Abstract
Maintaining homeostasis of the decidual immune microenvironment at the maternal–fetal interface is essential for placentation and reproductive success. Although distinct decidual immune cell subpopulations have been identified under normal conditions, systematic understanding of the spectrum and heterogeneity of leukocytes under recurrent miscarriage in human deciduas remains unclear. To address this, we profiled the respective transcriptomes of 18,646 primary human decidual immune cells isolated from patients with recurrent pregnancy loss (RPL) and healthy controls at single-cell resolution. We discovered dramatic differential distributions of immune cell subsets in RPL patients compared with the normal decidual immune microenvironment. Furthermore, we found a subset of decidual natural killer (NK) cells that support embryo growth were diminished in proportion due to abnormal NK cell development in RPL patients. We also elucidated the altered cellular interactions between the decidual immune cell subsets in the microenvironment and those of the immune cells with stromal cells and extravillous trophoblast under disease state. These results provided deeper insights into the RPL decidual immune microenvironment disorder that are potentially applicable to improve the diagnosis and therapeutics of this disease.
Subject terms: Immunology, Gene expression profiling
Introduction
Recurrent pregnancy loss (RPL), defined as loss of two or more consecutive pregnancies, affects up to 5% of women trying to conceive1,2. Known causes of RPL include genetic abnormalities, endocrine disorders, uterine malformations, as well as other influencing factors such as thrombophilia and maternal infections3. The etiology of RPL remains unknown in about 50% of cases. Studies have shown that abnormalities of the decidual immune microenvironment might associate with the pathogenesis of RPL4. However, the underlying mechanisms through which dysregulation of the decidual immune microenvironment causes RPL remain unclear.
The decidual immune cells at the maternal–fetal interface are mainly composed of natural killer (NK) cells, macrophages, T cells, and a variety of minority cell types (e.g., dendritic cells, NKT cells, etc.)5, and their proportion changes with gestational age6. Decidual NK (dNK) cells represent the largest population, comprising about 50–70% of the maternal immune cells during the first trimester of pregnancy7. Studies have shown that dNK cells exert multiple functions to maintain homeostasis of the decidual microenvironment. For instance, dNK cells can modulate trophoblast invasion8, promote fetal growth9, and regulate immune tolerance10. These cells can also exert effector functions upon exposure to exogenous stress11.
Decidual macrophages comprise approximately 10–20% of decidual leukocytes during the first trimester of pregnancy and have been reported to function as anti-inflammatory cells with M2-like phenotypes12–14. Moreover, crosstalk between decidual macrophages and other cells of the decidual immune microenvironment has been reported to maintain overall immune homeostasis at the maternal–fetal interface15,16. Decidual macrophages are known to have many functions similar to those of dNK cells, including remodeling of spiral arteries, trophoblast invasion, promotion of angiogenesis, hindering T cell activation, and mediating canonical responses to antimicrobial infections16–18. Decidual T cells also have functional roles in both normal and pathological pregnancies19. These maternal leukocytes, together with decidual stromal cells and extravillous trophoblasts (EVTs), interact with each other to form a highly complex immune microenvironment5,20.
A number of recent studies have employed single-cell RNA sequencing (scRNA-seq) technology to interrogate the cellular composition and inter-cellular communication events that occur at the maternal–fetal interface6,21–24. Building from these seminal studies of global cell types, subsequent focus studies of immune cell subsets have been conducted for normal decidua25–27. These foundational studies have defined the composition and distribution of various immune cell types of the immune microenvironment of healthy decidua at high resolution. However, the lack of similar high-resolution data for a dysregulated decidua context—as for example in RPL patients—has limited data-driven hypothesis generation about any immune-related patho-mechanisms underlying failed pregnancies or insights which may more fully elucidate the etiology of recurrent miscarriage.
Here, we profiled the decidual immune cells present at the maternal–fetal interface in RPL patients and healthy controls using scRNA-seq. We found dramatic differential distributions of decidual immune cell subsets in RPL patients. Furthermore, we demonstrated a subset of dNK cells that support embryo growth was reduced due to abnormal NK cell development in disease state revealed by Palantir28. We also discovered that macrophage subsets mediated inflammatory T cell chemotaxis in patients. Finally, we constructed a disease-specific interaction network between the major cell types in the decidual immune microenvironment. Our results provide guidance for RPL diagnosis and therapeutics in both cellular and molecular perspectives.
Results
An atlas of decidual immune cells in RPL patients
We obtained 24 human first-trimester decidual samples with normal embryonic karyotypes: 9 from RPL patients and 15 from healthy controls. Specifically, the patients and healthy controls were aged 21–39 years. The mean gestational ages are 7.24 weeks in controls and 8.50 weeks in RPL patients. Four of nine RPL patients experienced three pregnancy loss and five experienced two. The patients were advised to obtain an induced abortion after a diagnosis of pregnancy loss (see Materials and methods, Supplementary Table S1). The decidual sample tissues were digested into single-cell suspensions, and the CD45+ leukocytes were then sorted and subjected to scRNA-seq using the 10× platform (Fig. 1a). Low-quality cells were then filtered after rigorous quality control (QC) definition (Supplementary Fig. S1a–d), and we retained a total of 18,646 high-quality CD45+ single transcriptomes. Of these, 8504 cells were originated from RPL patients and 10,142 cells from normal controls.
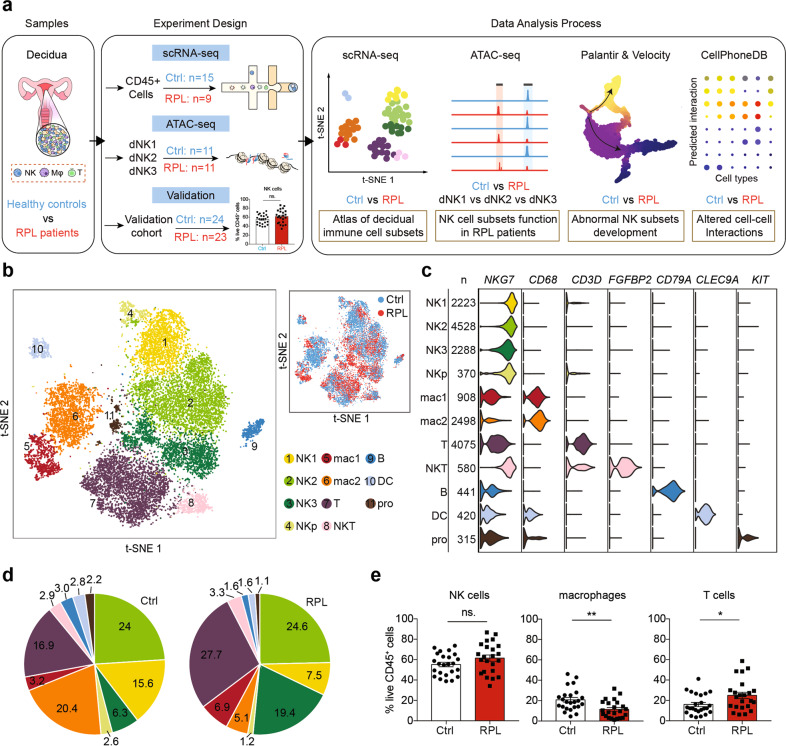
Fig. 1. An atlas of decidual immune cells in RPL patients.
a Flowchart depicting the overall design of the study. Numbers indicate the number of individuals analyzed. b A t-SNE projection of the 18,646 total CD45+ leukocytes from nine RPL patients and 15 healthy controls, indicating 11 main clusters. Different colors indicate cell clusters and disease status (see legend for key). NKp, proliferating natural killer cells; pro, progenitor cells. c Violin plots of selected marker genes (upper row) for multiple immune cell subsets. The first column shows the names of the decidual cell clusters, the second column represents the numbers of cells in each cluster. d Pie chart showing the proportion (i.e., % of total sequenced immune cell complement) for each of the 11 cell clusters in healthy controls and RPL patients. e Flow cytometry analysis of a larger cohort of healthy controls and RPL patients (including those sequenced in scRNA-seq), showing the proportions of the decidual NK cells, macrophages, and T cells among the gated CD45+ leukocytes. ns, P > 0.05; *P < 0.05; **P < 0.01; Student’s t-test. All points are shown and bars represent means with SEM (nc = 24, np = 23; np: number of RPL patients, nc: number of healthy controls).
We then applied Seurat29 to normalize and cluster the gene expression matrix and identified 11 unique immune cell subsets, which were visualized via t-distributed stochastic neighbor embedding (t-SNE) (Fig. 1b). We identified cell lineages, including NK cells, macrophages, T cells, dendritic cells, NKT cells, and immune progenitor cells based on the expression of known marker genes (Fig. 1c). To corroborate our results, we extracted the cells from the healthy controls and compared their transcriptomes with the recently reported scRNA-seq profiles from the decidual immune cells at the maternal–fetal interface25 (Supplementary Fig. S2a, b). The two datasets provided extensive overlap in cell identities and the cell-type distributions (Supplementary Fig. S2c, d), suggesting that we have obtained reliable cell atlas of the decidual immune microenvironment in RPL patients and healthy controls.
We also used another integration method, Harmony30, to help confirm the reliability of our cell clustering results from Seurat and visualized it with UMAP method (Supplementary Fig. S3a, b). We found strong similarities of the cell clusters processed by Seurat and Harmony (Supplementary Fig. S3c, d), supporting the robustness of our cell clustering results.
Our first general investigation of potential differences between the RPL patients and healthy controls focused on any divergence in the proportion of major immune cell types: there was no apparent difference in the ratio of NK cells, but the RPL patients had decreased macrophage populations and slightly increased T cell populations (Fig. 1d). To validate this result, we measured the distributions of NK, macrophage, and T cells in a larger cohort of RPL patients (n = 23) and healthy controls (n = 24) using flow cytometry (Supplementary Table S2). As expected, we found a slight but not significant increase in the proportion of NK cells (P = 0.0809), a remarkable and significant decrease in macrophages (P = 0.0051), and a slight but significant increase in T cells (P = 0.0307) (Fig. 1e). Thus, our scRNA-seq provides a useful atlas for identifying disease-associated differences and immune cell population divergence at the maternal–fetal interface during early pregnancy.
A subset of angiogenic dNK cells are decreased in RPL patients
Since dNK cells are the most abundant cell type in the decidual immune microenvironment, we initially explored the dNK cell subsets and their functions at the maternal–fetal interface. We identified three known subsets of dNK cells25: CD39+ CD18− CD103− (dNK1), CD18+ CD103− CD39− (dNK2), and CD18+ CD103+ CD39− (dNK3), as well as a group of proliferating natural killer cells (dNKp) (Fig. 2a, b; Supplementary Fig. S4a). Whereas our aforementioned analysis indicated no significant differences in the proportion of total dNK cells between RPL patients and healthy controls (Fig. 1d, e), there were obvious differences between patients and controls in the proportions of the dNK cell subsets (Fig. 2c). Specifically, dNK1 cells were significantly decreased in RPL patients, while dNK2 cells were slightly increased and dNK3 cells were significantly increased. We confirmed these findings using flow cytometry analysis of the larger RPL patient cohort (Fig. 2d, np = 23, nc = 24, P = 0.0004, P = 0.0001, P = 0.0019, respectively).
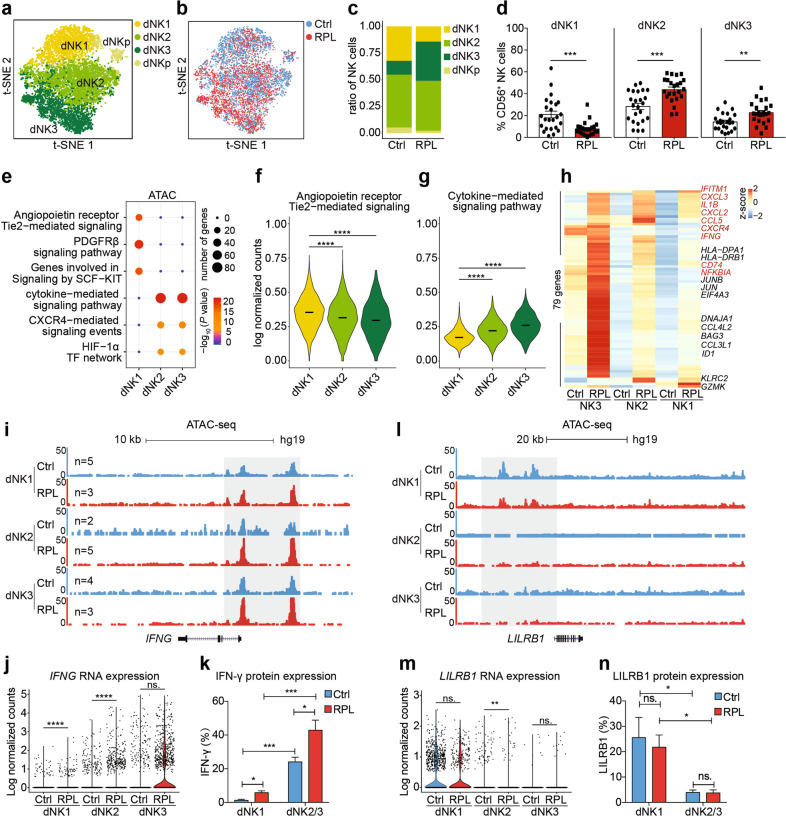
Fig. 2. A subset of angiogenic dNK cells are decreased in RPL patients.
a, b t-SNE plots of 9409 dNK cells, indicating four main clusters (a) from RPL patients and healthy controls (b). Colors indicate cell clusters and disease status. NKp, proliferating natural killer cells. c The proportion of dNK1, dNK2, dNK3, and dNKp cell subsets among the 9409 dNK cells from healthy controls and RPL patients. d Flow cytometry analysis of a larger cohort of healthy controls (nc = 24) and RPL patients (np = 23) to confirm the proportions of dNK1, dNK2, and dNK3 cell subsets among all gated dNK (CD45+CD56+CD3−) cells. e Scatter plot of the enriched genomic features in chromatin accessible sites of the three dNK cell subsets by ATAC-seq of the healthy controls (dNK1, nc = 5, np = 3; dNK2, nc = 2, np = 5; dNK3, nc = 4, np = 3). f, g Violin plots of the expression of genes involved in the signaling pathway “angiopoietin receptor Tie2-mediated signaling pathway” and “cytokine-mediated signaling pathway” in the three dNK cell subsets from healthy controls. h Heatmap for unsupervised clustering analysis of the differentially expressed genes between healthy controls and RPL patients, assessed for each of the three dNK cell subsets. Genes colored in red belong to the term of “cytokine-mediated signaling pathway”. i UCSC genome browser visualization of the chromatin accessibility profiling at the IFNG locus. j Violin plots of single-cell RNA expression of the IFNG gene in each dNK cell subset in healthy control and RPL patients. k Bar graph showing the percentage of IFN-γ expression in dNK1, dNK2, and dNK3 cell subsets from healthy controls (nc = 7) and RPL patients (np = 7). l UCSC genome browser visualization of the chromatin accessibility profiling at the LILRB1 locus. m Violin plots of single-cell expression of the LILRB gene in each dNK cell subset in healthy control and RPL patients. n Bar graph showing the percentage of LILRB1 expression in dNK1, dNK2, and dNK3 cell subsets from healthy controls (nc = 7) and RPL patients (np = 7). ns, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; Student’s t-test. Bars represent means with SEM.
To investigate any distinct functions for these three dNK cell subsets, we performed a differential analysis of all the transcriptomes obtained from our scRNA-seq experiment alongside chromatin accessibility profiles from bulk ATAC-seq analysis of sorted dNK subsets. We found that genes of the angiopoietin receptor Tie2-mediated signaling pathway were enriched in the dNK1 cell subset (Fig. 2e): dNK1 cells expressed genes including CDKN1A, RELA, and TNIP2 (Supplementary Fig. S4b). Further, we detected elevated expression levels for the 24 genes of this signaling pathway (Fig. 2f). Similar to recently reported findings, we found that dNK1 cells expressed high levels of KIR gene family members (encoding the killer immunoglobulin receptor proteins), suggesting that dNK1 cells are likely recognized by EVTs (Supplementary Fig. S4c, d). dNK1 cells also expressed LILRB1 (Supplementary Fig. S4e, f), which binds to HLA-G proteins expressed on trophoblast cells to increase the secretion of growth-supporting factors31.
Regarding the dNK2 and dNK3 cell subsets, these cells had similar extents of chromatin accessibility and had somewhat similar gene expression profiles. Both dNK2 and dNK3 cells were highly enriched for genes of cytokine-mediated signaling pathways (Fig. 2e, g), and the dNK3 cells expressed especially high levels of the immunomodulatory IFNG gene (encoding IFN-γ) (Supplementary Fig. S4g, h). These results highlight the functions of the three dNK cell subsets we detected at the maternal–fetal interface, and suggest that the dNK1 cells have embryo growth-supporting activity whereas the dNK2 and dNK3 cells are prone to the cytokine secretion.
Next, seeking etiopathogenic insights about RPL, we examined the functional divergence of the NK cell subsets in RPL patients and healthy controls. Unsupervised clustering of disease-associated differentially expressed genes in the dNK1, dNK2, and dNK3 cell subsets indicated an overall enhancement of cytokine-mediated signaling pathways in the three dNK cell subsets from RPL patients (Fig. 2h; Supplementary Fig. S4i and Table S3). Confirming these findings from our ATAC-seq, scRNA-seq data and flow cytometry analysis showed significantly increased accessibility, expression, and secretion of IFNG in RPL patients in all the three dNK cell subsets (Fig. 2i–k), further supporting that dNK cells function to promote an inflammatory environment in RPL decidua. In addition, we found that the expression of LILRB1 in dNK1 cells was slightly decreased, suggesting that interaction between dNK1 cells and EVTs was weakened under disease conditions (Fig. 2l–n). Collectively, these results indicate that, in RPL decidua, the normal angiogenic function of dNK cells is weakened, and this is accompanied by an enhancement of cells that exert pro-inflammatory dNK functions and an apparent reduction in receptivity for trophoblasts.
Aberrant differentiation trajectory impairs dNK1 cell subset accumulation in RPL patients
We then investigated the specific trajectories of the three dNK subsets throughout the course of dNK cell differentiation in the decidua. We applied a high-resolution pseudo-time prediction algorithm Palantir28 to construct the differentiation potential trajectory of all dNK cells from RPL patients and healthy controls (Fig. 3a–c and Supplementary Fig. S5a). We found three developmental branches where dNKp differentiate into dNK1 cells (Path 1) and into two distinct branches of dNK2 and dNK3 cells (Path 2 and Path 3) (Fig. 3c and Supplementary Fig. S5a). We also identified a differentiation pathway wherein dNK2-like cells with an apparent tendency to transform into dNK1 cells (Path T) (Fig. 3c). Our discovery of this differentiation pathway illustrates a previously unknown source of pregnancy-promoting dNK1 cells in the decidua.
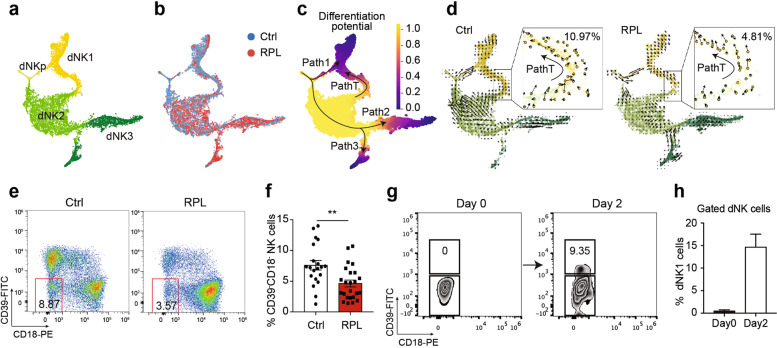
Fig. 3. Aberrant developmental trajectory impairs dNK1 cell subset accumulation in RPL patients.
a–c Palantir’s t-SNE map of the total dNKp, dNK1, dNK2, and dNK3 cell subsets (a), cells colored by the conditions of RPL patients and healthy controls (b), and cells colored by Palantir differentiation potential (c). d Velocity field projected into the t-SNE map of the dNKp, dNK1, dNK2, and dNK3 cell subsets in the healthy controls (left) and RPL patients (right). Arrows show the local average velocity evaluated on a regular grid. The right frame, velocities of Path T cells shown on the t-SNE map. e The proportion of CD39− CD18− dNK cells among gated dNK (CD45+CD56+CD3−) cells from healthy controls (left) and RPL patients (right). f Bar plot of the proportion of CD39− CD18− NK cells for multiple samples in healthy controls (nc = 20) and RPL patients (np = 24). g Percentage of CD39+ dNK1 cells before (left) and after (right) CD39−CD18− NK cells cultured in vitro for 2 days. h Percentage of CD39+ dNK1 cells before and after CD39−CD18− NK cells cultured in vitro for 2 days. Results in g were representative in five independent experiments. **P < 0.01. Significance was evaluated with Student’s t-test. All points are shown and bars represent means with SEM.
We next integrated our single-cell data for healthy individuals with the decidua profiles from a previous study25 and successfully confirmed that Path 1, Path 2, and Path T also exist in healthy individuals (Supplementary Fig. S5b). In contrast, this integrative analysis indicated that Path 3 is apparently RPL-specific. Further, by comparing the gene expression patterns of cells in Path 2 and Path 3, we noticed that genes related to inflammatory responses were enriched in Path 2 cells, indicated from the expression of TGFB1, NFKB1, and REL (Supplementary Fig. S5c, d). Path 3 cells featured enriched expression for genes of cytokine-mediated signaling pathways, such as IFNG, TNF (Supplementary Fig. S5c, e), consistent with the aforementioned increase in cytokine-mediated signaling pathways in the dNK3 cells of RPL patients.
In addition to such pseudo-time analyses, the future state of individual cells can be predicted based on the time derivative of the gene expression state in each cell in terms of RNA velocity32. We applied this approach to reveal the differentiation trend of every single cell at one stage to its future stage. As expected, RNA velocity predicted that dNKp cells differentiate and bifurcate into both dNK1 and dNK2 cells (Supplementary Fig. S5f). Interestingly, we found a clear difference in the proportion of Path T cells between RPL patients and healthy individuals, with patients having significantly smaller subsets of Path T cells (Fig. 3d). More specifically, healthy decidua had an average of 540 (540/4923, 10.97%) such cells, whereas RPL patients had an average of only 216 (216/4486, 4.81%) cells, suggesting a weaker tendency of cells transform into dNK1 cells. Note that we successfully confirmed the presence of Path T cells based on Smart-seq2 single-cell transcriptome data for healthy decidua25 (Supplementary Fig. S5g).
To characterize these Path T cells in greater detail, we examined the expression of dNK cell subset marker genes in our RNA-seq data and found that Path T cells expressed low levels of CD39 and CD18 genes (Supplementary Fig. S5h). A subsequent flow cytometry analysis showed that the number of CD39−CD18− dNK cells was significantly decreased in RPL patients compared to healthy controls (Fig. 3e, f, P = 0.0026), suggesting that this disease may be affected by a diminished source of dNK1 cells. We then sorted the CD39−CD18− dNK cells and cultured them in vitro and confirmed that these cells can indeed successfully transform into dNK1 cells (Fig. 3g, h). Thus, beyond empirically illustrating developmental plasticity for the dNK cell population, our findings suggest that impaired accumulation of the CD39−CD18− dNK cells may lead to a decreased number of dNK1 cells, and thereby insufficient support for fetal growth in RPL patients.
Disease characteristics and cell–cell interactions of macrophage and T cells
After dNK cells, macrophages are the most abundant leukocytes5, and we examined the cellular heterogeneity of macrophages in the decidual immune microenvironment. We detected two subsets of macrophages from a total of 3406 single-macrophage transcriptomes: mac1 and mac2 (Fig. 4a). Recalling our initial finding that RPL patients had dramatically reduced overall macrophage populations (Fig. 1e), we found that RPL patients had modestly elevated mac1 populations compared to healthy controls and had remarkably decreased mac2 populations (Fig. 4b, c). We then performed a differential analysis of gene expression between mac1 and mac2 subsets to help characterize any functional differences (Supplementary Fig. S6a). Gene Ontology (GO) enrichment of the differentially expressed genes suggested that mac1 cells apparently function in neutrophil-mediated immunity, while mac2 cells may be functionally associated with the regulation of NK cell chemotaxis (Supplementary Fig. S6b). In order to better characterize the phenotype of macrophage subsets, we performed protein interaction network analysis using differentially expressed genes of macrophage subsets by STRING33. We found S100A10, S100A6, S100B, and S100A4 genes were overexpressed in mac1 and PLAU, DAB2, EGR1, etc., were enriched in the mac2 subset (Supplementary Fig. S6c).
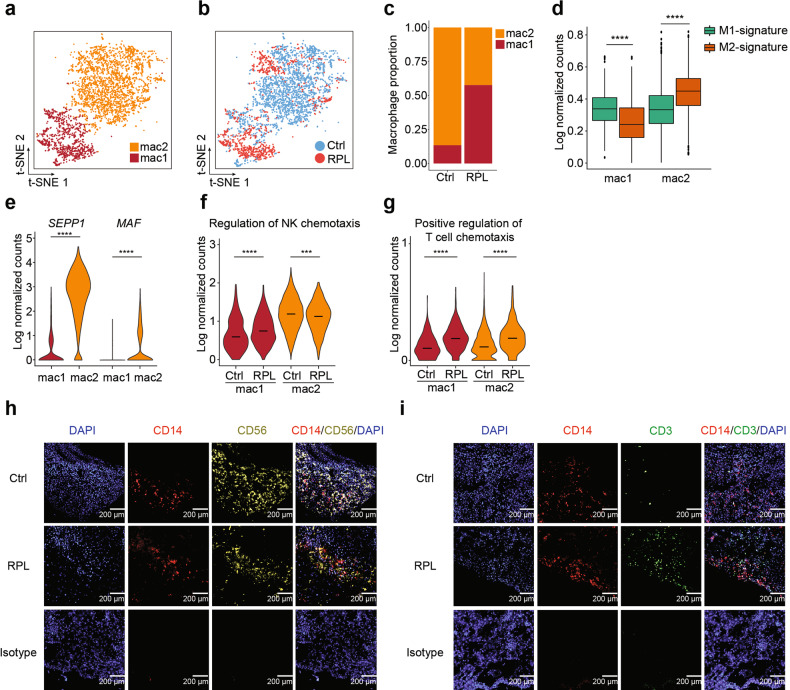
Fig. 4. Macrophages enhance cytokine-mediated pathways in RPL patients.
a, b t-SNE plots of macrophages from RPL patients and healthy controls, indicating the mac1 and mac2 subsets (a), and the distribution of RPL patients and healthy controls (b). Colors indicate cell clusters and disease status. c Bar plot showing the proportions of mac1 and mac2 cell subsets in healthy controls and RPL patients. d Box plots of the M1 and M2 macrophage signature genes’ expressions in mac1 and mac2 cell subsets. The box represents the second, third quartiles and median, whiskers each extend 1.5 times the interquartile range; dots represent outliers. e Violin plot of SEPP1 and MAF gene expression in mac1 and mac2 cells. f, g Violin plots of the expressions of genes involved in “regulation of NK chemotaxis” (f) and “positive regulation of T cell chemotaxis” (g) in mac1 and mac2 cell subsets from healthy controls and RPL patients. h, i Representative example of immunofluorescence staining of DAPI (h, i, blue), CD14 (h, i, red), CD56 (h, yellow), CD3 (i, green), and overlay from a decidual tissue region from healthy controls and RPL patients and from decidual tissue with negative control staining (bottom row). Results in h, i were representative in three independent experiments. Scale bars, 200 μm.
To assess the M1/M2 polarization potential of these two macrophage subsets, we examined the expression of M1 and M2 genes34 in mac1 and mac2 cells. We found that mac1 cells showed M1 polarization characteristics (Fig. 4d); however, mac2 cells were highly enriched with M2 specific genes (Fig. 4d). For example, SEPP1 and MAF (Fig. 4e)—which are important M2-like macrophage marker and regulator35—were both expressed in the mac2 cells. These results suggest that mac2 cells may functionally contribute to successful pregnancy by promoting normal M2 macrophage homeostasis in the decidua.
To investigate how the functions of mac1 and mac2 cells may be altered in RPL patients, we conducted a pairwise comparison of the differentially expressed genes of the macrophage subsets between RPL patients and healthy controls. GO analysis indicated that the functions of mac1 cells were enhanced in the disease state (Supplementary Fig. S6d, e). To our surprise, the enrichment analysis predicted that the functions of RPL patient mac2 cells were altered compared to healthy mac2 cells, for example switching from a “regulation of NK cell chemotaxis” predicted function in healthy mac2 cells to “positive regulation of T cell chemotaxis” in RPL patients (Fig. 4f, g and Supplementary Fig. S6f). We also observed the expressions of genes involved in “NK cell chemotaxis” were decreased in mac2 cells in RPL patients, and those with “T cell chemotaxis” were significantly higher expressed in both mac1 and mac2 cells in RPL patients (Fig. 4f, g). Thus, we applied the immunofluorescence assay to characterize the spatial co-localization between macrophages and NK cells and T cells. Our results indicated that macrophages aggregated with NK cells in the normal decidua, whereas under disease condition macrophages co-localized with T cells (Fig. 4h, i).
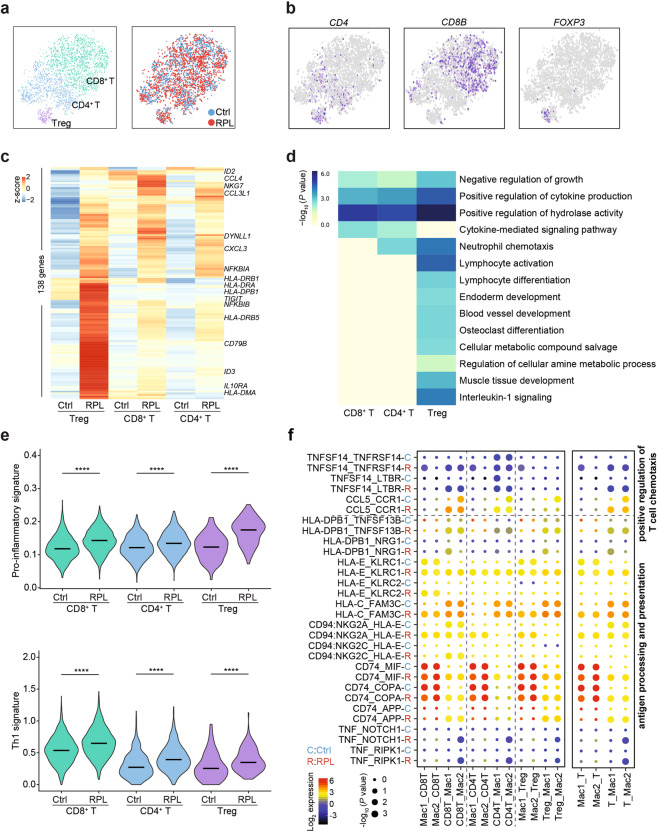
We next investigated the heterogeneity of T cells from RPL patients and healthy individuals. We re-clustered T cells and identified three T cell subsets including CD4+ T, CD8+ T, and FOXP3+ regulatory T cells (Fig. 5a, b). Unsupervised clustering of disease-associated differentially expressed genes in the CD4+ T, CD8+ T, and Treg cell subsets indicated an overall enhancement of cytokine-mediated signaling pathways in T cells from RPL patients (Fig. 5c, d). Th1 and pro-inflammatory signatures36 were significantly enhanced in T cells and each T subset of RPL patients (Fig. 5e). To further illustrate the molecular basis of the crosstalk between macrophages and T cells, we applied the CellPhoneDB25 algorithm to analyze the enrichment of interaction pairs. And we found TNFSF14-TNFRSF14, TNFSF14-LTBR, and CCL5-CCR1 receptor/ligand pairs involved in the “positive regulation of T cell chemotaxis” were enhanced in RPL patients (Fig. 5f).
Fig. 5. T cell subsets and their interactions with macrophages.
a t-SNE plots of 4071T cells, indicating three main clusters (left) from RPL patients and healthy controls (right). Colors indicate cell clusters and disease status. b t-SNE plots showing the expression of marker genes in each T cell subset. c Heatmap for unsupervised clustering of the differentially expressed genes between healthy controls and RPL patients, assessed for each of the three T cell subsets. d Heatmap of the enriched genomic features of the differentially expressed genes between healthy controls and RPL patients. e Violin plots of the pro-inflammatory and Th1 signature genes’ expressions in T cell subsets from healthy controls and RPL patients. ****P < 0.0001. Significance was evaluated with Student’s t-test. f Dot plot of the predicted interactions of macrophages with T cells in the RPL patients and healthy controls. P values were indicated by circle size. The expression levels of all the interacted genes were indicated by colors, scales on the right.
In summary, our analysis of the second and third most common immune cell types in the decidua revealed that RPL patients exhibit reductions in a homeostasis-promoting subset of macrophages. Instead of dNK cells, macrophages in the RPL disease state are prone to interact with Th1-like T cells.
Alteration of the decidual immune response in RPL patients
The decidual immune microenvironment is a complex system in which multiple immune cells function via interactions with other cells. In addition to our analyses which considered the RPL-associated changes in specific immune cell types including dNK, macrophages, and T cells, we also used the curated receptor/ligand interaction database CellPhoneDB to identify alterations of molecular interactions between the various immune cell subsets and EVT and stromal cells in the RPL patients (see Materials and methods). We found 959 pairs of interactions between immune cells were increased in RPL patients (Supplementary Table S4). Macrophages engage in extensive interactions with all the other immune cells in patients and in healthy controls and associated with 25.86% (248 pairs) of the overall alteration, suggesting a central role in the regulation of the disease (Supplementary Fig. S7a, b).
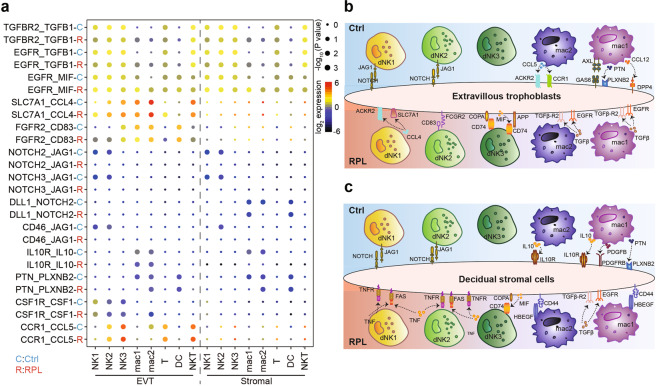
To further explore the immune microenvironment of the maternal–fetal interface in RPL patients, we integrated our single-cell decidua data with the scRNA-seq data from healthy EVTs and healthy stromal cells25. This dataset integration enabled predictions for the interactions of the eight major immune cell types from RPL patients with EVTs and stromal cells (Fig. 6a). As expected, several receptor/ligand pairs (e.g., CSF1R-CSF1 and CCR1-CCL5) connected the dNK subsets with EVTs in the healthy decidua. In contrast, several other receptor/ligand pairs in immune cell subsets with EVTs and stromal cells were dysregulated according to the RPL decidua data. The strength of interaction through the NOTCH signaling pathway (e.g., NOTCH3-JAG1, NOTCH2-JAG1) between dNK1 and dNK2 cells with EVTs and with stromal cells was significantly reduced in the RPL patients. We summarized the RPL-associated changes in receptor/ligand interactions between immune cells and both the EVTs and stromal cells (Fig. 6b, c and Supplementary Table S5). Collectively, these findings illustrated the molecular basis of cell–cell interactions at the maternal–fetal interface in an inflamed state, leading to a better understanding of the mechanisms of reproductive failure in RPL patients.
Fig. 6. Alteration of decidual immune responses in RPL patients.
a Dot plot of predicted interactions of immune cells with EVTs and with stromal cells in the RPL patients and healthy controls. P values were indicated by circle size. The expression levels of all the interacted genes were indicated by colors, scales on the right. b, c Summary illustrations depicting the altered interactions of decidual immune cells with EVTs (b) and stromal cells (c) in healthy controls (up) and RPL patients (down).
Discussion
The immune microenvironment at the maternal–fetal interface is essential for maintaining normal embryonic development37. Previous studies have illustrated the landscape of decidual immune cells and their intercellular interactions at the maternal–fetal interface in healthy people25. Here, we present single-cell transcriptomes of the decidual immune microenvironment in patients with RPL. We identified that RPL patients have a remarkably decreased number of a particular subset of dNK cells (dNK1), which may contribute to the secretion of growth-promoting factors to promote normal pregnancy38. Meanwhile, RPL patients harbor significantly increased numbers of a pro-inflammatory subset of dNK cells (dNK3) that produce cytokines. We also characterized a population of dNK2-like (Path T) cells which can transform into dNK1 cells, and discovered that the RPL patients have dramatically fewer Path T cells than healthy controls. In addition to dNK cells, we also found that macrophages function differently in normal pregnancy environments and disease states. The former tend to recruit NK cells to maintain immune tolerance, while the latter tend to recruit T cells with an inflammatory signature in RPL patients. Finally, we provide a series of detailed maps presenting the global interactions between our sequenced decidual immune cells and both EVTs and stromal cells, enabling ligand/receptor level hypothesis generation about the likely causes underlying pregnancy failure.
Studies have reported that the normal pregnancy decidual immune microenvironment presents a Th2-biased situation, while Th1-type immunity may lead to pregnancy failure39. For one, Th1 cells prevent growth and differentiation of trophoblast through a mechanism that depends on proinflammatory cytokines40,41. Indeed, our study found that the overall decidual immune microenvironment of RPL patients presented as primarily Th1-type: the number of dNK2 and dNK3 cell subsets with cytokine-secreting functions were increased in RPL patient decidua, and the proportion of inflammatory macrophage sub-population mac1 was also increased, consistent with recent publication42. This mac1 increase was accompanied by a substantial decrease in the mac2 macrophage subset, which was enriched with M2 signature genes and might function to maintain immune homeostasis43. However, how RPL macrophage subsets lead to increased cytokine secretion from dNK cells (e.g., IFNG) and promote the chemotaxis of inflammatory T cells still requires further investigation. Viewed collectively, these findings suggest that therapeutically adjusting the proportions of the decidual immune cell subsets may help restore normal pregnancy.
There are inevitable difficulties in obtaining the optimum tissues from patients with RPL who are unlikely to request termination44. Although we had taken special care and followed the published procedures10 when we collected decidua samples from RPL patients and healthy controls (see Materials and methods), there is a limitation that we obtained samples from women at the time of the pregnancy loss and not at an elective termination in women with this clinical history. Since the decidual samples from RPL patients were collected after fetal demise, it is still possible that some alterations of decidual immune cells we observed in this study could be the consequence of the fetal death. Our empirical data, while not fully satisfactory, delineate the dNK cell plasticity which is associated with the impaired accumulation of pregnancy-promoting dNK1 cells in patients, and improve our understanding of the heterogeneity and interaction of cells at the maternal–fetal interface in disease status, providing clues for future treatment of the disease.
Materials and methods
Human samples
Normal decidual samples with no previous pregnancy loss were obtained from elective terminations of apparently normal pregnancies. The elective termination was performed via dilation and curettage (D&C). For the decidua samples from abnormal pregnancies, the patients were advised to obtain an induced abortion after a clinical diagnosis of embryo demise. The diagnosis of embryo demise was following the guidelines from First Affiliated Hospital of University of Science and Technology of China, such as mean sac diameter of 25 mm or greater and no embryo or absence of an embryo with heartbeat 2 weeks or more after a scan that showed a gestational sac without a yolk sac. Fetal heart activity was assessed using Doppler ultrasound at 7–9 weeks of gestation. When abnormal fetal heart activity was observed, the patients were advised to be tested for serum β-human chorionic gonadotropin levels, with additional ultrasounds every other week. D&C was performed within 24 h of ultrasonographic documentation of fetal loss.
When decidua samples were obtained, chorionic villi and blood clots were identified and carefully separated from the maternal decidua. The chorionic villi were then sent for cytogenetic analysis. Normal embryo karyotypes were identified to enable the exclusion of genetic or mechanical causes for embryo demise. We also excluded patients with clinical symptoms of heavy bleeding and cramps prior to induced abortion. The mean gestational ages are 7.24 weeks in controls and 8.50 in RPL patients. All of the decidua samples were collected from the First Affiliated Hospital of the University of Science and Technology of China. Before surgery, informed consent was obtained from each patient. Ethical approvals were obtained from the ethics committee of the University of Science and Technology of China.
Cell isolation
Fresh decidual tissues were washed extensively in phosphate-buffered saline with 100 IU/mL penicillin/streptomycin and sheared into tiny pieces. Mononuclear lymphocytes were released by digesting the tissues with 1 mg/mL collagenase IV (Sigma) in RPMI 1640 at 37 °C for 1 h, with shaking at 250× r.p.m. The suspensions were filtered via 70 μm nylon mesh cell strainers (i-Quip) and then loaded on a Ficoll-Paque density gradient for separation of decidual cells, which include CD45+ immune cells and stromal cells. Next, decidual CD45+ cells were sorted and cryopreserved according to the official recommendations from 10× Genomics for scRNA-seq analysis. For ATAC-seq analysis of the dNK cell subsets, dNK1 cells (CD45+CD3−CD56+CD39+CD103−CD18−), dNK2 cells (CD45+CD3−CD56+CD39−CD103−CD18+), and dNK3 cells (CD45+CD3−CD56+CD39−CD103+CD18+) were sorted, and the ATAC-seq libraries were prepared immediately.
Antibodies and reagents
The following antibodies were used for the analysis of decidual immune cells with FACS, intracellular staining, and cell sorting: PerCP/Cy5.5 anti-human CD45 (HI30), Brilliant VioletTM 421 anti-human CD56 (HCD56), APC/Cy7 anti-human CD3 (SK7), PE anti-human CD14 Antibody (63D3), FITC anti-human CD39 (A1), PE anti-human CD18 (TS1/18), Alexa Fluor® 647 anti-human CD103 (Integrin αE) Antibody (Ber-ACT8), APC anti-human IFN-γ (4S.B3), and APC anti-human CD85j (LILRB1) Antibody (GHI/75). All antibodies above were purchased from BioLegend.
Flow cytometry
Decidual mononuclear cells were prepared and stained using the aforementioned human mAbs. Homologous IgGs served as negative controls. FACS surface marker staining was performed according to BioLegend antibody instructions. For intracellular staining of cytokines, decidual cells were stimulated with PMA (50 ng/mL, Sigma) and ionomycin (1 μg/mL, Sigma) in the presence of brefeldin (5 μg/mL, BioLegend) for 4 h. After stimulation, the cells were then collected, stained with fluorescein-labeled antibody, washed, and blocked according to the product instructions for BD Cytofix/Cytoperm Cell Permeabilization/Fixation Solution (BD Biosciences).
ScRNA-seq
We sorted viable CD45+ cells from each decidual sample of RPL patients and healthy controls. The cells were then counted and resuspended at a concentration of 1000 cells/μL, aiming for an estimated 8000 cells per library, following the instructions of single-cell 3ʹ solution v2 reagent kit (10× Genomics). Briefly, the cell suspensions were loaded onto a chromium single-cell chip along with reverse transcription master mix and 3ʹ gel beads. After the generation of single-cell gel bead-in-emulsions (GEMs), reverse transcription was performed using a C1000 TouchTM Thermal Cycler (Bio-Rad). The amplified cDNA molecules were then purified with SPRIselect beads (Beckman Coulter). Single-cell libraries were then constructed following fragmentation, end repair, polyA-tailing, adaptor ligation, and size selection according to the manufacturer’s standard protocols. Each sequencing library was generated with a unique sample index. Libraries were sequenced on the Illumina HiSeq X Ten platform.
Transposome generation
To generate Tn5 transposomes for ATAC-seq library preparation on dNK cell subsets, two oligos (R1, R2) were annealed separately to common pMENTs oligos (5Phos/CTGTCTCTTATACACATCT) at 95 °C for 2 min, with cooling until 14 °C at a rate of 0.1 °C/s. After annealing, the annealed R1 and R2 oligos were mixed at a 1:1 molar ratio, incubated with unloaded transposase Tn5 at 25 °C for 30 min, and then stored at −20 °C for ATAC-seq library construction.
The nucleotide sequences of the two oligos were as follows.
R1: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG.
R2: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG.
ATAC-seq library preparation and sequencing
ATAC-seq of dNK cell subsets was performed as previously described45, with minor modifications. Briefly, dNK1, dNK2, and dNK3 subsets were sorted using the SH800S sorter (Sony). Samples were obtained from a distinct cohort from those were used for the scRNA-seq. Approximately 50k cells were used per library. Samples were lysed in cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% NP-40 (Roche) for 3 min on ice to prepare the nuclei. Immediately after cell lysis, nuclei were centrifuged at 500× g for 5 min and the supernatant was discarded. Nuclei extracts were then incubated with the generated Tn5 transposomes and 5× Tris-DMF tagmentation buffer (pH 8.0, 50 mM Tris-HCl, 25 mM MgCl2, 50% DMF) at 37 °C for 30 min. After DNA purification with a MinElute Kit (Qiagen), PCR was performed to amplify the library for 12–15 cycles according to a quantitative PCR reaction for optimum cycles. The PCR thermocycling program was as follows: 98 °C for 30 s; then 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 1 min for the appropriate number of cycles. Following PCR, sample libraries were purified and sequenced using the Illumina HiSeq X Ten platform with the 150-bp paired-end configuration.
NK cell culturing in vitro
To characterize the “Path T” cells, we sorted CD39−CD18− dNK cells, which we cultured in RPMI-1640 with 10% fetal bovine serum. After 2 days, we collected the cultured cells and stained them with fluorescently labeled antibodies as follows: Brilliant VioletTM 421 anti-human CD56 (HCD56), APC/Cy7 anti-human CD3 (SK7), FITC anti-human CD39 (A1), PE anti-human CD18 (TS1/18), and Alexa Fluor® 647 anti-human CD103 (Integrin αE) Antibody (Ber-ACT8). We then performed FACS for the detection of CD39 and CD18 expression in cultured dNK cells.
Immunofluorescence assay
Decidual tissues from RPL patients and healthy controls were embedded in Optimal Cutting Temperature Compound (O.C.T.) and snap-frozen. The cryostat sections were fixed with 4% PFA and incubated in the blocking buffer at room temperature for 1 h. The fluorescent-labeled antibodies were then added in the dark at 37 °C for 1 h, followed by DAPI staining. PBS + 0.1%BSA was used for washing unlabeled antibodies. The following antibodies were used: FITC anti-human CD3 Antibody (HIT3a, 1:100, Biolegend), PE anti-human CD14 Antibody (63D3, 1:200, Biolegend), and Alexa Fluor® 647 anti-human CD56 Antibody (5.1H11, 1:200, Biolegend). Finally, the decidual tissue sections were evaluated with a confocal microscope (IXplore SpinSR, Olympus). We used Fiji/ImageJ version 2.1.0 to analyze our immunofluorescence images.
ScRNA-seq data processing
Droplet-based raw data were processed using Cell Ranger (Version 3.0.0)46 against the GRCh37 human reference genome with default parameters. First, data from each batch was normalized separately using the NormalizeData function and scaled with the ScaleData function implemented in the Seurat pipeline29. Then data from different batches were integrated using the canonical correlation analysis (CCA) method implemented in Seurat29. For each subset of immune cells, NK cells and macrophage subtypes were identified based on the clustering analysis of the single-cell profiles from RPL patients and healthy control individuals shown in Fig. 1. T cells were extracted, normalized, re-clustered, and analyzed separately. We retained cells with detected gene numbers between 500 and 3000 and less than 10% mitochondrial UMIs. Moreover, genes expressed in fewer than three cells were also excluded. Downstream data processing and analysis steps, including filtering, normalization, batch removal, dimension reduction, were performed using the Seurat package version 2.3.1. We clustered all the cells based on the integrated gene expression matrix using Seurat with a parameter Resolution=0.6 and generated 11 clusters. In addition, we combined the Droplet-based dataset from Vento-Tormo et al.25 with our transcriptome data from healthy controls (1:20 subspaces) using CCA in the Seurat pipeline. To quantify the similarity of the two datasets, we applied matchSCore247 to calculate the Jaccard index of clusters using the top 100 ranking cell-type-specific marker genes. In addition to Seurat, we also used Harmony30 to integrate different batches from healthy controls and RPL patients to verify our integration reliability. We used the same gene expression matrix as used in Seurat and performed RunHarmony function in Hamony with default parameters to perform data integration. We then used the same clustering algorithm as that used in Seurat to cluster the cells with resolution 0.3 to generate distinct cell type clusters.
Differential expression analysis
We performed differential gene expression analysis for our identified cell subsets using the Wilcoxon rank-sum test within the Seurat “FindAllMarkers” command. We also performed differential gene expression analysis between normal individuals and patients using the “FindMarkers” function in the Seurat package. We applied multiple thresholds (fold-change > 1.5, P value < 0.01 for NK cells, fold-change > 2, P value < 0.01 for macrophage cells) to identify marker genes expressed in each cell subset and differentially expressed genes between normal and patients. We then performed GO analysis using the KEGG pathway and Biological Process database through the GSEApy python package. Protein–protein association network analysis was performed using differentially expressed genes of macrophage subsets by STRING v1133 with setting k-means clustering method (k = 5).
Single-cell data imputation analysis
We applied SAVER (Version 1.1.1)48 to denoise our single-cell transcriptome data with the NK subsets and macrophages. Raw data were processed using the “saver” function with the setting “estimates.only = TRUE”. Finally, we used the imputation data to visualize the differentially expressed genes that were enriched in the RPL patients.
ATAC-seq data processing
Primary data were processed as described previously49. Briefly, we removed adapter sequences and then mapped reads to the hg19 using Bowtie250. The PCR duplicates and reads mapped to chromosome M were removed. The uniquely mapped reads were shifted +4/−5 bp according to the strand of the read. All mapped reads were then extended to 50 bp centered through the cleavage position. Peak calling was performed using MACS251 with the options - f BED -g hs, -q 0.01, --nomodel, and --shift 0. The number of raw reads mapped to each peak at each condition was quantified using the intersectBed function in BedTools52. Raw counts in peaks were normalized using the DESeq53 package in R. Peak intensity was defined as the log2 of the normalized counts. Significance analysis was then performed by pair-wise comparison using DESeq with P value < 0.01 and log2|fold-change| > 1. GO analysis of cis-regulatory regions was performed with GREAT54.
Developmental trajectory analysis using Palantir
Palantir28 is a high-resolution algorithm, which allows computing a continuous probabilistic process to model cell fate choice by applying multiple diffusion components. Here, Palantir was applied to NK subsets in our data and to the integrated dataset. Basically, we used the CCA-aligned subspaces generated from Seurat to replace the low-dimensional principal components subspaces to reduce the batch effects. Twenty diffusion components were selected and computed with default parameters in Palantir. Diffusion components scaled by an Eigen gap were used as inputs and perplexity was set to 200 to generate the t-SNE maps. To accurately define the initial cell state, we imputed a pseudo cell as the start cell by calculating the average gene expression of our identified dNKp cells. A waypoints = 1200 value was applied, and the parameter k was set to 50 for datasets.
Cell velocity analysis
RNA velocity32 analysis was performed using the “velocyto run” and “velocyto run_smartseq2” commands (velocyto, version 0.17.17), following this pipeline (https://github.com/velocyto-team/velocyto-notebooks/tree/master/python/DentateGyrus.ipynb) as described. First, cells with the lowest 5 percentile spliced and unspliced counts were filtered out. Genes with less than 50 read counts or were detected in fewer than 20 cells were excluded for spliced molecules and high variation genes set to 3000 were selected. Genes with less than 25 read counts or detected in fewer than 10 cells were also filtered out for unspliced molecules. We also filtered out genes based on a cluster-wise expression with thresholds (unspliced = 0.05, spliced = 0.1). Then, spliced and unspliced molecules counts were normalized separately using the default parameters implemented in the pipeline. To reduce dimensionality, we selected the top 30 principal components to construct a k-nearest neighbors algorithm (KNN, k = 200) graph, applying the Euclidean distance metric. Finally, velocity-based extrapolation was computed using the assumption of constant velocity under model I. To visualize the predicted results on low-dimensional maps, we projected the velocities onto the t-SNE embedding space generated from Palantir by using the recommended procedures in the pipeline.
Receptor–ligand interaction analysis
CellPhoneDB25 enables analysis of cell–cell communication networks by predicting ligands, receptors, and interactions. To identify potential interactions between NK cells, macrophages, T cells, and other cell types, we applied the CellPhoneDB algorithm to our transcriptome data for normal individuals and patients. Briefly, receptor–ligand interactions were only considered based on the expression of a ligand by one specific cluster and a receptor by another cluster (as least 10% cells expressed). Pairwise comparisons between all cell subsets were performed by randomly permuting cluster labels for all cells 1000 times automatically. And a P value for each receptor–ligand in every cluster–cluster interaction was computed using a null distribution. For the EVTs and stromal cells, we finally prioritized interactions with more significant (P < 0.05) cell–cell interaction pairs in healthy controls than that in RPL patients, while the selection criteria of interactions between macrophages and T cells was the opposite. We also selected interactions based on biological relevance. Networks were created using Cytoscape (version 3.7.1).
Statistics
We used unpaired Student’s t-tests to assess differences in the proportions of NK cells, macrophage cells, and T cells in healthy controls versus RPL patients. We also used unpaired Student’s t-tests to analyze the proportions of dNK cell subsets (dNK1, dNK2, dNK3) and CD39−CD18− NK cells (percentage of CD56+ NK cells) between the two sample groups. Gene expression and gene set expression between healthy controls and RPL patients were analyzed using unpaired Student’s t-tests. To identify marker genes expressed in each subset and the differentially expressed genes between healthy controls and RPL patients, we used the Wilcoxon rank-sum tests implemented in Seurat. A permutation test for the CellphoneDB analysis was used to evaluate the significance of a receptor/ligand pair.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China grants (31970858, 91940306, 31771428, and 91640113 to K.Q., 31700796 to C.G., and 81871479 to J.L.), the National Key R&D Program of China (2017YFA0102900 to K.Q.), the Fundamental Research Funds for the Central Universities (WK2070000158, YD2070002019, and WK9110000141 to K.Q. and WK9100000001 to J.L.). We thank the USTC supercomputing center and the School of Life Science Bioinformatics Center for providing supercomputing resources for this project. We thank CAS Interdisciplinary Innovation Team for helpful discussion.
Author contributions
K.Q. conceived and supervised the project. K.Q. and C.G. designed the experiments. C.G. and L.J. performed the experiments and conducted all the sample preparation for NGS with help from Q.S., C.J., D.Z., J.L., Z.Z., and L.L. P.C. performed the data analysis with help from L.J., Q.Y., W.Z., Q.L., K.L., and J.F. X.T, D.L., Y.W., and F.L. provided normal and RPL decidua samples. K.Q. and C.G. wrote the manuscript with the helps from H.W. and all the other authors.
Data availability
Raw data for single-cell RNA-seq and ATAC-seq samples are available in the Genome Sequence Archive (GSA) database as accession number CRA002181. Reviewer access link: https://bigd.big.ac.cn/gsa/s/nBQxzt82.
Conflict of interest
Jingwen Fang is the chief executive officer of HanGen Biotech. The other authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chuang Guo, Pengfei Cai, Liying Jin
Supplementary information
Supplementary Information accompanies the paper at (10.1038/s41421-020-00236-z).
References
- 1.Practice Committee of American Society for Reproductive Medicine. Definitions of infertility and recurrent pregnancy loss: a committee opinion. Fertil. Steril. 2013;99:63. doi: 10.1016/j.fertnstert.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 2.El Hachem H, et al. Recurrent pregnancy loss: current perspectives. Int. J. Women’s Health. 2017;9:331–345. doi: 10.2147/IJWH.S100817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido-Gimenez C, Alijotas-Reig J. Recurrent miscarriage: causes, evaluation and management. Postgrad. Med. J. 2015;91:151–162. doi: 10.1136/postgradmedj-2014-132672. [DOI] [PubMed] [Google Scholar]
- 4.Triggianese P, Perricone C, Chimenti MS, De Carolis C, Perricone R. Innate immune system at the maternal–fetal interface: mechanisms of disease and targets of therapy in pregnancy syndromes. Am. J. Reprod. Immunol. 2016;76:245–257. doi: 10.1111/aji.12509. [DOI] [PubMed] [Google Scholar]
- 5.Erlebacher A. Immunology of the maternal–fetal interface. Annu. Rev. Immunol. 2013;31:387–411. doi: 10.1146/annurev-immunol-032712-100003. [DOI] [PubMed] [Google Scholar]
- 6.Pique-Regi R, et al. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife. 2019;8:e52004. doi: 10.7554/eLife.52004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koopman LA, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanna J, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat. Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 9.Fu B, et al. Natural killer cells promote fetal development through the secretion of growth-promoting factors. Immunity. 2017;47:1100–1113. doi: 10.1016/j.immuni.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Fu B, et al. Natural killer cells promote immune tolerance by regulating inflammatory TH17 cells at the human maternal–fetal interface. Proc. Natl Acad. Sci. USA. 2013;110:E231–E240. doi: 10.1073/pnas.1206322110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siewiera J, et al. Human cytomegalovirus infection elicits new decidual natural killer cell effector functions. PLoS Pathog. 2013;9:e1003257. doi: 10.1371/journal.ppat.1003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ning F, Liu H, Lash GE. The role of decidual macrophages during normal and pathological pregnancy. Am. J. Reprod. Immunol. 2016;75:298–309. doi: 10.1111/aji.12477. [DOI] [PubMed] [Google Scholar]
- 13.Svensson J, et al. Macrophages at the fetal-maternal interface express markers of alternative activation and are induced by M-CSF and IL-10. J. Immunol. 2011;187:3671–3682. doi: 10.4049/jimmunol.1100130. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y, et al. An M1-like macrophage polarization in decidual tissue during spontaneous preterm labor that is attenuated by rosiglitazone treatment. J. Immunol. 2016;196:2476–2491. doi: 10.4049/jimmunol.1502055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vacca P, et al. Crosstalk between decidual NK and CD14+ myelomonocytic cells results in induction of Tregs and immunosuppression. Proc. Natl Acad. Sci. USA. 2010;107:11918–11923. doi: 10.1073/pnas.1001749107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houser BL. Decidual macrophages and their roles at the maternal–fetal interface. Yale. J. Biol. Med. 2012;85:105–118. [PMC free article] [PubMed] [Google Scholar]
- 17.Tang MX, Hu XH, Liu ZZ, Kwak-Kim J, Liao AH. What are the roles of macrophages and monocytes in human pregnancy? J. Reprod. Immunol. 2015;112:73–80. doi: 10.1016/j.jri.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Thiruchelvam U, Dransfield I, Saunders PT, Critchley HO. The importance of the macrophage within the human endometrium. J. Leukoc. Biol. 2013;93:217–225. doi: 10.1189/jlb.0712327. [DOI] [PubMed] [Google Scholar]
- 19.Nancy P, Erlebacher A. T cell behavior at the maternal–fetal interface. Int. J. Dev. Biol. 2014;58:189–198. doi: 10.1387/ijdb.140054ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ander SE, Diamond MS, Coyne CB. Immune responses at the maternal–fetal interface. Sci. Immunol. 2019;4:31. doi: 10.1126/sciimmunol.aat6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson AC, Mould AW, Bikoff EK, Robertson EJ. Single-cell RNA-seq reveals cell type-specific transcriptional signatures at the maternal-foetal interface during pregnancy. Nat. Commun. 2016;7:11414. doi: 10.1038/ncomms11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlicev M, et al. Single-cell transcriptomics of the human placenta: inferring the cell communication network of the maternal–fetal interface. Genome Res. 2017;27:349–361. doi: 10.1101/gr.207597.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, et al. Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res. 2018;28:819–832. doi: 10.1038/s41422-018-0066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsang JCH, et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl. Acad. Sci. USA. 2017;114:E7786–E7795. doi: 10.1073/pnas.1710470114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suryawanshi H, et al. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018;4:eaau4788. doi: 10.1126/sciadv.aau4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huhn O, et al. Distinctive phenotypes and functions of innate lymphoid cells in human decidua during early pregnancy. Nat. Commun. 2020;11:381. doi: 10.1038/s41467-019-14123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setty M, et al. Characterization of cell fate probabilities in single-cell data with Palantir. Nat. Biotechnol. 2019;37:451–460. doi: 10.1038/s41587-019-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajagopalan S, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Manno G, et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 35.Kang K, et al. Interferon-gamma represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity. 2017;47:235–250. doi: 10.1016/j.immuni.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mor G, Aldo P, Alvero AB. The unique immunological and microbial aspects of pregnancy. Nat. Rev. Immunol. 2017;17:469–482. doi: 10.1038/nri.2017.64. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y, et al. PBX1 expression in uterine natural killer cells drives fetal growth. Sci. Transl. Med. 2020;12:537. doi: 10.1126/scitranslmed.aax1798. [DOI] [PubMed] [Google Scholar]
- 39.Raghupathy R, et al. Cytokine production by maternal lymphocytes during normal human pregnancy and in unexplained recurrent spontaneous abortion. Hum. Reprod. 2000;15:713–718. doi: 10.1093/humrep/15.3.713. [DOI] [PubMed] [Google Scholar]
- 40.Saito S, Nakashima A, Shima T, Ito M. Th1/Th2/Th17 and regulatory T-cell paradigm in pregnancy. Am. J. Reprod. Immunol. 2010;63:601–610. doi: 10.1111/j.1600-0897.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Sung N, Gilman-Sachs A, Kwak-Kim J. T helper (Th) cell profiles in pregnancy and recurrent pregnancy losses: Th1/Th2/Th9/Th17/Th22/Tfh cells. Front. Immunol. 2020;11:2025. doi: 10.3389/fimmu.2020.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada S, Ebina Y, Iijima N, Deguchi M, Yamada H. Decidual CD68(+) HLA-DR(+) CD163(-) M1 macrophages increase in miscarriages with normal fetal chromosome. Am. J. Reprod. Immunol. 2018;79:e12791. doi: 10.1111/aji.12791. [DOI] [PubMed] [Google Scholar]
- 43.Faas MM, Spaans F, De Vos P. Monocytes and macrophages in pregnancy and pre-eclampsia. Front. Immunol. 2014;5:298. doi: 10.3389/fimmu.2014.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laird SM, et al. A review of immune cells and molecules in women with recurrent miscarriage. Hum. Reprod. Update. 2003;9:163–174. doi: 10.1093/humupd/dmg013. [DOI] [PubMed] [Google Scholar]
- 45.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng GX, et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mereu E, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020;38:747–755. doi: 10.1038/s41587-020-0469-4. [DOI] [PubMed] [Google Scholar]
- 48.Huang M, et al. SAVER: gene expression recovery for single-cell RNA sequencing. Nat. Methods. 2018;15:539–542. doi: 10.1038/s41592-018-0033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo Z, et al. ATAC-pipe: general analysis of genome-wide chromatin accessibility. Brief. Bioinform. 2018;20:1934–1943. doi: 10.1093/bib/bby056. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for single-cell RNA-seq and ATAC-seq samples are available in the Genome Sequence Archive (GSA) database as accession number CRA002181. Reviewer access link: https://bigd.big.ac.cn/gsa/s/nBQxzt82.