Abstract
Purpose
Factors associated with invasive recurrence (REC) of ductal carcinoma in situ (DCIS) are less known. This study was aimed at identifying better biomarkers to predict the prognosis of DCIS.
Methods
RNA extracted from formalin-fixed paraffin-embedded blocks of twenty-four pure DCIS cases was subjected to differential gene expression analysis. The DCIS cases were selected by matching age and estrogen receptor status. Sixteen REC-free and 8 invasive-REC cases with disease-free interval of > 5 years were analyzed. Immunohistochemistry (IHC) staining was used to validate sixty-one independent pure DCIS cases, including invasive-REC (n = 16) and REC-free (n = 45) cases.
Results
Eight differentially expressed genes (DEGs) were statistically significant (log 2-fold change [FC] < –1 or > 1 and p < 0.001). Less than ½ fold expression of CUL1, androgen receptor (AR), RPS27A, CTNNB1, MAP3K1, PRKACA, GNG12, MGMT genes was observed in the REC group compared to the no evidence of disease group. AR and histone deacetylase 1 (HDAC1) genes were selected for external validation (AR: log 2-FC − 1.35, p < 0.001, and HDAC1: log 2-FC − 0.774, p < 0.001). External validation showed that the absence of AR and high HDAC1 expression were independent risk factors for invasive REC (hazard ratio [HR], 5.04; 95% confidence interval [CI], 1.24–20.4; p = 0.023 and HR, 3.07; 95% CI, 1.04–9.04; p = 0.042). High nuclear grade 3 was also associated with long-term invasive REC.
Conclusion
Comparative gene expression analysis of pure DCIS revealed 8 DEGs among recurring cases. External validation with IHC suggested that the absence of AR and overexpression of HDAC1 are associated with a greater risk of long-term invasive REC of pure DCIS.
Keywords: Receptors, androgen; Carcinoma, intraductal, noninfiltrating; Histone deacetylase 1; Prognosis
INTRODUCTION
With cancer screening becoming more widespread, up to 20% of breast cancer patients are now being diagnosed with ductal carcinoma in situ (DCIS). It is a “non-obligate precursor to invasive ductal carcinoma (IDC)” with an excellent prognosis. While surgical resection is the primary therapy for DCIS, adjuvant radiotherapy (RT) after lumpectomy has long been a controversial issue. Trials have shown that the local recurrence (REC) rate has been relatively reduced by 50% in the irradiated group. However, these studies did not show any benefit on distant metastases or overall survival [1]. RT may induce cardiac toxicity, fibrosis of lung, and skin changes. It also requires daily visits to the radiation center and is expensive. Its risks could outweigh the benefits in patients who are originally at low-risk. This implies that patient groups must be accurately classified according to their risk of subsequent invasive RECs. The absolute benefit of chemoprevention is being investigated, with a consideration of possible side effects such as uterine carcinoma and menopausal symptoms. However, owing to the lack of tools to accurately classify risk and benefit of such treatments, current guidelines recommend surgical resection followed by radiation and/or chemoprevention with tamoxifen, despite the indolent clinical course of the disease.
Young age, high histologic grade, large tumor size, and absence of hormone receptor are the historical prognostic markers of DCIS [2,3,4]. Van Nuys prognostic index classifies the risk of invasive REC into low/intermediate/high-risk group by adding scores of age, margin width, nuclear grade/necrosis, and tumor size [5]. Oncotype DCIS predicts prognosis using gene expression of DCIS tumor samples [6]. DCISionRT® is a recently developed radio-genomic tool that predicts the benefit of radiation [7]. However, both of these methods require a high level of evidence and overall feasibility, including cost effectiveness [8]. For DCIS, it is important to assess the risk of long-term REC because almost 98% of cases are reported to be disease-free up to five years after diagnosis. The aim of the study was to investigate prognostic biomarkers of DCIS to better predict long-term invasive REC. Immunochemistry was used to identify markers that could be easily applied in clinic.
METHODS
Patient selection
We selected patients diagnosed with pure DCIS with no invasive component after definitive surgery. We chose only those who had undergone surgery before 2008 to investigate the long-term prognosis. We used messenger RNA (mRNA) expression profiling as the primary analysis method and immunohistochemistry (IHC) staining for external validation. We selected a discovery cohort for gene expression analysis and a validation cohort for IHC staining.
Thirty-six pure DCIS patients were selected for the discovery cohort, including twelve with invasive REC and twenty-four with no REC (NED). The NED cases were selected by 1:2 ratio matching for age and estrogen receptor (ER). All patients were diagnosed with DCIS and showed a negative tumor margin after undergoing a breast-conserving surgery or mastectomy between 1995 and 2004. Sequencing was successfully performed in eight REC cases and sixteen NED cases. Sixty-one pure DCIS patients were selected for the validation cohort. REC and NED cases were selected as done for discovery cohort. All selected patients underwent surgery at Asan Medical Center between 1995 and 2008.
RNA extraction and quantification
Surgical specimens used in the discovery phase were initially examined pathologically to confirm the diagnosis. Areas of the tumor component were dissected from the formalin-fixed paraffin-embedded (FFPE) tissue. Total RNA was extracted from the sections using MasterPure™ Complete DNA & RNA Purification Kit (Lucigen-Epicentre, Middleton, USA). Purity and yield of the extracted RNA were assessed using a DS 11 Spectrophotometer (Denovix Inc., Wilmington, USA). Total RNA (300 ng) was added to the sample preparation reaction in the available 5-μL volume. Quality check was performed with Fragment Analyzer (Advanced Analytical Technologies, Ankeny, USA). All the procedures were performed according to the manufacturer's instructions.
The mRNA expression profiling by nanoString nCounter system and data analysis
Expression assay was performed with 300 ng total RNA isolates. Digital multiplexed nanoString nCounter human mRNA expression assay was used. For evaluation, nCounter PanCancer Pathways Panel that can target 730 major cancer pathway genes was used. Two kinds of probe sets—8 μL of nCounter Reporter probes in hybridization buffer, and 2 μL of nCounter Capture probes—were added in the 5 μL of each RNA sample. The 2 combinations were kept overnight at 65°C for 16–20 hours for the reaction to occur. Two-step magnetic bead-based purification system in the nCounter Prep Station was used to remove the excess probes (nanoString Technologies, Seattle, USA).
Specific target molecules were quantified using the nCounter Digital Analyzer, which counted the individual fluorescent barcodes and assessed each target molecule. For each assay, a high-density scan that encompassed 280 fields of view was performed. Images of the immobilized fluorescent reporters in the sample cartridge were obtained using a charged-couple device camera nSolver software analysis, a freely available software from nanString Technology, was used for data analysis and R software (R Core Team [2013]. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/.) was used for the normalization of the mRNA profiling data.
Biomarker determination
Surgical specimens used in the validation phase were initially examined pathologically to confirm the diagnosis. FFPE tissue sections were immunohistochemically stained for anti-androgen receptor (AR) (host-Rabbit, clone SP107, 1:100, 200R-16; CELL MARQUE, Roklin, USA) using a BenchMark XT automatic immunostaining device (Ventana Medical Systems, Tucson, USA) with OptiView DAB IHC Detection Kit (Ventana Medical Systems). Tissues were sliced with a microtome into 4-µm-thick sections. These sections were transferred to silanized charged slides and were allowed to dry for 10 minutes at room temperature, followed by 20 minutes in an incubator at 65°C. Antigen-antibody reactions were triggered by heat-induced epitope retrieval method. The slides were kept in the Cell Conditioning 1 buffer for 32 minutes and then incubated for 16 minutes with antibodies in the autoimmunostainer. Reactions were visualized using Ventana OptiView DAB IHC Detection Kit (Optiview HQ Linker 8 minutes, Optiview HRP Multimer 8 minutes, Optiview H2O2/DAB 8 minutes, Optiview Copper 4 minutes). The slides were then counterstained with Ventana Hematoxylin II for 12 minutes and Ventana Bluing reagent for 4 minutes. Finally, the slides were removed from the stainer. They were dehydrated and then coverslipped for microscopic examination. All the procedures were performed according to the manufacturer's instructions.
The conventional biomarker ER expression was quantified by the Allred score and classified based on the St. Gallen and American Society of Clinical Oncology/College of American Pathologists guidelines (2013). Cases with Allred score 0 were considered AR-negative. An Allred score > 5 was defined as histone deacetylase 1 (HDAC1)-high, and an Allred score < 6 was defined as HDAC1-low.
Statistical analyses
Independent Student's t-test and Mann-Whitney test were performed for all continuous variables. χ2 test and Fisher's exact test was used to evaluate the relationship between the categorical clinical characteristics and relapse status. Kaplan-Meier method was used to estimate the REC-free survival. The prognostic role of risk factors was analyzed using a Cox proportional hazards model. All the p-values were based on 2-sided testing, and values lower than 0.05 were considered statistically significant. Analyses were performed with SPSS 20.0 (IBM Corp., Armonk, USA).
Ethics approval
Informed consent was obtained from all the discovery cohort patients for RNA analyses. Samples from validation cohort were retrieved anonymously according to the institutional system (Asan Biomedical Research Environment) Among the validation cohort, informed consent was retrieved for cases after February 2013. The study was approved by the Institutional Review Board (IRB) of Asan Medical Center (IRB No. 2016-0976).
RESULTS
Differentially expressed genes (DEGs) among the discovery cohort
Among thirty-six pure DCIS patients selected for comparative mRNA expression profiling, sequencing was successfully performed in eight REC cases and sixteen NED cases. Clinicopathological characteristics of these twenty-four cases (8 REC, 16 NED) are displayed in Table 1. The characteristics of the REC and NED group showed no statistically significant differences. Six of the REC cases showed local REC, while 2 showed regional REC. The median follow-up was 149 months in total: 56 months for the REC group and 162 months for the NED group. The mRNA expression levels for these 2 groups were compared. Fold change (FC) was calculated as the ratio of gene expression level REC/NED. Genes with |log2_FC| > 1 and p-value < 0.001 were selected as DEGs. Cases in the REC group had significantly lower expression of CUL1, AR, RPS27A, CTNNB1, MAP3K1, PRKACA, GNG12, and MGMT than that observed in the NED group. No gene was significantly overexpressed among the REC cases. The DEGs are listed in Table 2. All the data of analyzed genes are shown in Supplementary Table 1.
Table 1. Patient characteristics of discovery cohort.
| Characteristics | Overall (n = 24) | NED (n = 16) | REC (n = 8) | p-value | |
|---|---|---|---|---|---|
| Average age (yr) | 40.3 (25–56) | 41.1 (28–56) | 38.7 (25–50) | 0.667* | |
| ≥ 40 | 12 (50.00) | 9 (56.25) | 3 (37.50) | ||
| < 40 | 12 (50.00) | 7 (43.75) | 5 (62.50) | ||
| NG | 0.333† | ||||
| 1 | 2 (8.33) | 2 (12.50) | 0 (0.00) | ||
| 2 | 21 (87.50) | 14 (87.50) | 7 (87.50) | ||
| 3 | 1 (4.17) | 0 (0.00) | 1 (12.50) | ||
| Total | 24 | 16 | 8 | ||
| Tumor size (cm) | 0.189 | ||||
| < 2 | 8 (33.33) | 7 (43.75) | 1 (12.50) | ||
| ≥ 2 | 16 (66.67) | 9 (56.25) | 7 (87.50) | ||
| Total | 24 | 16 | 8 | ||
| ER | 0.578 | ||||
| + | 20 (83.33) | 14 (87.50) | 6 (75.00) | ||
| − | 4 (16.67) | 2 (12.50) | 2 (25.00) | ||
| Total | 24 | 16 | 8 | ||
| PR | 1.000 | ||||
| + | 19 (79.17) | 13 (81.25) | 6 (75.00) | ||
| − | 5 (20.83) | 3 (18.75) | 2 (25.00) | ||
| Total | 24 | 16 | 8 | ||
| HER2 | 0.137 | ||||
| + | 7 (29.17) | 3 (18.75) | 4 (50.00) | ||
| − | 16 (66.67) | 13 (81.25) | 3 (37.50) | ||
| Total | 23 | 16 | 7 | ||
| HT (n = 10) | 1.000 | ||||
| + | 6 (25.00) | 5 (31.25) | 1 (12.50) | ||
| − | 4 (16.67) | 3 (18.75) | 1 (12.50) | ||
| Total | 10 | 8 | 2 | ||
| RT | 0.509 | ||||
| + | 9 (37.50) | 6 (37.50) | 3 (37.50) | ||
| − | 3 (12.50) | 3 (18.75) | 0 (0.00) | ||
| Total | 12 | 9 | 3 | ||
| RM | 1.000 | ||||
| + | 3 (12.50) | 2 (12.50) | 1 (12.50) | ||
| − | 21 (87.50) | 14 (87.50) | 7 (87.50) | ||
| Total | 24 | 16 | 8 | ||
Values are presented as median (interquartile range) or number (%).
NED = no evidence of disease; REC = recurrence; ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2; HT = hormone therapy; RT = radiotherapy; RM = resection margin; NG = nuclear grade.
*Fisher's exact test for ≥ 40 group vs. < 40 group; †Fisher's exact test for NG 1, 2 group vs. NG 3 group.
Table 2. List of differentially expressed genes.
| Gene | Gene name | Log2 FC | Standard error | Lower CL | Upper CL | p-value |
|---|---|---|---|---|---|---|
| CUL1 | Culin1 | −1.13 | 0.189 | −1.49 | −0.755 | < 0.001 |
| AR | Androgen receptor | −1.35 | 0.306 | −1.95 | −0.746 | < 0.001 |
| RPS27A | Ribosomal protein S27a | −1.43 | 0.330 | −2.07 | −0.780 | < 0.001 |
| CTNNB1 | Catenin beta 1 | −1.34 | 0.333 | −1.99 | −0.689 | < 0.001 |
| MAP3K1 | Mitogen-activated protein | −1.54 | 0.383 | −2.30 | −0.792 | < 0.001 |
| Kinase kinase kinase 1 | ||||||
| PRKACA | Protein kinase CAMP-activated | −1.68 | 0.433 | −2.53 | −0.831 | < 0.001 |
| Catalytic subunit alpha | ||||||
| GNG12 | G protein subunit gamma 12 | −1.27 | 0.331 | −1.92 | −0.620 | < 0.001 |
| MGMT | O-6-methylguanine-DNA | −1.04 | 0.272 | −1.57 | −0.505 | < 0.001 |
| Methyltransferase |
Cutoff score: log2-FC < −1 or > 1 and p < 0.001.
FC = fold change; CL = confidence limit.
Among the DEGs, AR (log2_FC − 1.35, p < 0.001) and HDAC1 (log2_FC − 0.774, p < 0.001) were chosen as candidate prognostic biomarkers. We evaluated the prognostic impact of both AR and HDAC1 within an independent validation cohort by IHC.
Validation of prognostic biomarker among independent cases
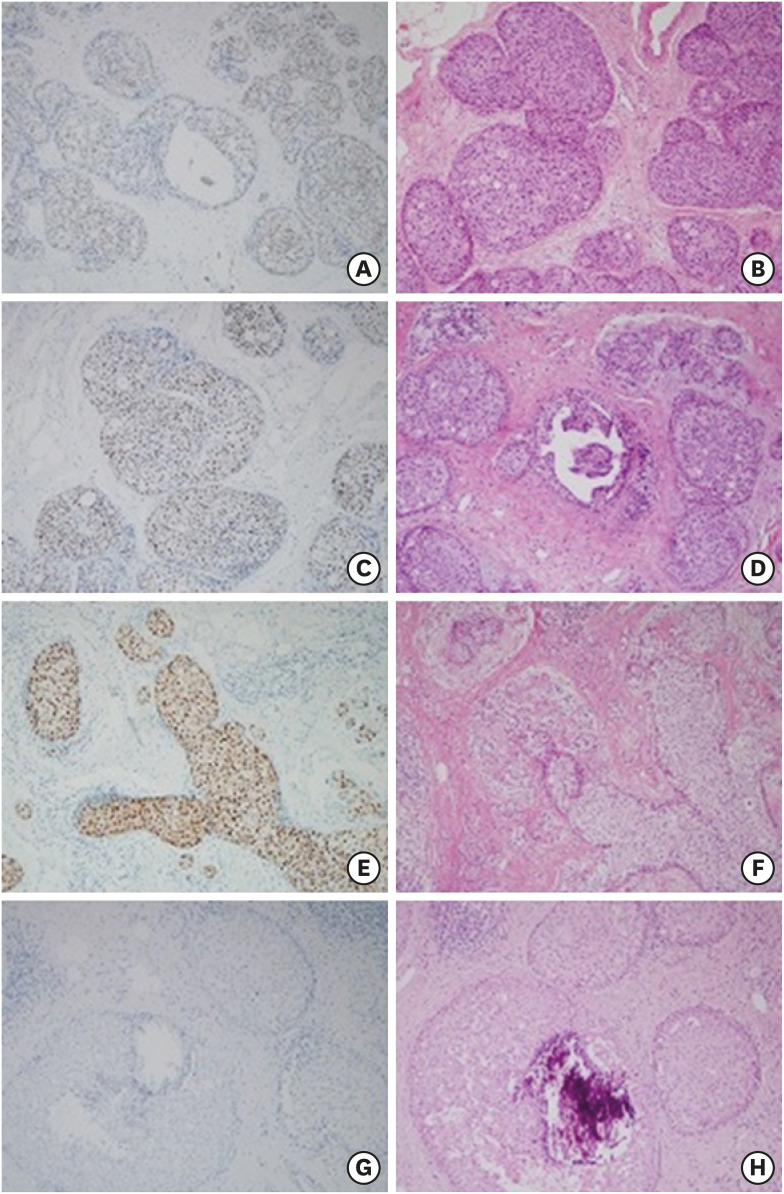
An independent cohort was selected for validation of AR and HDAC1 as prognostic biomarkers. Samples from sixty-one pure DCIS surgical specimens were obtained for IHC staining. Of these, sixteen were REC patients and forty-five were NED patients. Clinicopathological characteristics of the validation cohort are described in Table 3. In the REC group, thirteen local REC cases and 3 regional lymph node REC cases were included. The median follow-up time was 110 months in total: 65 months for the REC group and 114 months for the NED group. Figure 1 shows the IHC staining results for AR. To evaluate factors associated with AR negativity (vs. AR-positive) and high HDAC1 expression (vs. HDAC1-low), the validation cohort was divided according to AR-positive vs. AR-negative group, and a HDAC1-high vs. HDAC1-low group. Each pair of groups was statistically similar for other conventional risk factors (Table 4).
Table 3. Patient characteristics for the validation cohort.
| Characteristics | Overall (n = 61) | NED (n = 45) | REC (n = 16) | p-value | |
|---|---|---|---|---|---|
| Average age (yr) | 43.1 (27–65) | 43.6 (27–65) | 41.9 (28–59) | 0.674 | |
| ≥ 40 | 37 (60.66) | 28 (62.22) | 9 (56.25) | ||
| < 40 | 24 (39.34) | 17 (37.78) | 7 (43.75) | ||
| Nuclear grade | 0.063 | ||||
| 1 | 13 (21.31) | 9 (20.00) | 4 (25.00) | ||
| 2 | 36 (59.02) | 30 (66.67) | 6 (37.50) | ||
| 3 | 12 (19.67) | 6 (13.33) | 6 (37.50) | ||
| Total | 61 | 45 | 16 | ||
| Tumor size (cm) | 0.576 | ||||
| Multiple | 7 (11.48) | 4 (8.89) | 3 (18.75) | ||
| ≥ 2 | 17 (27.87) | 13 (28.89) | 4 (25.00) | ||
| < 2 | 37 (60.66) | 28 (62.22) | 9 (56.25) | ||
| Total | 61 | 45 | 16 | ||
| ER | 1.000 | ||||
| + | 46 (76.67) | 34 (75.56) | 12 (80.00) | ||
| − | 14 (23.33) | 11 (24.44) | 3 (20.00) | ||
| Total | 60 | 45 | 15 | ||
| PR | 1.000 | ||||
| + | 45 (75.00) | 34 (75.56) | 11 (73.33) | ||
| − | 15 (25.00) | 11 (24.44) | 4 (26.67) | ||
| Total | 60 | 45 | 15 | ||
| HER | 0.803 | ||||
| + | 27 (45.76) | 21 (46.67) | 6 (42.86) | ||
| − | 32 (54.24) | 24 (53.33) | 8 (57.14) | ||
| Total | 59 | 45 | 14 | ||
| RT (n = 57) | 0.333 | ||||
| + | 50 (87.72) | 40 (90.91) | 10 (76.92) | ||
| − | 7 (12.28) | 4 (9.09) | 3 (23.08) | ||
| Total | 57 | 44 | 13 | ||
| HT (n = 43) | 0.295 | ||||
| + | 24 (55.81) | 20 (60.61) | 4 (40.00) | ||
| − | 19 (44.19) | 13 (39.39) | 6 (60.00) | ||
| Total | 43 | 33 | 10 | ||
| AR | 0.108 | ||||
| + | 56 (91.80) | 43 (95.56) | 13 (81.25) | ||
| − | 5 (8.20) | 2 (4.44) | 3 (18.75) | ||
| Total | 61 | 45 | 16 | ||
| HDAC1 | 0.164 | ||||
| 0–6 | 47 (77.05) | 37 (82.22) | 10 (62.50) | ||
| 7–8 | 14 (22.95) | 8 (17.78) | 6 (37.50) | ||
| Total | 61 | 45 | 16 | ||
| RM | 1.000 | ||||
| + | 5 (8.20) | 4 (8.89) | 1 (6.25) | ||
| − | 56 (91.80) | 41 (91.11) | 15 (93.75) | ||
| Total | 61 | 45 | 16 | ||
Values are presented as median (interquartile range) or number (%).
NED = no evidence of disease; REC = recurrence; ER = estrogen receptor; PR = progesterone receptor; HER = human epidermal growth factor receptor; RT = radiotherapy; HT = hormone therapy; AR = androgen receptor; HDAC1 = histone deacetylase 1; RM = resection margin.
Figure 1. Immunohistochemistry and H&E staining findings for AR. The presented slides are classified according to the intensity of AR: (A), (C), (E) show cases with intensity 1, 2, 3, respectively. (G) shows a case with negative AR intensity. (B), (D), (F), (H) are the H&E results.
H&E = hematoxylin and eosin stain; AR = androgen receptor.
Table 4. Univariate analysis of clinicopathologic factors.
| Characteristics | AR+ (n = 56) | AR*− (n = 5) | p-value | HDAC1 high (n = 14) | HDAC1 low (n = 47) | p-value | |
|---|---|---|---|---|---|---|---|
| Average age (yr) | 42 (27–65) | 46 (32–55) | 0.640 | 39 (28–59) | 44 (27–65) | 0.120 | |
| ≥ 40 | 33 (58.9) | 4 (80.0) | 6 (42.9) | 31 (66.0) | |||
| < 40 | 23 (41.1) | 1 (20.0) | 8 (57.1) | 16 (34.0) | |||
| Nuclear grade | 0.573 | 0.124 | |||||
| 1 | 10 (17.9) | 3 (60.0) | 3 (21.4) | 10 (21.3) | |||
| 2 | 34 (60.7) | 2 (40.0) | 6 (42.9) | 30 (63.8) | |||
| 3 | 12 (21.4) | 0 (0.0) | 5 (35.7) | 7 (14.9) | |||
| Total | 56 | 5 | 14 | 47 | |||
| Tumor size (cm) | 0.634 | 0.492 | |||||
| Multiple | 6 (10.7) | 1 (20.0) | 3 (21.4) | 4 (8.5) | |||
| ≥ 2 | 16 (28.6) | 1 (20.0) | 3 (21.4) | 14 (29.8) | |||
| < 2 | 34 (60.7) | 3 (60.0) | 8 (57.1) | 29 (61.7) | |||
| Total | 56 | 5 | 14 | 47 | |||
| ER | 1.000 | 1.000 | |||||
| + | 43 (76.8) | 3 (75.0) | 11 (78.6) | 35 (76.1) | |||
| − | 13 (23.2) | 1 (25.0) | 3 (21.4) | 11 (23.9) | |||
| Total | 56 | 4 | 14 | 46 | |||
| PR | 0.258 | 0.734 | |||||
| + | 43 (76.8) | 2 (50.0) | 10 (71.4) | 35 (76.1) | |||
| − | 13 (23.2) | 2 (50.0) | 4 (28.6) | 11 (23.9) | |||
| Total | 56 | 4 | 14 | 46 | |||
| HER2 | 0.617 | 1.000 | |||||
| + | 26 (47.3) | 1 (25.0) | 6 (42.9) | 21 (46.7) | |||
| − | 29 (52.7) | 3 (75.0) | 8 (57.1) | 24 (53.3) | |||
| Total | 55 | 4 | 14 | 45 | |||
| RT (n=53) | 0.070 | 0.333 | |||||
| + | 48 (90.6) | 2 (50.0) | 10 (76.9) | 40 (90.9) | |||
| − | 5 (9.4) | 2 (50.0) | 3 (23.1) | 4 (9.1) | |||
| Total | 53 | 4 | 13 | 44 | |||
| HT (n=43) | 0.079 | 0.295 | |||||
| + | 24 (60.0) | 0 (0.0) | 4 (40.0) | 20 (60.6) | |||
| − | 16 (40.0) | 3 (100.0) | 6 (60.0) | 13 (39.4) | |||
| Total | 40 | 3 | 10 | 33 | |||
Values are presented as median (interquartile range) or number (%).
AR = androgen receptor; HDAC1 = histone deacetylase 1; ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2; RT = radiotherapy; HT = hormone therapy.
*Absence of AR-positive cells.
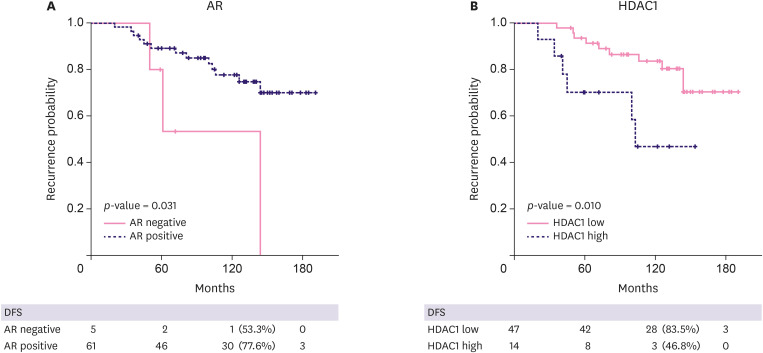
Survival analysis was performed to evaluate AR and HDAC1 protein expression and the risk of invasive REC. Figure 2 illustrates a Kaplan-Meier survival curve for the AR-negative vs. AR-positive group, and the HDAC1-high and HDAC1-low group. Ten-year disease-free survival rates were 53.3% vs. 77.6% for the AR-negative vs. AR-positive group, and 46.8% vs. 83.5% for the HDAC1-high vs. HDAC1-low group. Factors associated with REC, identified by univariate and multivariate Cox regression analysis, are described in Table 5. Conventional risk factors such as age and size were not significantly associated. NG 3 cases had a higher REC rate than that of NG 1 or 2 cases, indicating high-grade (HG) DCIS as an independent risk factor (HR, 4.89; p = 0.005). Both AR negativity and high HDAC1 expression were independently associated with invasive REC (HR, 5.04; p = 0.031 and HR, 3.07; p = 0.010, respectively). We then expanded the survival analysis to the IHC result of the discovery cohort. Again, both AR negativity and high expression of HDAC1 were independent risk factors (p = 0.004 and p = 0.038, data not shown).
Figure 2. Recurrence-free survival comparing AR-negative group vs. AR-positive group, and HDAC1-low group vs. HDAC1-high group: (A) AR-negative group showed poor prognosis (p-value 0.031, 10 years DFS 53.3%, 77.6% for AR-negative and positive groups, respectively). (B) Group with high HDAC1 showed higher recurrence rate (p-value 0.010, 10 years DFS 46.8%, 83.5% for HDAC1-high and low groups, respectively).
DFS = disease-free survival; AR = androgen receptor; HDAC1 = histone deacetylase 1.
Table 5. Survival analysis of independent validation cohort.
| Variables | Univariate analysis† | Multivariate analysis‡ | ||
|---|---|---|---|---|
| p-value | HR | 95% CI | p-value | |
| NG (1, 2 vs. 3) | 0.006 | 4.89 | 1.60–14.9 | 0.005 |
| Size (cut off 2 cm) | 0.617 | 1.18 | 0.42–3.37 | 0.749 |
| AR (negative*) | 0.031 | 5.04 | 1.24–20.4 | 0.023 |
| HDAC1 (high) | 0.010 | 3.07 | 1.04–9.04 | 0.042 |
NG = nuclear grade; AR = androgen receptor; HDAC1 = histone deacetylase 1; HR = hazard ratio; CI = confidence interval.
*Absence of AR-positive cells; †Kaplan-Meier test was conducted for univariate analysis; ‡Cox regression test was conducted for multivariate analysis.
DISCUSSION
To identify biomarkers for long-term invasive REC in pure DCIS, we performed differential mRNA expression profiling of cancer gene panel, followed by external validation of AR and HDAC1 with IHC. Our study showed that DCIS cases that progress to invasive carcinoma have different gene expression features compared to DCIS cases that remain in the NED state. Differential gene expression profiling found eight genes that were less expressed in REC cases (|log2_FC| > 1, p < 0.001). Among them, AR and HDAC1 were selected for external validation by IHC. Survival analysis revealed that the group lacking AR cells and the group with high HDAC1 expression exhibited a high-risk of invasive REC. These results were statistically significant, suggesting that both AR and HDAC1 may serve as potential prognostic biomarkers for pure DCIS patients.
Unlike ER, the role of AR in the carcinogenesis of breast cancer is not well-established. AR is co-expressed with ER in 70%–90% of cases and is also expressed in 40% of ER-negative cancers [9]. The effect of AR on ER-negative breast cancer is mixed, but crosstalk between ER signaling and AR signaling exists in several ER-positive invasive breast cancers. Several studies have showed that AR expression is associated with favorable characteristics such as older age, smaller size, well-differentiated tumors, lower proliferation index, and higher positivity of hormone receptors and as a consequence, better overall survival [10,11,12,13]. Results from in vitro models support the findings that the overexpression of AR in ER-positive cell lines inhibits the proliferative activity of ER [14]. AR seems to be a competitive inhibitor of ER in its binding to the estrogen-response element of ER target genes [15]. Therefore, it is receiving attention as a biomarker for prognosis prediction and as a potential therapeutic target [16].
However, the prognostic role of AR in DCIS is more ambiguous. IHC staining of hormone receptors showed that the rate of AR expression was lower in HG-IDC than in HG-DCIS [17]. Other studies have shown that AR expression level is higher in DCIS adjacent to IDC than in pure DCIS [18,19]. In statistical analyses of the expression of several biomarkers of breast cancer, AR did not show statistical significance as a risk factor for REC [20]. Recently, based on the concept of competition between AR and ER, Ravaioli et al. [21] suggested AR/ER ratio as a prognostic marker for DCIS. AR was associated with a favorable prognosis in this study population, which correlates with the previously reported biological effect of AR as an ER signal inhibitor. As the cases were selected according to hormone receptor positivity rather than in a consecutive manner, we could not directly observe the effect of AR confined to hormone receptor-positive DCIS cases. Moreover, the numbers analyzed were small, especially those of the REC cases.
HDAC1 is a member of the protein family HDAC, which modifies the chromatin by removing acetyl groups from histones. The HDAC family is an epigenetic regulator of gene expression that plays an important role in both normal cell development and carcinogenesis. In cancer cells, HDAC removes an acetyl group from histones, causing chromatin condensation that suppresses gene expression [22]. Tumor suppressor genes, and genes related to cell cycle inhibitors, apoptosis inducers, or differentiation factors are affected by global hypoacetylation. HDAC1 regulates the expression of key proteins involved in the cell cycle, such as p21, p53, and cyclin D1. Overexpression of HDAC1 upregulates the expression of vascular endothelial growth factor and hypoxia-inducible factor-1α, and increases angiogenesis. Besides specific gene regulation, global loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 are the hallmarks of human cancer cells, including breast cancer cells [23]. Moreover, global loss of histone modification is related to poor breast cancer-specific survival and disease-free survival [24].
Little is known about the correlation between HDAC1 and breast cancer. In an in vitro study, overexpression of HDAC1 induced the loss of ER and increased cell proliferation, and an HDAC inhibitor induced ER re-expression [25]. In breast cancer cells, HDAC1 inhibitors induce cell cycle arrest and apoptosis. Therefore, tumor cells are differentiated, and the growth is inhibited. [26]. Suzuki et al. [27] reported DCIS showing reduced acetylation compared to normal breast epithelial cells, regardless of HDAC1 expression. Several retrospective studies have shown HDAC1 to be associated with favorable prognosis [28]. However, a recent meta-analysis argued that in invasive breast cancer, HDAC1 overexpression does not correlate with disease-free survival and overall survival [29]. Thus, the expression of HDAC1 in DCIS remains controversial.
This study implies the potential role of AR and HDAC1 as prognostic biomarkers for pure DCIS. The strength of this study is that the cohort comprised pure DCIS cases with long-term follow-up, as most of DCIS patients do not undergo > 5–10 years surveillance. Observation of pure DCIS lesions over a long follow-up period, compared to analyzing synchronous DCIS lesions, enables investigating the natural history more directly. Synchronous DCIS lesions may not be a high-risk DCIS and may possess different biological features. Moreover, to represent true high-risk group, in situ RECs were excluded and only invasive REC were included for the REC group. However, there are several limitations to this study. The number of cases analyzed in both the cohorts were small. The study was not a consecutive case-series analysis. Rather, each REC and NED case was selected according to age and hormone receptor status in a 1:2 ratio. However, as DCIS has an exceptionally favorable outcome, the number of cases with invasive REC within certain timeframe is small. The REC and NED cases were matched by age and hormone receptor status to minimize the effect of these known risk factors. The cutoff value of AR and HDAC1 should be further addressed as there is no current standard cutoff.
In conclusion, we performed a matched comparative gene expression analysis of pure DCIS cases to identify prognostic indicators of long-term invasive REC. Our study revealed that the absence of AR and overexpression of HDAC1 are associated with a greater risk of invasive REC. Further validation within a larger series is needed to confirm this research.
Footnotes
Funding: This work was supported by a grant (2018-783) from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
Conflict of Interest: The authors declare that they have no competing interests.
Author Contributions: Conceptualization: Kim J; Data curation: Lee CM, Jo HG; Formal analysis: Lee CM, Lee SB, Kim HJ, Kim J; Funding acquisition: Kim J; Investigation: Chung IY; Methodology: Park S, Yun KW, Lee HJ; Resources: Park HJ; Supervision: Son BH, Ahn SH; Validation: Ko BS, Lee JW, Kim J; Writing - original draft: Lee CM; Writing - review & editing: Chung IY, Kim J.
SUPPLEMENTARY MATERIAL
Differential gene expression list
References
- 1.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) Correa C, McGale P, Taylor C, Wang Y, Clarke M, et al. Overview of the randomized trials of radiotherapy in ductal carcinoma in situ of the breast. J Natl Cancer Inst Monogr. 2010;2010:162–177. doi: 10.1093/jncimonographs/lgq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCormick B, Winter K, Hudis C, Kuerer HM, Rakovitch E, Smith BL, et al. RTOG 9804: a prospective randomized trial for good-risk ductal carcinoma in situ comparing radiotherapy with observation. J Clin Oncol. 2015;33:709–715. doi: 10.1200/JCO.2014.57.9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solin LJ, Gray R, Hughes LL, Wood WC, Lowen MA, Badve SS, et al. Surgical excision without radiation for ductal carcinoma in situ of the breast: 12-year results from the ECOG-ACRIN E5194 study. J Clin Oncol. 2015;33:3938–3944. doi: 10.1200/JCO.2015.60.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JS, Chen YH, Gadd MA, Gelman R, Lester SC, Schnitt SJ, et al. Eight-year update of a prospective study of wide excision alone for small low- or intermediate-grade ductal carcinoma in situ (DCIS) Breast Cancer Res Treat. 2014;143:343–350. doi: 10.1007/s10549-013-2813-6. [DOI] [PubMed] [Google Scholar]
- 5.Gilleard O, Goodman A, Cooper M, Davies M, Dunn J. The significance of the Van Nuys prognostic index in the management of ductal carcinoma in situ . World J Surg Oncol. 2008;6:61. doi: 10.1186/1477-7819-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solin LJ, Gray R, Baehner FL, Butler SM, Hughes LL, Yoshizawa C, et al. A multigene expression assay to predict local recurrence risk for ductal carcinoma in situ of the breast. J Natl Cancer Inst. 2013;105:701–710. doi: 10.1093/jnci/djt067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres MA. Genomic assays to assess local recurrence risk and predict radiation therapy benefit in patients with ductal carcinoma in situ. Int J Radiat Oncol Biol Phys. 2019;103:1021–1025. doi: 10.1016/j.ijrobp.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Raldow AC, Sher D, Chen AB, Recht A, Punglia RS. Cost effectiveness of the oncotype DX DCIS score for guiding treatment of patients with ductal carcinoma in situ . J Clin Oncol. 2016;34:3963–3968. doi: 10.1200/JCO.2016.67.8532. [DOI] [PubMed] [Google Scholar]
- 9.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Mod Pathol. 2011;24:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kensler KH, Poole EM, Heng YJ, Collins LC, Glass B, Beck AH, et al. Androgen receptor expression and breast cancer survival: results from the Nurses' Health Studies. J Natl Cancer Inst. 2019;111:700–708. doi: 10.1093/jnci/djy173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu Q, Mao Y, Fei XC, Shen KW. The impact of androgen receptor expression on breast cancer survival: a retrospective study and meta-analysis. PLoS One. 2013;8:e82650. doi: 10.1371/journal.pone.0082650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castellano I, Allia E, Accortanzo V, Vandone AM, Chiusa L, Arisio R, et al. Androgen receptor expression is a significant prognostic factor in estrogen receptor positive breast cancers. Breast Cancer Res Treat. 2010;124:607–617. doi: 10.1007/s10549-010-0761-y. [DOI] [PubMed] [Google Scholar]
- 13.Park S, Koo JS, Kim MS, Park HS, Lee JS, Lee JS, et al. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann Oncol. 2011;22:1755–1762. doi: 10.1093/annonc/mdq678. [DOI] [PubMed] [Google Scholar]
- 14.Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009;69:6131–6140. doi: 10.1158/0008-5472.CAN-09-0452. [DOI] [PubMed] [Google Scholar]
- 15.Need EF, Selth LA, Harris TJ, Birrell SN, Tilley WD, Buchanan G. Research resource: interplay between the genomic and transcriptional networks of androgen receptor and estrogen receptor α in luminal breast cancer cells. Mol Endocrinol. 2012;26:1941–1952. doi: 10.1210/me.2011-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kono M, Fujii T, Lim B, Karuturi MS, Tripathy D, Ueno NT. Androgen receptor function and androgen receptor-targeted therapies in breast cancer: a review. JAMA Oncol. 2017;3:1266–1273. doi: 10.1001/jamaoncol.2016.4975. [DOI] [PubMed] [Google Scholar]
- 17.Hanley K, Wang J, Bourne P, Yang Q, Gao AC, Lyman G, et al. Lack of expression of androgen receptor may play a critical role in transformation from in situ to invasive basal subtype of high-grade ductal carcinoma of the breast. Hum Pathol. 2008;39:386–392. doi: 10.1016/j.humpath.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Yu Q, Niu Y, Liu N, Zhang JZ, Liu TJ, Zhang RJ, et al. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann Oncol. 2011;22:1288–1294. doi: 10.1093/annonc/mdq586. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez LO, Corte MD, Junquera S, Bongera M, Rodriguez JC, Vizoso FJ. Expression of androgen receptor and two androgen-induced proteins (apolipoprotein D and pepsinogen C) in ductal carcinoma in situ of the breast. Histopathology. 2007;50:866–874. doi: 10.1111/j.1365-2559.2007.02687.x. [DOI] [PubMed] [Google Scholar]
- 20.Provenzano E, Hopper JL, Giles GG, Marr G, Venter DJ, Armes JE. Biological markers that predict clinical recurrence in ductal carcinoma in situ of the breast. Eur J Cancer. 2003;39:622–630. doi: 10.1016/s0959-8049(02)00666-4. [DOI] [PubMed] [Google Scholar]
- 21.Ravaioli S, Tumedei MM, Foca F, Maltoni R, Rocca A, Massa I, et al. Androgen and oestrogen receptors as potential prognostic markers for patients with ductal carcinoma in situ treated with surgery and radiotherapy. Int J Exp Pathol. 2017;98:289–295. doi: 10.1111/iep.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 23.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 24.Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 25.Kawai H, Li H, Avraham S, Jiang S, Avraham HK. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int J Cancer. 2003;107:353–358. doi: 10.1002/ijc.11403. [DOI] [PubMed] [Google Scholar]
- 26.Lagger G, Doetzlhofer A, Schuettengruber B, Haidweger E, Simboeck E, Tischler J, et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol. 2003;23:2669–2679. doi: 10.1128/MCB.23.8.2669-2679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki J, Chen YY, Scott GK, Devries S, Chin K, Benz CC, et al. Protein acetylation and histone deacetylase expression associated with malignant breast cancer progression. Clin Cancer Res. 2009;15:3163–3171. doi: 10.1158/1078-0432.CCR-08-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eom M, Oh SS, Lkhagvadorj S, Han A, Park KH. HDAC1 expression in invasive ductal carcinoma of the breast and its value as a good prognostic factor. Korean J Pathol. 2012;46:311–317. doi: 10.4132/KoreanJPathol.2012.46.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao W, Liu H, Liu R, Liu Q, Zhang T, Guo W, et al. Prognostic and clinical significance of histone deacetylase 1 expression in breast cancer: a meta-analysis. Clin Chim Acta. 2018;483:209–215. doi: 10.1016/j.cca.2018.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential gene expression list