and fully developed mature leaves (electronic supplementary material, figure S5). Our findings pose the first evidence of Avr9 recognition in S. chilense and suggest that detailed testing of other species under different conditions might yield novel interesting results.
Avr9 is recognized by gene products of two Cf-9 homologues, referred to as Cf-9 and 9DC [21], Avr4 is recognized by Cf-4. We evaluated the presence of known canonical domains that define Cf-9, 9DC, and Cf-4 and found that the tested plants in all our populations do have the Cf-9 domain but lack the 9DC domain. The presence of the Cf-9 domain is an interesting contrast with previous findings [20] that showed the presence of 9DC to be predominant in another wild tomato species, S. pimpinellifolium, and a complete loss of Cf-9 in southern populations of that species. The southernmost S. pimpinellifolium populations are geographically relatively close to the most northern S. chilense populations, yet they populate different ecological niches [7]. The canonical Cf-4 domain is present in some individuals, but its presence does not correlate with our phenotypic data on the development of an HR upon infiltration with the Avr4 protein. The recognizing region might be missing in the gene coding for the Avr4-responsive Cf protein. This would not be surprising as recombination events have been reported for genes at the Hcr9 locus, albeit on a phylogenetic rather than population scale [21]. Cf-4 and Cf-9 were assumed to be mutually exclusive or even allelic. Crossings between MM lines carrying Cf-4 and Cf-9 resulted in extreme genetic instability in the offspring [19,34]. We show the presence of the canonical regions of Cf-4 and Cf-9 as well as dual recognition of Avr4 and Avr9 in a subset of plants (electronic supplementary material, table S3), indicating the presence of both genes previously considered allelic. The genetic basis of this is yet unclear but possibly due to heterozygosity at the locus, the presence of new recombinant genes with dual functionality, or the presence of multiple recombined paralogs.
Recombination and gene conversion has been shown to play a major role in gene family evolution for the RLP family as well as other resistance gene families [34–36] and in Arabidopsis thaliana defence-associated RLPs tend to show various degrees of copy number variation between accessions [37]. Gene conversions, or recombination, of an NLR gene can also be observed between accessions of another wild Solanum sp., where it appears to be an alternative mechanism to maintain different functional alleles [38]. Thus, we conclude that the Cf gene family is also likely not conserved sensu stricto and speculate that a large number of possibly functional alleles are formed and maintained through intragenic micro recombinations, not just on a phylogenetic scale, but also between or even within populations of the same species.
The loss of Avr recognition in southern populations could be because these plants recognize different Avrs which are absent in our C. fulvum race 5. Yet, northern and central populations of S. chilense do possess resistance against C. fulvum race 5. Our apoplastic extract contains at least 70 secreted effectors, which are all potential Avrs [39]. The chance of losing recognition for all of these effectors by mutations in the matching receptors is highly unlikely. Moreover, Cf-9 homologues appear to be expressed in both Avr9-recognizing and non-recognizing plants. Thus, the accumulation of deleterious mutations in genes encoding essential co-receptors, like SOBIR1 or BAK1 might have caused the loss of Cf recognition in general. It is shown that single point mutations in such co-receptors lead to a loss in the ability to induce HR when expressed in heterologous systems and in A. thaliana [40,41]. Genomic analyses for non-responding plants from the southern populations revealed several non-synonymous mutations, as well as indels in multiple Cf co-receptor-encoding genes. Follow-up studies should determine whether and which one of the many altered genes are causal. Since the co-receptors have multiple functions apart from Cf protein signalling, such experiments might simultaneously shed light on how the different components of the signalling network interact.
Loss of resistance over time at the population level is poorly understood, with some theories possibly explaining this mechanism [42]. For instance, evolutionary loss of resistance in certain populations might happen as a result of genetic drift, e.g. after a severe bottleneck. However, if these populations encounter any pathogen pressure afterwards, this would be detrimental. Loss of resistance can also be the result of selection due to the much-debated, assumed fitness costs of carrying obsolete resistance genes [43,44]. Loss of resistance, as observed in our system, further suggests that a mechanism of ecological feedback might take place, where the ecology of the population becomes a driving force to lose resistance or to maintain it. Therefore, the absence of the C. fulvum pathogen in the southern locations, due to extremely dry climatic conditions, which are not suitable for infection by C. fulvum, would be a plausible explanation. Such a role of the distribution of C. fulvum in shaping the evolutionary distribution of Cf-2 homologues has also been proposed [23]. Interestingly, higher susceptibility to other pathogens also requiring relatively high humidity for successful infection, like Phytophthora infestans and Alternaria sp., was already described for southern populations of S. chilense [12]. Seeing that some co-receptors potentially play a role in abiotic stress responses, another possibility could be that the loss of resistance comes as a trade-off for environmental adaptation. BAK1/SERK3a is a co-regulator, in not only defence responses, but also plays a role in general cell regulatory processes and is drought-responsive [45]. Beneficial mutations related to drought adaptation might generate a trade-off in resistance. Yet, this is likely to be intrinsically coupled to a decreased pathogen pressure. Another possibility in regard to the loss of resistance to C. fulvum in the south can be a trade-off mechanism due to the presence of different pathogens in these regions which are capable of exploiting the resistance allele for C. fulvum present in these populations as a susceptible allele as shown before in a different host-pathogen system [46].
In conclusion, we show that the function of the Hcr9 locus is much more complex than was thought before [20,21,34]. This might in part be due to the stronger niche differentiation, or the larger heterozygosity of S. chilense specifically [47]. However, it seems more likely that there is a lot of undiscovered diversity in the Cf gene family. Furthermore, we provide an example of the loss of resistance in a wild tomato species at the edge of its geographical distribution, possibly explained by changes in the underlying immune receptor complexes. Overall, this study places major gene-mediated molecular defence mechanisms in a geographical context. Thereby, it is tempting to propose that the corresponding regional pattern of responses is explained by a loss of resistance that evolved when the plant species expanded its distribution area southwards where pathogen pressure was absent due to climatic constraints.
Supplementary Material
Acknowledgements
We would like to thank Liza Keitel and Lina Muñoz for help with the experiments, Prof. Aurélien Tellier for sharing the S. chilense populations and Sabine Zuber, Bärbel Breulmann, and Anneliese Keil for maintaining them.
Data accessibility
Raw data for infection, infiltration assays, and qPCR, extracted alignment files for the genes mentioned in this study and scripts used for graphs and calculations are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.866t1g1p9 [48]. Raw sequence datasets have been deposited to NCBI as reported before [10].
Authors' contributions
R.S., R.H., P.S.K., and M.J. were involved in conceptualization; P.S.K., M.S.S., G.Z., and D.S. performed the experiments; M.J. contributed materials; R.S. acquired funding; P.S.K. and R.S. wrote the paper.
Competing interests
The authors declare that no competing interests exist.
Funding
This work was funded through the German Science Foundation (DFG), SFB924. Bioinformatics analyses were performed on the BMBF-funded de.NBI Cloud within the German Network for Bioinformatics Infrastructure (de.NBI) (031A537B, 031A533A, 031A538A, 031A533B, 031A535A, 031A537C, 031A534A, 031A532B).
References
- 1.Thrall PH, Burdon JJ, Bever JD. 2002. Local adaptation in the Linum marginale-Melampsora lini host-pathogen interaction. Evolution 56, 1340–1351. ( 10.1111/j.0014-3820.2002.tb01448.x) [DOI] [PubMed] [Google Scholar]
- 2.Thrall PH. 2003. Evolution of virulence in a plant host-pathogen metapopulation. Science 299, 1735–1737. ( 10.1126/science.1080070) [DOI] [PubMed] [Google Scholar]
- 3.Laine A-L. 2005. Spatial scale of local adaptation in a plant-pathogen metapopulation. J. Evol. Biol. 18, 930–938. ( 10.1111/j.1420-9101.2005.00933.x) [DOI] [PubMed] [Google Scholar]
- 4.Soubeyrand S, Laine AL, Hanski I, Penttinen A. 2009. Spatiotemporal structure of host-pathogen interactions in a metapopulation. Am. Nat. 174, 308–320. ( 10.1086/603624) [DOI] [PubMed] [Google Scholar]
- 5.Petit E, et al. 2017. Co-occurrence and hybridization of anther-smut pathogens specialized on Dianthus hosts. Mol. Ecol. 26, 1877–1890. ( 10.1111/mec.14073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flor HH. 1971. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9, 275–296. ( 10.1146/annurev.py.09.090171.001423) [DOI] [Google Scholar]
- 7.Peralta IE, Spooner DM, Knapp S. 2008. Taxonomy of wild tomatoes and their relatives (Solanum sect. Lycopersicoides, sect. Juglandifolia, sect. Lycopersicon; Solanaceae). American; Society of Plant Taxonomists; See https://agris.fao.org/agris-search/search.do?recordID=US201300127979. [Google Scholar]
- 8.Chetelat RT, Pertuzé RA, Faúndez L, Graham EB, Jones CM. 2009. Distribution, ecology and reproductive biology of wild tomatoes and related nightshades from the Atacama Desert region of northern Chile. Euphytica 167, 77–93. ( 10.1007/s10681-008-9863-6) [DOI] [Google Scholar]
- 9.Böndel KB, Lainer H, Nosenko T, Mboup M, Tellier A, Stephan W. 2015. North–South colonization associated with local adaptation of the wild tomato species Solanum chilense. Mol. Biol. Evol. 32, 2932–2943. ( 10.1093/molbev/msv166) [DOI] [PubMed] [Google Scholar]
- 10.Stam R, Silva-Arias GA, Tellier A. 2019. Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats New Phytol. 224, 367–379. ( 10.1111/nph.16017) [DOI] [PubMed] [Google Scholar]
- 11.Böndel KB, Nosenko T, Stephan W. 2018 Signatures of natural selection in abiotic stress-responsive genes of Solanum chilense. R. Soc. Open Sci. 5, 171198 ( 10.1098/rsos.171198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stam R, Scheikl D, Tellier A. 2017. The wild tomato species Solanum chilense shows variation in pathogen resistance between geographically distinct populations. PeerJ 5, e2910 ( 10.7717/peerj.2910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stam R, Nosenko T, Hörger AC, Stephan W, Seidel M, Kuhn JMM, Haberer G, Tellier A. 2019. The de Novo reference genome and transcriptome assemblies of the wild tomato species Solanum chilense highlights birth and death of NLR genes between tomato species. G3: Genes, Genomes, Genetics 9, 3933–3941. ( 10.1534/g3.119.400529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iida Y, et al. 2015. Novel mutations detected in Avirulence genes overcoming tomato Cf resistance genes in isolates of a Japanese population of Cladosporium fulvum. PLoS ONE 10, e0123271 ( 10.1371/journal.pone.0123271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wit PJGM, et al. 2012. The genomes of the fungal plant pathogens Cladosporium fulvum and Dothistroma septosporum reveal adaptation to different hosts and lifestyles but also signatures of common ancestry. PLoS Genet. 8, e1003088 ( 10.1371/journal.pgen.1003088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joosten M, de Wit P. 1999. The tomato–Cladosporium fulvum interaction: a versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37, 335–367. ( 10.1146/annurev.phyto.37.1.335) [DOI] [PubMed] [Google Scholar]
- 17.Thomas CM, Dixon MS, Parniske M, Golstein C, Jones JDG. 1998. Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Phil. Trans. R. Soc. Lond. B 353, 1413–1424. ( 10.1098/rstb.1998.0296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones D, Thomas C, Hammond-Kosack K, Balint-Kurti P, Jones J. 1994. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. ( 10.1126/science.7973631) [DOI] [PubMed] [Google Scholar]
- 19.Thomas CM, Jones DA, Parniske M, Harrison K, Balint-Kurti PJ, Hatzixanthis K, Jones JD. 1997. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. ( 10.1105/tpc.9.12.2209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruijt M, Kip DJ, Joosten MHAJ, Brandwagt BF, de Wit PJGM. 2005. The Cf-4 and Cf-9 resistance genes against Cladosporium fulvum are conserved in wild tomato species. MPMI 18, 1011–1021. ( 10.1094/MPMI-18-1011) [DOI] [PubMed] [Google Scholar]
- 21.der Hoorn RALV, Kruijt M, Roth R, Brandwagt BF, Joosten MHAJ, Wit PJGMD. 2001. Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. PNAS 98, 10 493–10 498. ( 10.1073/pnas.181241798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caicedo AL, Schaal BA. 2004. Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. PNAS 101, 17 444–17 449. ( 10.1073/pnas.0407899101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caicedo AL. 2008. Geographic diversity cline of R gene homologs in wild populations of Solanum pimpinellifolium (Solanaceae). Am. J. Botany 95, 393–398. ( 10.3732/ajb.95.3.393) [DOI] [PubMed] [Google Scholar]
- 24.Tigchelaar EC. 1984. Collections of isogenic tomato stocks. Rep. Tomato Genet. Coop. 34, 55–57. [Google Scholar]
- 25.Yan L, Zhang C, Ding L, Ma Z. 2008. Development of a real-time PCR assay for the detection of Cladosporium fulvum in tomato leaves. J. Appl. Microbiol. 104, 1417–1424. ( 10.1111/j.1365-2672.2007.03660.x) [DOI] [PubMed] [Google Scholar]
- 26.van den Burg H. 2019. Studies towards the intrinsic function of the AVR4 and AVR9 elicitors of the fungal tomato pathogen Cladosporium fulvum. PhD Thesis Wageningen University, The Netherlands: See https://edepot.wur.nl/121419. [Google Scholar]
- 27.Wu J. 2020. Regulation and activation of SOBIR1-containing receptor complexes involved in plant immune signalling. The Netherlands: Wageningen University, ISBN 9789463951821. [Google Scholar]
- 28.Halter T, et al. 2014. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. ( 10.1016/j.cub.2013.11.047) [DOI] [PubMed] [Google Scholar]
- 29.Li H, et al. 2009. The sequence alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. ( 10.1093/bioinformatics/btr509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertea G, Pertea M. 2020. GFF utilities: GffRead and GffCompare. F1000Res 9, 304 ( 10.12688/f1000research.23297.1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30, 3276–3278. ( 10.1093/bioinformatics/btu531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mesarich CH, Griffiths SA, van der Burgt A, Ökmen B, Beenen HG, Etalo DW, Joosten MHAJ, de Wit PJGM. 2014. Transcriptome sequencing uncovers the Avr5 avirulence gene of the tomato leaf mold pathogen Cladosporium fulvum. MPMI 27, 846–857. ( 10.1094/MPMI-02-14-0050-R) [DOI] [PubMed] [Google Scholar]
- 34.Parniske M, Hammond-Kosack KE, Golstein C, Thomas CM, Jones DA, Harrison K, Wulff BBH, Jones JDG. 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. ( 10.1016/S0092-8674(00)80470-5) [DOI] [PubMed] [Google Scholar]
- 35.Mondragon-Palomino M, Gaut BS. 2005. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 22, 2444–2456. ( 10.1093/molbev/msi241) [DOI] [PubMed] [Google Scholar]
- 36.Mondragón-Palomino M, Stam R, John-Arputharaj A, Dresselhaus T. 2017. Diversification of defensins and NLRs in Arabidopsis species by different evolutionary mechanisms. BMC Evol. Biol. 17, 255 ( 10.1186/s12862-017-1099-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steidele C, Stam R. 2020. Multi-omics approach highlights differences between functional RLP classes in Arabidopsis thaliana ( 10.1101/2020.08.07.240911) [DOI]
- 38.Witek K, et al. 2020. A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. bioRxiv, 2020.05.15.095497 ( 10.1101/2020.05.15.095497) [DOI]
- 39.Mesarich CH, et al. 2017. Specific hypersensitive response–associated recognition of new apoplastic effectors from Cladosporium fulvum in wild tomato. MPMI 31, 145–162. ( 10.1094/MPMI-05-17-0114-FI) [DOI] [PubMed] [Google Scholar]
- 40.Bi G, Liebrand TWH, Bye RR, Postma J, van der Burgh AM, Robatzek S, Xu X, Joosten MHAJ. 2016. SOBIR1 requires the GxxxG dimerization motif in its transmembrane domain to form constitutive complexes with receptor-like proteins: functional analysis of SOBIR1. Mol. Plant Pathol. 17, 96–107. ( 10.1111/mpp.12266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albert I, Zhang L, Bemm H, Nürnberger T. 2019. Structure-function analysis of immune receptor AtRLP23 with its ligand nlp20 and coreceptors AtSOBIR1 and AtBAK1. MPMI 32, 1038–1046. ( 10.1094/MPMI-09-18-0263-R) [DOI] [PubMed] [Google Scholar]
- 42.Koskella B. 2018. Resistance gained, resistance lost: an explanation for host–parasite coexistence. PLoS Biol. 16, e3000013 ( 10.1371/journal.pbio.3000013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheldon BC, Verhulst S. 1996. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol. Evol. 11, 317–321. ( 10.1016/0169-5347(96)10039-2) [DOI] [PubMed] [Google Scholar]
- 44.Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423, 74–77. ( 10.1038/nature01588) [DOI] [PubMed] [Google Scholar]
- 45.Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C. 2011. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046 ( 10.1371/journal.pgen.1002046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorang JM, Sweat TA, Wolpert TJ. 2007. Plant disease susceptibility conferred by a ‘resistance’ gene. PNAS 104, 14 861–14 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moyle LC. 2008. Ecological and evolutionary genomics in the wild tomatoes (Solanum SECT. Lycopersicon). Evolution 62, 2995–3013. ( 10.1111/j.1558-5646.2008.00487.x) [DOI] [PubMed] [Google Scholar]
- 48.Kahlon PS, Seta SM, Zander G, Scheikl D, Hückelhoven R, Joosten MHAJ, Stam R. 2020. Data from: Population studies of the wild tomato species Solanum chilense reveal geographically structured major gene-mediated pathogen resistance. Dryad Digital Repository ( 10.5061/dryad.866t1g1p9) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Kahlon PS, Seta SM, Zander G, Scheikl D, Hückelhoven R, Joosten MHAJ, Stam R. 2020. Data from: Population studies of the wild tomato species Solanum chilense reveal geographically structured major gene-mediated pathogen resistance. Dryad Digital Repository ( 10.5061/dryad.866t1g1p9) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw data for infection, infiltration assays, and qPCR, extracted alignment files for the genes mentioned in this study and scripts used for graphs and calculations are available from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.866t1g1p9 [48]. Raw sequence datasets have been deposited to NCBI as reported before [10].