Highlights
-
•
Cell-free (in vitro) method is ideal for short bioactive peptides rapid expression.
-
•
Camel lactoferricin is the most active among natural lactoferricin peptides.
-
•
Consensus lactoferricin is a candidate for further development into therapeutic use.
Abbreviations: bLFcin, bovine lactoferricin; CAMH, cation-adjusted Mueller-Hinton broth; CFPS, cell-free protein synthesis; cLf, camel lactoferrin; cLFcin, camel lactoferricin; ConLFcin, consensus lactoferricin; 3D, three dimensional structures; ELISA, enzyme-linked immunosorbent assay; hLf, human lactoferrin; hLFcin, human lactoferricin; HSV, herpes simplex virus; LC50, concentration lethal to 50 % of the cells; LFcin, lactoferricin; Lf, lactoferrin; MIC, minimum inhibitory concentration; MICs, minimum inhibitory concentrations; MRSA, methicillin-resistant Staphylococcus aureus; PBMCs, peripheral blood mononuclear cells; p-NPP, p-Nitrophenyl phosphate; SD, Shine-Dalgarno sequence; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis
Keywords: Lactoferrin, Lactoferricin, Bioactive peptides, In vitro protein synthesis, Antimicrobial
Abstract
For the first time, we produced four lactoferricin (LFcin) peptides by a cell-free (in vitro) method. These short antimicrobial peptides were expressed in an E. coli cell-free protein synthesis (CFPS) system and the bioactivity of the produced peptides was demonstrated. Additionally, we designed a novel synthetic consensus peptide (ConLFcin). The genes of bovine Lfcin (bLFcin), human Lfcin (hLFcin), camel Lfcin (cLFcin), and ConLFcin were cloned into pET101/D-TOPO vector then peptides were synthesized in vitro by E. coli CFPS system. The antibacterial activity of these synthesized peptides was evaluated against Escherichia coli, Salmonella typhi, Pseudomonas aeruginosa, Staphylococcus aureus, and methicillin-resistant Staphylococcus aureus (MRSA). The four cell-free synthesized peptides showed significant antibacterial potency at minimum inhibitory concentration (MIC) values between 1.25 and 10 μg/mL. cLFcin and ConLFcin showed higher antibacterial effects than bLFcin and hLFcin. Thus, cell-free expression system is an ideal system for rapid expression of functionally active short bioactive peptides.
1. Introduction
Natural antimicrobial peptides are possibly one of the successful forms of innate eukaryotic cells chemical defenses against microorganisms. Various antimicrobial peptides were isolated from nature and thousands of synthetic variants with broad-spectrum antimicrobial activities were also identified. A considerable number of these peptides demonstrated bactericidal, antiviral, and anticancer activities [1,2]. Antimicrobial peptides are commonly cationic and amphipathic, this allows interaction between the peptides and lipid cell membranes leading to lipid membranes disruption.
Lactoferricin is one of the most potent cationic antimicrobial peptides, which represents a small peptide fragment from N-terminal region of lactoferrin released by gastric pepsin digestion and comprises the highly positively charged sequence of Lf N-terminal region [3]. Lactoferrin is a multifunctional agent that acts as a potent innate immune defense mechanism in the host [[4], [5], [6]]. Camel lactoferrin (cLf) was reported to be the most active lactoferrin against various pathogenic bacteria, whereas human lactoferrin (hLf) was the least active [[7], [8], [9], [10]]. In addition, several studies proved the superior inhibitory effect of native or recombinant cLf and even its N- and C-lobes on viral infectious diseases [[11], [12], [13], [14], [15]]. These differences in biological activities between cLf and other lactoferrins were partly related to variance in sequence and levels of intrinsic disorder of the amino acid region 17–41 of the Lf N-terminal that corresponds to lactoferricin [8,14,16,17].
Most of the previous research has focused only on the production and bioactivity evaluation of bLFcin. It was separated from milk of cows and its sequence comprises 25 amino acid residues; (FKCRRWQWRMKKLGAPSITCVRRAF), with 2 Cys residues which form a disulfide bond between N-terminal region (highly positively charged) and C-terminal region of the peptide [18,19]. bLFcin displayed significant cytotoxic effect in vitro on several types of human and mouse cancer cell lines [20,21], at concentrations not toxic for normal cells as erythrocytes, lymphocytes, or fibroblasts. Additionally, bLFcin showed bactericidal effect on a wide range of bacteria as well as antiviral activity against many viruses such as herpes simplex virus (HSV) [22,23]. On the contrary, only a few studies about the production and in vitro bactericidal activity of hLFcin and cLFcin were published [24,25].
Production of lactoferricin by pepsin digestion of lactoferrin or artificial synthesis is difficult and expensive which hurdles research and application of this peptide. Recombinant production of LFcin has the advantages of being easy, quick, and scalable. For example, recombinant camel lactoferricin with suitable antimicrobial activity was produced at high yield by Pichia pastoris [25]. However, the toxicity of recombinant cationic antimicrobial peptides to E. coli and Saccharomyces cerevisiae is still an obstacle for recombinant in vivo expression. Besides, this short amino acid sequence is susceptible to degradation by host proteases. Commonly, both problems were solved by expression of recombinant LFcin linked with a fusion protein [26,27].
A new wave of interest in CFPS systems has shown their utility for protein engineering, including therapeutic proteins production, protein microarrays, and in vitro protein evolution. This technology uses an in vitro approach (using the necessary biological machinery for protein synthesis without the utilization of living host organisms), avoiding many host-protein interactions to solve problematic issues associated with using living host cells for recombinant protein expression, for example protein cytotoxicity and insolubility [[28], [29], [30], [31], [32], [33], [34], [35], [36]]. Currently used CFPS model systems are either prokaryotic (prepared from E. coli lysate) or eukaryotic (prepared from wheat germ, rabbit reticulocytes, and insect or human cells lysates). Unfortunately, the focus of previous efforts has been centered on utility and optimization of CFPS systems for producing various proteins at high titers not their capability of producing short bioactive peptides. Only a few studies addressed the utility of CFPS systems in antimicrobial peptide synthesis [37,38].
This study is the first report where the cell-free (in vitro) antimicrobial peptide synthesis approach was employed to express four LFcin peptides of about 6 kDa weight; LFcin peptides linked with fusion tags in the expression vector. Recombinant bLFcin, hLFcin, cLFcin, and ConLFcin were rapidly synthesized using E. coli CFPS system in active form. The in vitro antibacterial effects of these peptides were compared, for first time, against some pathogens (E. coli, S. typhi, P. aeruginosa, S. aureus, and MRSA).
2. Materials and methods
2.1. Strains and vectors
pET101/D-TOPO linear expression vector (Invitrogen, Carlsbad, USA) was used for cell-free protein production of LFcin peptides under control of T7 promoter.
E. coli XL2-Blue MRF’ strain (Stratagene. Heidelberg, Germany) was used as a cloning host for vector construction, amplification and storage.
Escherichia coli (Migula) Castellani and Chalmers (ATCC 25922) and Salmonella enterica subsp. enterica serovar Typhi (ATCC 19430) were obtained from American type culture collection (ATCC, USA). Pseudomonas aeruginosa was collected from Botany and Microbiology Department, Faculty of Science, Al-Azhar University, Assiut Branch (Egypt). Staphylococcus aureus subsp. aureus Rosenbach (ATCC 25923) was purchased from Becton Dickinson (France). MRSA clinical isolate was obtained from blood of a patient at Almery University Hospital (Alexandria, Egypt) and subjected to the confirming BD GeneOhm™ MRSA assay. All of these bacterial strains were used for the antimicrobial assay.
2.2. Alignment of ten N-terminal amino acids sequences of lactoferrin from different animal species to build a consensus lactoferricin peptide (ConLFcin) and consensus sequence analysis
The multiple sequence alignment program ClustalW version 1.4 (www.ebi.ac.uk/Tools/clustalw/) [39] was used to identify conserved sequence pattern in the amino acid sequences of hLFcin, bLFcin, cLFcin, and seven N-terminal sequences (the amino acid region 17–41) of lactoferrin from different animal species that correspond to other characterized lactoferricin peptides, then building a consensus sequence (ConLFcin). These aligned amino acid sequences of the lactoferrin gene from different animal species were obtained from the NCBI database (www.ncbi.nlm.nih.gov). The alignment was checked carefully for biological relevance and edited manually by the multifunctional sequence alignment editor BioEdit version 7 (www.mbio.ncsu.edu/BioEdit/bioedit.html).
The consensus sequence; ConLFcin was built by calculated variations on the natural N-terminal amino acid sequences of different lactoferrins. BioEdit was used to analyze the degree of similarity of the designed ConLFcin amino acid sequence to cLFcin, hLFcin, and bLFcin peptides using pairwise alignments. The physico-chemical properties of the bLFcin, hLFcin, cLFcin, and ConLFcin peptides in untagged and polyhistidine tagged forms were computed by the ProtParam tool – ExPASy.
The hydrophobic, hydrophilic, and charge properties of primary structures of bLFcin, hLFcin, cLFcin, and ConLFcin peptides were analyzed by the Helical Wheel tool (https://heliquest.ipmc.cnrs.fr/). This analysis includes the addition of the polyhistidine tag to the recombinant peptides.
The putative three dimensional (3D) structures of the four recombinant peptides were modeled by an automated protein homology-modeling server; SWISS-MODEL (http://swissmodel.expasy.org) [40]. The modeled structure was subjected to 1 ns MD simulation with GROMACS 5.0.2 in gromos43a1 and SPC water model to obtain correct structure [41].
2.3. Oligonucleotides design for synthesis of the hLFcin, bLFcin, cLFcin, and ConLFcin genes
The DNA sequences obtained by reverse translation of the amino acid sequences of hLFcin, bLFcin, cLFcin, and designed ConLFcin were codon optimized for optimal expression in E. coli-based CFPS system. To synthesize the bLFcin, hLFcin, cLFcin, and ConLFcin genes, overlapped oligonucleotides that span most of the sequence of both strands of these genes were designed based on E. coli codon usage. Each gene was divided into four oligonucleotides ranging from 30 to 35 nucleotides in length. The oligonucleotides were designed to have partial overlaps of 5–20 bp. The gene-specific forward PCR primers designed for gene amplification and subsequent pET101/D-TOPO expression vector directional cloning contains the sequence CACC at the 5′ end of the primers. Oligonucleotides and pairs of gene-specific primers for amplification and subsequent cloning were synthesized at Thermo Fisher Scientific (Bedford, MA, USA) and were purified using desalting.
2.4. Two-step PCR-mediated construction of the synthetic hLFcin, bLFcin, cLFcin, and ConLFcin genes
The bLFcin, hLFcin, cLFcin, and ConLFcin genes were assembled and amplified in two-step PCR [42], using the designed overlapped oligonucleotides and Expand high fidelity PCR system (Roche Molecular Biochemicals, Mannheim, Germany). PCR reactions were carried out using the Techne TC-3000 DNA thermal cycler (Bibby Scientific Limited, Staffordshire, UK).
The optimal reaction conditions for this two-step PCR were as follows: the oligonucleotide primers were added to a final concentration of 10 pM/each primer in assembling step, while the amplification step was run with 1 μL of the assembled products (now serving as the template) and 20 pM of the outermost oligonucleotide primers of each strand. The reaction cocktail was; 1x reaction buffer, 0.5 mM dNTPs Mix, 2.5 mM MgCl2, and 3.5 U Expand high fidelity polymerase mix in a final reaction volume of 25 μL.
Reactions were run using the company-recommended profile as follows: two stages; 10 cycles for first stage and 20 cycles for second stage, each cycle conditions were 94 °C for 30 s, 59 °C for 2 min, 72 °C for 2 min, followed by a final extension of 72 °C for 10 min.
2.5. Plasmids construction for cell-free expression
The blunt-ended PCR products were cloned into pET101/D-TOPO vector; the forward PCR primer for the amplification had CACC at the 5′ end that complements the pET101 vector GTGG overhang sequence. The ligated pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin plasmids were transformed into E. coli XL2-Blue MRF’ strain as a cloning host for vector construction, amplification and storage. The pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin constructs were confirmed with PCR analysis using pairs of designed gene-specific forward primers and sequencing reverse primers provided with the vector. Constructs that have the correct sized gene bands were subsequently sent for nucleotide sequencing. Circular constructs contain a T7 promoter, a Shine-Dalgarno (SD) sequence before the ATG start site, bLFcin, hLFcin, cLFcin, or ConLFcin gene, and fusion tags (V5 epitope and polyhistidine tag) controlling the cell-free peptide expression.
2.6. Cell-free peptide synthesis
Cell-free peptide synthesis was performed using the rapid translation system RTS100 E. coli HY (5 PRIME, Hamburg, Germany) for optimum (yield of about 400 μg/mL) T7 promoter-controlled in vitro expression from the constructed pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin templates. The cell-free synthesis reactions were run following the company instruction manual with some modifications. Briefly, in each reaction tube these components were pipetted: 12 μL E. coli S30 lysate, 10 μL reaction mix, 12 μL amino acids, 1 μL Methionine (3 mM), 5 μL reconstitution buffer, and 0.5 μg of the circular DNA templates (pET-bLFcin, pET-hLFcin, pET-cLFcin, or pET-ConLFcin) dissolved in 10 μL water or TE buffer. The reaction tubes were incubated at 25 °C for 4 h.
2.7. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) under non-denaturing conditions (native gel electrophoresis)
The soluble synthesized peptides were harvested from 50 μL cell-free reactions by centrifugation for 5 min at 12,000 xg and precipitation using 100 μL of −20 °C cold acetone. The synthesized peptides were separated on 18 % SDS-PAGE under non-denaturing conditions (native gel electrophoresis) to avoid SDS interaction with these strong cationic peptides. The protocol followed was very similar to that for denaturing SDS-PAGE but native or non-denaturing gel electrophoresis was run in the absence of SDS and reducing agent (β-mercaptoethanol or DTT). The precipitates were dissolved in 50 μL sample buffer, heated for 5 min at 95 °C, and then separated by 18 % native gel electrophoresis using an Invitrogen Novex Mini-Cell (USA). Gel was stained with Coomassie brilliant blue (Sigma, St. Louis, Mo., USA).
2.8. Detection of synthesized peptides by enzyme-linked immunosorbent assay (ELISA)
The assay was carried out as formerly described [33]. Costar ELISA plate (Cambridge, USA) was coated with 50 μL of each in vitro reaction. After an incubation period of 24 h at 4 °C; the plate was washed five times with 0.12 M NaCl, 0.04 M sodium phosphate pH 7.2 buffer (PBS), and then 100 μL of blocking buffer (2% gelatin in PBS) were added for 1 h at 37 °C. Then 50 μL of rabbit polyclonal anti-lactoferrin (Abcam, Cambridge, MA, USA) diluted 1:100 in 2% gelatin-PBS were added to each well. After 1 h of incubation at 37 °C, the plate was washed five times with PBS, and 50 μL of alkaline phosphatase–conjugated anti-rabbit IgG (BIO-RAD, Alfred Nobel, Hercules, USA) diluted 1:1000 with 2% gelatin–PBS were added, followed by an incubation of 1 h at 37 °C. p-Nitrophenyl phosphate (p-NPP) was added for color development, and optical density was measured at 405 nm.
2.9. Bioactivity evaluation of cell-free synthesized peptides by agar disc diffusion assay
Cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides were tested for antibacterial potency against Gram-positive bacteria (S. aureus, and MRSA) and Gram-negative bacteria (E. coli, S. typhi, and P. aeruginosa) by using the agar disc diffusion technique on Mueller-Hinton agar [43].
Test bacterial strains were cultured overnight in LB broth medium with shaking at 150 rpm and 37 °C. Mueller-Hinton agar plates were overlaid with 100 μL of 1 × 106 CFU/mL inoculum from these overnight cultures, then wells were bored into agar by using a sterile 6 mm cork borer, and 100 μL of cell-free reactions containing synthesized peptides at concentration of 40 μg were added into each well. Vancomycin antibacterial standard disc at concentration of 30 μg/mL was used as positive control in case of MRSA while chloramphenicol at concentration of 50 μg/mL was used as positive control in case of S. aureus, E. coli, S. typhi, and P. aeruginosa. Cell-free reaction run under the same conditions but in the absence of plasmid constructs was used as negative control. The culture plates were incubated at 4 °C for 2 h to allow proper diffusion of tested antimicrobials before being incubated at 37 °C for 24 h. Then plates were examined for the presence of the inhibition zones. The diameters of inhibition zones were determined by measuring the radius of the zone (from the center of the antimicrobial well or antibiotic disc to a point on the circumference of the zone where a distinct edge is present) then multiplying this measurement by 2. Experiment was done in triplicate and the results were presented as mean diameter of inhibition zone ± SD of triplicate.
Activity of cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides at different concentrations against test bacterial strains was also determined using this assay. Wells of inoculated Mueller-Hinton agar plates were incubated with serial dilution of cell-free reactions containing synthesized peptides (40, 20, 10, 5, 2.5, and 1.25 μg/mL). The culture plates were incubated at 4 °C for 2 h to allow proper diffusion of tested antimicrobials before being incubated at 37 °C for 24 h. Then plates were examined for the presence of the inhibition zones.
2.10. Purification of synthesized peptides
The peptides encoded by pET101 carry six histidine (6xHis) residues at their C-terminus, which allows their purification with a metal-chelating resin such as Ni–NTA [33]. After the reactions scaling up, the soluble fraction samples were loaded onto 1 mL Ni–NTA spin columns (Qiagen, Hilden, Germany), which had been equilibrated with binding buffer (50 mM NaH2PO4; 300 mM NaCl; 10 mM imidazole, pH 8.0) and the bound peptides were eluted with elution buffer (50 mM NaH2PO4; 300 mM NaCl; 500 mM imidazole, pH 8.0). The eluted peptides concentrations were determined using Lowry assay [44], and were analyzed by 18 % native gel electrophoresis.
2.11. Determination of minimum inhibitory concentrations (MICs) of cell-free synthesized peptides
The minimum inhibitory concentrations of cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin against MRSA, S. aureus, E. coli, S. typhi, and P. aerueginosa were determined by broth micro dilution method [45]. The 96-well micro titer plates (Greiner, Frickenhausen, Germany) were inoculated with test microorganisms (100 μL of 1 × 106 CFU/mL inoculum), and then 100 μL of cation-adjusted Mueller-Hinton (CAMH) broth containing synthesized and purified peptides in serial dilution (40, 20, 10, 5, 2.5, and 1.25 μg/mL) were added. Plates inoculated with tested bacteria were incubated at 37 °C for 24 h. The MICs were determined by measuring the absorbance at 600 nm for test bacterial strains. The MIC was defined as the lowest concentration at which growth was completely inhibited. All MIC determinations were performed in triplicate. Bacteria in CAMH broth were used as control of growth.
2.12. Endotoxin content
The endotoxin content (bacterial lipopolysaccharide (LPS)) was checked as described in [46] to avoid its mitogenic effects on the cell-culture system. All peptides synthesized by the cell-free system were free of endotoxin (data not shown).
2.13. Isolation of human blood lymphocytes and detection of cytotoxicity of cell-free synthesized peptides against these normal human cells
Peripheral blood mononuclear cells (PBMCs) were isolated as reported by Lohr et al. [47] and Liao et al. [11]. Briefly, peripheral blood samples collected from a single healthy volunteer were diluted with five volumes of a freshly prepared red blood cell (RBC) lysis buffer (38.8 mM NH4Cl, 2.5 mM KHCO3, and 1 mM EDTA, pH 8.0). After incubation at room temperature for 10 min, the mixture was centrifuged at 272 xg for 5 min. The nucleated cells were precipitated and washed with phosphate-buffered saline. Cytotoxicity of the cell-free synthesized peptides against human lymphocytes was detected by MTT assay. PBMCs were plated in 96-well tissue culture plate (Greiner, Frickenhausen, Germany) in triplicate at the density 25 × 104 cell/mL, and cultured in RPMI-1640 medium supplemented with 20 % fetal bovine serum (FBS), 1% penicillin–streptomycin for 24 h at 37 °C in 5% CO2, 95 % air. The medium was refreshed with new RPMI-1640 supplemented medium, and then cells were treated with various dilutions of purified peptides in total volume of 200 μL/well for 24 h. At 24 h, cells were centrifuged at 484 xg for 10 min and resuspended with 180 μL RPMI-1640 medium to rinse treated samples. Twenty microliters of 5 mg/mL MTT solution (Sigma, St. Louis, MO, USA) were added to each well and incubated at 37 °C for 3 h. The formed formazan crystals were dissolved with 180 μL of DMSO (Sigma, St. Louis, MO, USA). Optical density was measured at 560 nm. The percentage of cytotoxicity compared with the untreated cells as a control was determined. The plot of % cytotoxicity versus peptide concentration was used to calculate the concentration lethal to 50 % of the cells (LC50).
2.14. Statistical analysis
All experiments were done in triplicate and the results were presented as mean ± SD of triplicate. Differences between the variants were tested using Student’s t-test and McNemar’s test; p values of less than 0.01 were considered statistically significant.
3. Results
3.1. Amino acid sequence alignment of N-terminal amino acid sequences of lactoferrin from different animal species
ClustalW first accomplished pairwise alignments for bLFcin, hLFcin, cLFcin, and seven N-terminal amino acid sequences of lactoferrin from different animal species that have sequence homology with these characterized LFcin peptides. The alignment result was further refined manually by alignment editor BioEdit. This editor has a coloring scheme for amino acids facilitating manual editing as presented in Fig. 1A. The alignment identified six conserved amino acid residues of the 30 amino acids among all LFcin peptides indicated by * (asterisk) in consensus key (Fig. 1A). A consensus sequence of the amino acid residues most abundant in the alignment at each position was built (Fig. 1B).
Fig. 1.
(A) Clustal W multiple sequence alignment of bLFcin, hLFcin, cLFcin, and seven N-terminal amino acid sequences of lactoferrin from different animal species, edited by BioEdit. (B) The designed ConLFcin amino acid sequence. Consensus key: * (asterisk) - positions have a single, fully conserved residue, : (colon) - conservation between groups of strongly similar properties, . (period) - conservation between groups of weakly similar properties, and blank spaces mean no consensus. Accession numbers of lactoferrin amino acid sequences of various species in NCBI GenBank are as follows: AAA30610.1 (Bos taurus; bLFcin), CAB53387.1 (Camelus dromedarius; cLFcin), AAA59511.1 (Homo sapiens; hLFcin), CAA06441.1 (Bubalus bubalis), ACT53713.1 (Capra hircus), NP_001157446.1 (Equus caballus), ACB11584.1 (Macaca cyclopis), ACL80331.1 (Mus musculus), ACT76166.1 (Ovis aries), and AAA31059.1 (Sus scrofa).
The results of BioEdit analysis to the degree of similarity of the designed ConLFcin amino acid sequence to cLFcin, hLFcin, and bLFcin peptides using pairwise alignments suggested that ConLFcin showed the highest degree of similarity (76.66 % identity) to cLFcin (Table 1).
Table 1.
Sequence similarity of the designed ConLFcin amino acid sequence to cLFcin, hLFcin, and bLFcin.
| Sequence ID | Accession Number | Identities | % Identity |
|---|---|---|---|
| Camelus dromedarius (cLFcin) | CAB53387.1 | 0.7666667 | 76.66% |
| Homo sapiens (hLFcin) | AAA59511.1 | 0.6333333 | 63.33% |
| Bos taurus (bLFcin) | AAA30610.1 | 0.5666667 | 56.66% |
The physico-chemical properties of the bLFcin, hLFcin, cLFcin, and ConLFcin peptides computed by the ProtParam tool – ExPASy are presented in Table 2. The results represented in Table 2 reveal that the designed ConLFcin has the highest cationicity followed by cLFcin then bLFcin while hLFcin showed the lowest cationicity. This high cationicity of ConLFcin and cLFcin is expected to allow these peptides to interact more readily than other peptides with anionic phospholipids and/or LPS in addition to peptidoglycans in bacterial membranes leading to enhanced disruption of the structure integrity of bacterial cell membranes, which promotes and accelerates bacterial cell death.
Table 2.
Physico-chemical properties of the bLFcin, hLFcin, cLFcin, and ConLFcin peptides.
| Property | bLFcin | hLFcin | cLFcin | ConLFcin |
|---|---|---|---|---|
| Number of amino acids | 30 | 30 | 30 | 30 |
| 36* | 36* | 36* | 36* | |
| Molecular weight (Daltons) | 3655.47 | 3576.28 | 3542.26 | 3737.58 |
| 4478.31* | 4399.12* | 4365.11* | 4560.42* | |
| Theoretical pI | 10.90 | 10.33 | 10.57 | 11.63 |
| Total number of positively charged residues (Aspartic acid (D) + Glutamic acid (E)) | 8 | 7 | 9 | 11 |
| Total number of negatively charged residues (Arginine (R) + Lysine (K)) | 1 | 1 | 1 | 1 |
| Total number of atoms | 520 | 504 | 503 | 535 |
| 622* | 606* | 605* | 637* | |
| Aliphatic index | 71.67 | 58.33 | 45.33 | 48.67 |
| 59.72* | 48.61* | 37.78* | 40.56* | |
| Grand average of hydropathicity (GRAVY) | −0.177 | −0.717 | −0.787 | −0.863 |
| −0.681* | −1.131* | −1.189* | −1.253* | |
| +Estimated half-life | −1.1 h (mammalian reticulocytes, in vitro). | −7.2 h (mammalian reticulocytes, in vitro). | −1.9 h (mammalian reticulocytes, in vitro). | −1.9 h (mammalian reticulocytes, in vitro). |
| −3 min (yeast, in vivo). | - >20 h (yeast, in vivo). | - >20 h (yeast, in vivo). | - >20 h (yeast, in vivo). | |
| −2 min (Escherichia coli, in vivo). | - >10 h (Escherichia coli, in vivo) | - >10 h (Escherichia coli, in vivo). | - >10 h (Escherichia coli, in vivo). | |
| ∼Instability index | 81.25 | 55.90 | 44.28 | 80.60 |
| 72.80* | 51.68* | 42.00* | 72.26* | |
| Atomic Formula | C164H265N51O36S4 | C154H256N50O40S4 | C151H258N50O40S4 | C163H275N57O36S4 |
| C200H307N69O42S4* | C190H298N68O46S4* | C187H300N68O46S4* | C199H317N75O42S4* |
+Estimated half-life: N-terminal of the hLFcin, bLFcin, cLFcin, and ConLFcin sequences are C (Cysteine).
∼Instability index of the hLFcin, bLFcin, cLFcin, and ConLFcin peptides is computed to be 55.90, 81.25, 44.28, and 80.60, respectively. This classifies the peptides as unstable.
Calculated values for peptides tagged with 6xHis-tag. The remaining data did not change after tagging of peptides.
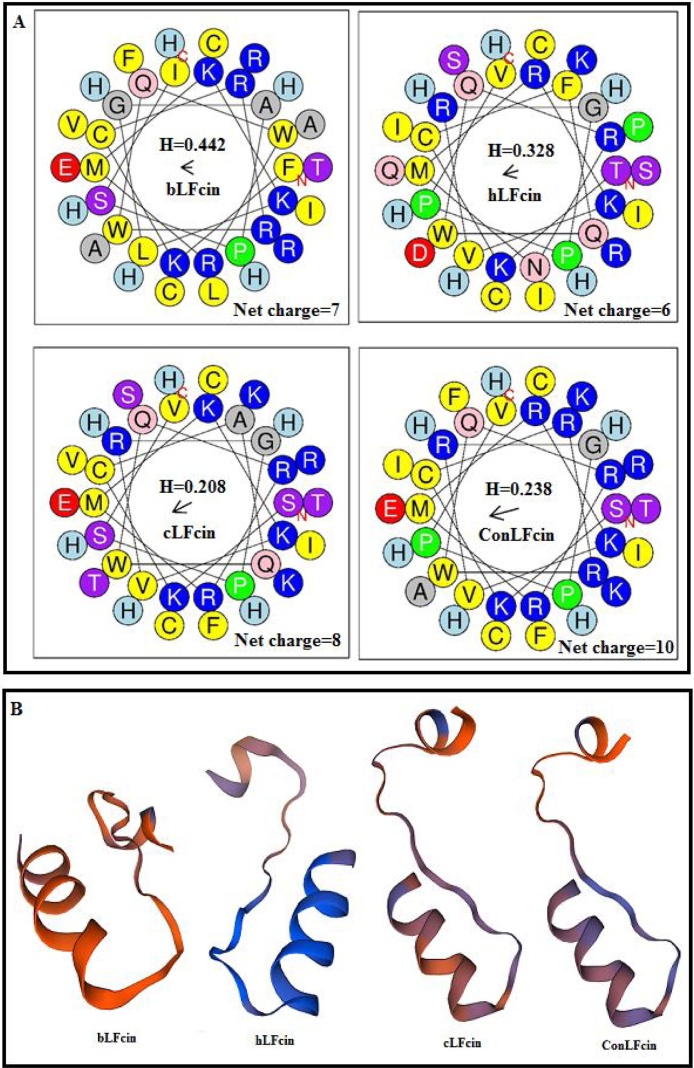
Helical Wheel representation of the properties of amphipathic cationic hLFcin, bLFcin, cLFcin, and ConLFcin peptides is showed in Fig. 2A. The presented results revealed that hLFcin has the lowest cationicity among the four peptides, while ConLFcin shows the highest cationicity. The 3D structures of the four recombinant peptides showed that bLFcin, hLFcin, cLFcin, and ConLFcin have a conserved amphipathic central α-helical structure (Fig. 2B). Furthermore, the addition of His-tag did not have any effect on the peptides 3D-functional structures (Fig. 2B).
Fig. 2.
(A) Helical Wheel representation of the properties of primary structures of amphipathic cationic bLFcin, hLFcin, cLFcin, and ConLFcin peptides tagged with His-tag. The color codes are as follows: yellow (nonpolar hydrophobic residues), red (acidic hydrophilic residues), violet (polar, uncharged hydrophilic residues), and blue (basic hydrophilic residues). H is abbreviation for hydrophobicity. No change was found in net charge values of peptides calculated by the Wheel after tagging of peptides with 6xHis-tag. On the other hand, hydrophobicity values for untagged bLFcin, hLFcin, cLFcin, and ConLFcin are 0.505, 0.368, 0.224, and 0.260, respectively, which are higher than those of tagged peptides. (B) 3D-structures of bLFcin, hLFcin, cLFcin, and ConLFcin peptides made by SWISS-MODEL. All peptides 3D-functional structures remain the same after the addition of His-tag.
3.2. Oligonucleotides and gene-specific primers for synthesis of the hLFcin, bLFcin, cLFcin, and ConLFcin genes
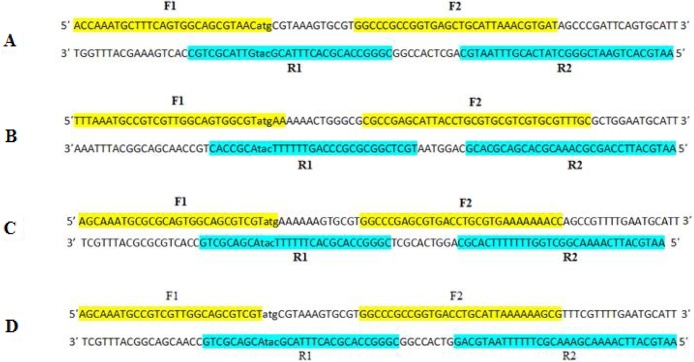
Overlapped oligonucleotides that span most of the sequence of both strands of the bLFcin, hLFcin, cLFcin, and ConLFcin genes were synthesized. The sequences were codon optimized for optimal expression in E. coli CFPS system by changing the original sequences to highly frequent codons used by E. coli translation machinery. Each gene was divided into four oligonucleotides ranging in length from 30 to 35 nucleotides. The overlap between these oligonucleotides was 5–20 bp. The relative positions and nucleotide sequences of the oligonucleotides are depicted in Fig. 3. The gene-specific forward PCR primers used in gene amplification and subsequent cloning in pET101/D-TOPO expression vector contains the sequence CACC at the 5′ end of the primers followed by start codon ATG.
Fig. 3.
Relative positions and nucleotide sequences of overlapped oligonucleotides used for the synthesis of; (A) hLFcin, (B) bLFcin, (C) cLFcin, and (D) ConLFcin genes by PCR assembling and amplification. Forward oligonucleotides are highlighted in yellow color while reverse oligonucleotides are highlighted in turquoise color. Gaps left between the designed oligonucleotides were not highlighted.
3.3. Construction of the synthetic genes encoding hLFcin, bLFcin, cLFcin, and ConLFcin peptides
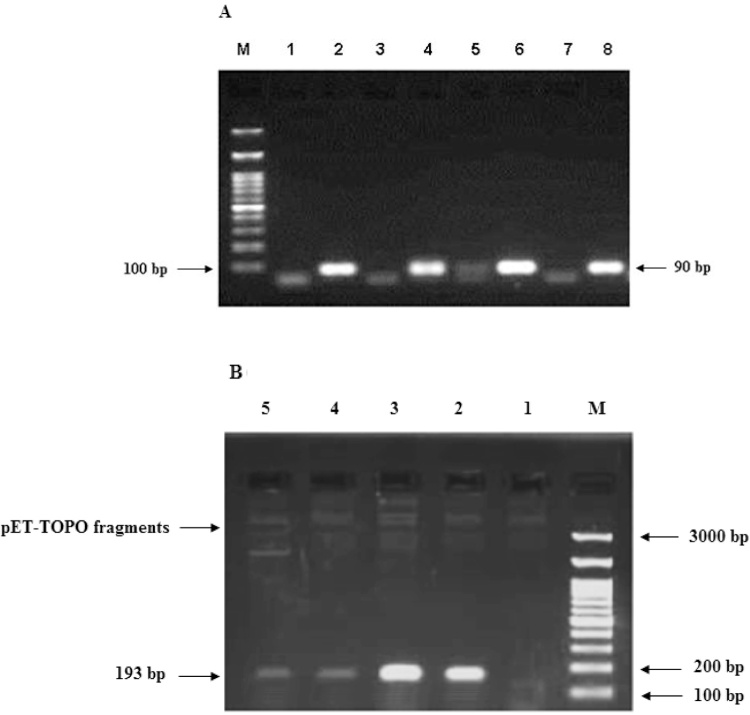
The PCR method was used to assemble and synthesize hLFcin, bLFcin, cLFcin, and ConLFcin genes of 90 bp using four overlapped oligonucleotides in two steps (Fig. 4A). All primers were mixed in equal molar concentrations in the assembling step to create short DNA duplex, thereby priming the elongation by DNA polymerase. These short duplexes serve as substrates for formation of longer duplexes, eventually resulting in the synthesis of the full-length genes, which were amplified using outermost primers of each strand. The Expand high fidelity PCR system successfully amplified the hLFcin, bLFcin, cLFcin, and ConLFcin genes in the correct size (90 bp) and sequences (Fig. 4A).
Fig. 4.
(A) Analysis of PCR products from the assembly and amplification of bLFcin, hLFcin, cLFcin, and ConLFcin genes on 2% agarose gel electrophoresis. Lane M points to GeneRuler 100 bp DNA marker, Lane 1 is assembly product of bLFcin, Lane 2 is amplified product of bLFcin (90 bp), Lane 3 is assembly product of hLFcin, Lane 4 is amplified product of hLFcin (90 bp), Lane 5 is assembly product of cLFcin, Lane 6 is amplified product of cLFcin (90 bp), Lane 7 is assembly product of ConLFcin, Lane 8 is amplified product of ConLFcin (90 bp). (B) Analysis of PCR products for positive transformants of pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin on 1.5 % agarose gel electrophoresis. Lane M points to GeneRuler 100 bp DNA marker, Lane 1 is negative transformant, Lane 2 is PCR product for positive transformant of pET-bLFcin, Lane 3 is PCR product for positive transformant of pET-hLFcin, Lane 4 is PCR product for positive transformant of pET-cLFcin, and Lane 5 is PCR product for positive transformant of pET-ConLFcin.
3.4. Construction of pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin Plasmids
The blunt-ended PCR products of the hLFcin, bLFcin, cLFcin, and ConLFcin genes were cloned into pET101/D-TOPO vector for high-level, T7-regulated expression in the cell-free system. Four bases; CACC were added to the forward PCR primer of each gene which complements the GTGG overhang in the pET101/D-TOPO vector. After transformation of the constructed pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin plasmids into E. coli XL2-Blue MRF’ strain, one transformant harboring each recombinant plasmid was subjected to PCR analysis and nucleotide sequencing. As indicated in Fig. 4B, the constructs have the correct sized hLFcin, bLFcin, cLFcin, and ConLFcin genes (193 bp, with 103 bp added to the actual size of the genes from the vector cloning site). The sequencing data strongly indicated that the transformants carried the correct constructs with no error-born mutation found (data not shown). The template plasmids were added to the in vitro protein synthesis reactions to be transcribed and translated simultaneously.
3.5. Expression of bLFcin, hLFcin, cLFcin, and ConLFcin peptides by cell-free protein synthesis system
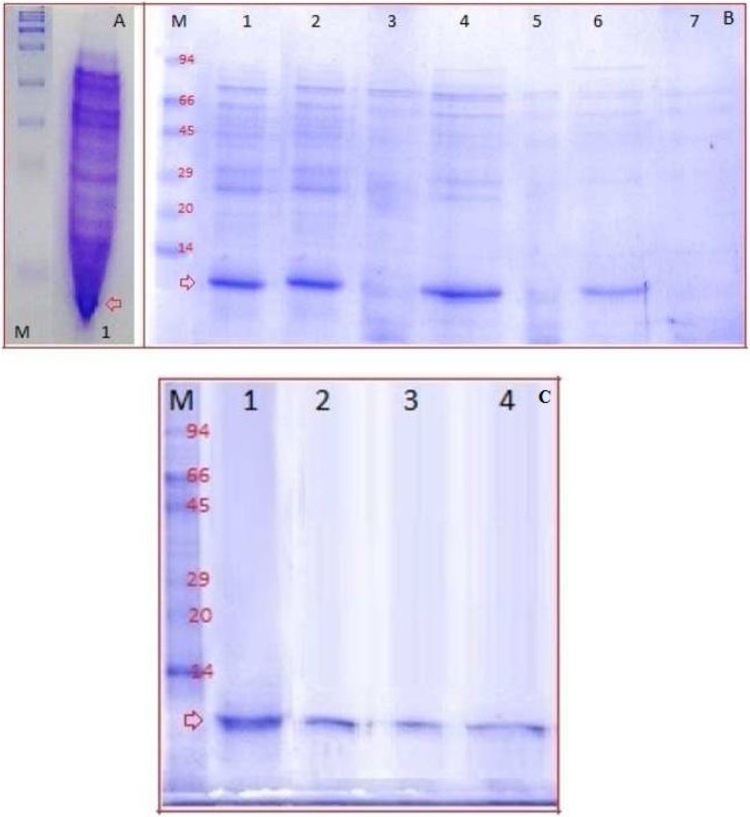
In order to ascertain that the translation products were definitely bLFcin, hLFcin, cLFcin, and ConLFcin peptides, samples obtained after 4 h incubation of in vitro reactions at 25 °C were analyzed on 18 % SDS-PAGE (Fig. 5A). From several separation trials, we obtained clear bands until the samples come to the position of these expressed highly positively charged hydrophobic LFcin peptides at which a sort of "squeezing in" of the gel lanes occurred (Fig. 5A). Thus, we separated the synthesized peptides on 18 % native gel electrophoresis as shown in Fig. 5B. Soluble proteins obtained from the cell-free synthesis reactions of bLFcin, hLFcin, cLFcin, and ConLFcin peptides contain discrete dense bands (overexpression) with molecular weight of about 6 kDa corresponding to the calculated molecular weight of bLFcin, hLFcin, cLFcin, and ConLFcin peptides plus added fusion tags (V5 epitope and polyhistidine tag) from the pET-TOPO vector (Fig. 5B). These bands were only present with the pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin templates added to in vitro protein synthesis reactions.
Fig. 5.
Separation of the LFcin peptides produced in the cell-free system on SDS-PAGE and native gel. (A) SDS-PAGE analysis of cell-free synthesized ConLFcin. Lane M: protein molecular weight marker. Lane 1: Soluble proteins obtained from the cell-free synthesis reaction of ConLFcin. The arrow points to "squeezing in" of the gel lane occurred at the peptide position. (B) Native gel analysis of cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin. Lane M: protein molecular weight marker. Lanes 1, 2, 4, and 6 are soluble proteins obtained from the cell-free synthesis reactions of bLFcin, hLFcin, cLFcin, and ConLFcin, respectively. Lanes 3, 5, and 7 are cell-free reactions run under the same conditions but without adding plasmid constructs as negative control. The arrow points to cell-free synthesized peptides. (C) Native gel electrophoresis analysis of purified cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin (at concentration of 15 μg/50 μL). Lane M: protein molecular weight marker. Lanes 1-4 are purified cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin, respectively.
Expression of bLFcin, hLFcin, cLFcin, and ConLFcin peptides was also verified by ELISA using rabbit polyclonal anti-lactoferrin antibody. The peptides activity of the reaction mixture of RTS100 with pET-bLFcin, pET-hLFcin, pET-cLFcin, and pET-ConLFcin as templates DNA for 4 h incubation was presented as the mean ± SD of three replicates. As it shown in Table 3, the mean absorbance values for cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides and human lactoferrin were respectively 0.375 ± 0.01, 0.519 ± 0.008, 0.351 ± 0.012, 0.284 ± 0.042, and 1.504 ± 0.16 compared with the value of reaction mixture of RTS100 as negative control of 0.042 ± 0.003. A statistical significant difference was found between human lactoferrin or cell-free synthesized peptides and the negative control in the mean absorbance value (p < 0.001). It was observed that there is a significant difference (p < 0.01) in ELISA activities between human lactoferrin and cell-free synthesized peptides; it is possibly due to that the epitopes density on human lactoferrin (full length) are greater than epitopes density on synthesized peptides.
Table 3.
Reactivity of rabbit polyclonal anti-lactoferrin antibody against cell-free synthesized peptides.
| Sample | OD at 405 nm (mean ± SD) |
|---|---|
| Human lactoferrin | 1.504 ± 0.16 |
| Negative control | 0.042 ± 0.003a |
| Cell-free synthesized bLFcin | 0.375 ± 0.01a |
| Cell-free synthesized hLFcin | 0.519 ± 0.008a |
| Cell-free synthesized cLFcin | 0.351 ± 0.012a |
| Cell-free synthesized ConLFcin | 0.284 ± 0.042a |
Polyclonal anti-lactoferrin antibody reacted significantly (p < 0.001) with cell-free synthesized peptides.
3.6. Purification of synthesized peptides
Synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides were purified by passage through Ni–NTA columns and the eluted peptides were analyzed by 18 % native gel electrophoresis (Fig. 5C). Single bands of about 6 kDa molecular weight were detected and approximately 400 μg of purified peptides were obtained from 1 mL of each cell-free peptide synthesis reaction solution as estimated by Lowry method.
3.7. Antibacterial activity of cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides
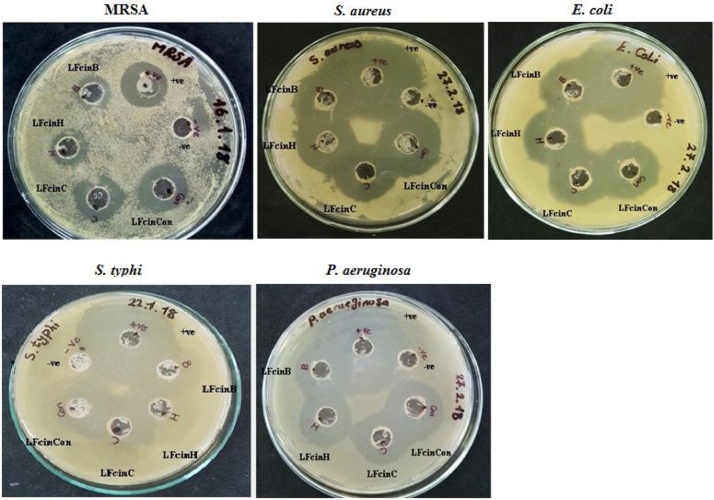
Cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides were proved to be bioactive by exhibiting antibacterial effects on tested Gram-positive bacteria (S. aureus, and MRSA) and Gram-negative bacteria (E. coli, S. typhi, and P. aeruginosa) (Fig. 6 and Table 4). At amount added of 40 μg, all peptides showed the lowest antibacterial activities against MRSA (the smallest inhibition zones in size) compared to their effects on S. aureus, E. coli, S. typhi, and P. aeruginosa (Fig. 6 and Table 4). In addition, at this concentration, synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides produced large overlapped inhibition zones against S. aureus, E. coli, S. typhi, and P. aeruginosa that cannot be used to compare their antimicrobial activity (Fig. 6 and Table 4). Results also showed inhibition zones around the wells containing cell-free synthesized peptides or vancomycin and chloramphenicol, and inhibition zones were not found when cell-free reaction run under the same conditions but without adding plasmid constructs was incubated into wells.
Fig. 6.
Assay of cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides antibacterial activity. +ve: represents positive control (chloramphenicol for S. aureus, E. coli, S. typhi, and P. aeruginosa and vancomycin for MRSA), -ve: represents negative control (cell-free reaction run under the same conditions but without adding plasmid constructs).
Table 4.
Antibacterial activity of cell-free synthesized peptides against tested Gram-positive and Gram-negative bacteria.
| Mean diameter of inhibition zonea (±1 mm) | ||||||
|---|---|---|---|---|---|---|
| Test strains | bLFcin (40 μg/100 μL) | hLFcin (40 μg/100 μL) | cLFcin (40 μg/100 μL) | ConLFcin (40 μg/100 μL) | C | VA |
| MRSA | 15 | 16 | 21 | 22 | R | 20 |
| S. aureus | 28 | 27 | 29 | 29 | 40 | NT |
| E. coli | 27 | 25 | 27 | 28 | 42 | NT |
| S. typhi | 25 | 25 | 27 | 27 | 45 | NT |
| P. aeruginosa | 26 | 25 | 28 | 28 | 45 | NT |
Mean of three assays; C- Chloramphenicol and VA – Vancomycin antibacterial standards at concentrations of 50 and 30 μg/mL, respectively; R - Resistant (no inhibition zone); NT – Not tested.
Furthermore, we found that cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin peptides inhibited MRSA, S. aureus, E. coli, S. typhi, and P. aeruginosa growth in a dose-dependent manner (produced concentration-dependent inhibition zones) at concentrations of 1.25−40 μg/mL (see Table 5). cLFcin and ConLFcin were able to inhibit the MRSA growth at concentrations ranging from 5 to 40 μg/mL. On the other hand, bLFcin and hLFcin inhibited the MRSA growth at concentrations ranging from 10 to 40 μg/mL (Table 5). These results suggested that cLFcin and ConLFcin demonstrated superior inhibitory activity (two times higher) against MRSA than bLFcin and hLFcin. In case of S. aureus and E. coli, cLFcin and ConLFcin could inhibit their growth at concentrations ranging from 1.25 to 40 μg/mL, while bLFcin and hLFcin at concentrations ranging from 2.5 to 40 μg/mL (Table 5). This confirmed superiority of cLFcin and ConLFcin antibacterial activity. cLFcin and ConLFcin could inhibit S. typhi growth at concentrations of 1.25−40 μg/mL, while hLFcin at concentrations of 5−40 μg/mL and bLFcin at concentrations of 2.5−40 μg/mL (Table 5). These observations suggested that cLFcin and ConLFcin demonstrated two times higher inhibitory activity against this pathogen than bLFcin and four times higher inhibitory activity than hLFcin. Additionally, cLFcin and ConLFcin inhibited P. aeruginosa growth at concentrations of 2.5−40 μg/mL, while bLFcin and hLFcin at concentrations of 5−40 μg/mL (Table 5), indicating superiority of antibacterial potency of cLFcin and ConLFcin (two times higher) against this pathogen than bLFcin and hLFcin.
Table 5.
Effect of different concentrations of cell-free synthesized peptides on tested Gram-positive and Gram-negative bacteria.
| Agent | Concentration (μg/mL) | Mean diameter of inhibition zonea (±1 mm) |
|||||
|---|---|---|---|---|---|---|---|
| MRSA | S. aureus | E. coli | S. typhi | P. aeruginosa | |||
| bLFcin | 40 | 16 | 28 | 27 | 25 | 26 | |
| 20 | 13 | 25 | 25 | 24 | 20 | ||
| 10 | 9 | 20 | 22 | 20 | 15 | ||
| 5 | R | 17 | 19 | 18 | 9 | ||
| 2.5 | R | 11 | 14 | 15 | R | ||
| 1.25 | R | R | R | R | R | ||
| hLFcin | 40 | 16 | 26 | 25 | 24 | 24 | |
| 20 | 13 | 24 | 21 | 21 | 20 | ||
| 10 | 8 | 20 | 18 | 16 | 16 | ||
| 5 | R | 15 | 14 | 11 | 8 | ||
| 2.5 | R | 9 | 10 | R | R | ||
| 1.25 | R | R | R | R | R | ||
| cLFcin | 40 | 20 | 28 | 26 | 27 | 28 | |
| 20 | 17 | 26 | 25 | 25 | 25 | ||
| 10 | 14 | 24 | 23 | 24 | 20 | ||
| 5 | 8 | 21 | 21 | 20 | 16 | ||
| 2.5 | R | 17 | 17 | 18 | 10 | ||
| 1.25 | R | 10 | 12 | 13 | R | ||
| ConLFcin | 40 | 21 | 29 | 28 | 28 | 29 | |
| 20 | 17 | 28 | 26 | 27 | 25 | ||
| 10 | 15 | 26 | 25 | 24 | 21 | ||
| 5 | 9 | 24 | 23 | 20 | 16 | ||
| 2.5 | R | 19 | 20 | 18 | 10 | ||
| 1.25 | R | 12 | 14 | 15 | R | ||
R - Resistant (no inhibition zone).
Mean of three assays.
3.8. The MIC values of the LFcin peptides
Both cLFcin and ConLFcin showed MIC values of 5, 1.25, 1.25, 1.25, and 2.5 μg/mL for MRSA, S. aureus, E. coli, S. typhi, and P. aeruginosa, respectively, while MIC values of bLFcin and hLFcin were 10, 2.5, 2.5, 2.5 and 5, and 5 μg/mL, respectively. This indicates superiority of antibacterial activities of cLFcin and ConLFcin over those of bLFcin and hLFcin against these pathogens (Table 6). cLFcin and ConLFcin achieved twice the inhibitory activity of bLFcin and hLFcin against test bacterial strains and even 4 times higher than that of hLFcin against S. typhi (Table 6).
Table 6.
MIC values of cell-free synthesized peptides against tested Gram-positive and Gram-negative bacteria.
| Test strains | MIC values (μg/mL) |
|||||
|---|---|---|---|---|---|---|
| BLFcin | hLFcin | cLFcin | ConLFcin | C | VA | |
| MRSA | 10 | 10 | 5 | 5 | R | 1.87 |
| S. aureus | 2.5 | 2.5 | 1.25 | 1.25 | 0.75 | NT |
| E. coli | 2.5 | 2.5 | 1.25 | 1.25 | 1.56 | NT |
| S. typhi | 2.5 | 5 | 1.25 | 1.25 | 1.56 | NT |
| P. aeruginosa | 5 | 5 | 2.5 | 2.5 | 1.56 | NT |
C – Chloramphenicol; VA – Vancomycin; R – Resistant (no inhibition zone); NT – Not tested.
3.9. Detection of cytotoxicity of cell-free synthesized peptides against normal human cells
The cytotoxic effect of purified cell-free synthesized bLFcin, hLFcin, cLFcin, and ConLFcin on human lymphocytes was evaluated after 24 h incubation by MTT assay. The LC50 values of the four recombinant peptides against normal human PBMCs presented in Table 7 were significantly (P < 0.01) higher than their MIC values against test bacterial strains indicating their toxicity to be selective toward bacterial cells but not toward the human cell line.
Table 7.
The LC50 values of cell-free synthesized peptides against PBMCs.
| Sample | LC50 (μg/mL) |
|---|---|
| bLFcin | 27.46 ± 0.53 |
| hLFcin | 25.49 ± 0.27 |
| cLFcin | 27.18 ± 0.22 |
| ConLFcin | 28.52 ± 0.38 |
All values were expressed as mean ± SD of three replicates.
4. Discussion
Research has shown that lactoferrin exhibits its antimicrobial activity via two mechanisms, iron sequestering and iron-independent pathways [24,48]. Iron-independent antimicrobial mechanisms involve LF direct interaction with different cell surfaces. The positively charged N-terminal of LF performs this direct interaction. Gastric pepsin digestion in the body releases a cationic peptide from LF N-terminal region that comprises the highly positively charged sequence of this region; called lactoferricin. LFcin was identified as the bactericidal domain of lactoferrin. LFcin has many biological functions of the lactoferrin complete protein and sometimes it can be even stronger than its parent protein [49]. It displays broad antibacterial spectrum against Gram-positive and Gram-negative bacteria, and has antiviral, antifungal, antiparasitic, and even antitumor activities [18,[49], [50], [51], [52], [53], [54], [55], [56]].
So far, lactoferricin peptides have been isolated and characterized from numerous animal species but most research studies focused on bLFcin [3,[19], [20], [21], [22], [23],50,[52], [53], [54], [55], [56]]. bLFcin comprises 25 amino acids; amino acids 17–41 from the N-terminal of bovine lactoferrin and has been shown to be more potent than its parent protein in terms of biological activity. In contrast, only a few studies about the production and in vitro antimicrobial activity of hLFcin and cLFcin were published. hLFcin was derived from N-terminus of human lactoferrin and was found to be highly effective against infections with antibiotic-resistant bacteria [24]. The first study aimed at the identification and production of cLFcin was published in 2016, where Chahardooli et al. [25] for the first time characterized and expressed recombinant cLFcin in Pichia pastoris and investigated its antimicrobial activity. They confirmed that it had suitable antimicrobial activity and its production by Pichia pastoris was successful. Tanhaeian et al. [57] have fused a codon-optimized partial cLFcin and camel lactoferrampin DNA sequences to construct a fused peptide. They used human embryonic kidney 293 (HEK-293) cells for synthesizing this recombinant peptide. This recombinant chimera inhibited the growth of test Gram-negative and Gram-positive bacterial plant pathogens at MIC values between 0.39 and 25.07 μg/mL for different bacterial isolates [57].
Production of lactoferricin peptides from different organisms by either pepsin digestion of lactoferrin parent protein or artificial synthesis is laborious and costly. Since the cost and yield of antimicrobial peptides production are vital for their use in pharmaceutical applications, recombinant production presents a favorable alternative method in which peptides can be produced quickly, easily, and on a large scale for research and further applications. Nevertheless, recombinant expression of LFcin peptides in E. coli and Saccharomyces cerevisiae is limited by toxicity of these recombinant cationic antimicrobial peptides to host expression cells besides the sensitivity to degradation of these short amino acid sequences by host proteases during in vivo expression. Mostly, recombinant LFcin was expressed linked with a fusion protein to overcome these obstacles [26,27].
This peptide has not been produced by a cell-free protein production method so far, and this is the first study to report in vitro expression of three natural recombinant lactoferricin peptides; bLFcin, hLFcin, and cLFcin. Moreover, we designed a novel synthetic consensus peptide (ConLFcin) which might prove more effective than its natural counterparts. In addition, this is the first report to compare antibacterial potency of natural bLFcin, hLFcin, and cLFcin peptides as well as including a designed synthetic lactoferricin peptide. The genes of bLFcin, hLFcin, cLFcin, and ConLFcin were cloned into pET101/D-TOPO expression vector then peptides were synthesized in vitro by prokaryotic extract-based CFPS system; prepared from E. coli cells lysate. After 4 h of incubation at 25 °C, bLFcin, hLFcin, cLFcin, and ConLFcin peptides were expressed in the cell-free reactions with a yield of about 20 μg/50 μL in a soluble active form. Expressed peptides were separated by 18 % native gel electrophoresis, strong bands of the expected size (about 6 kDa) were detected after 4 h of the reactions. Polyclonal anti-human lactoferrin antibody reacted significantly (p < 0.001) with cell-free synthesized peptides, showing strong signals in ELISA. pET101 vector carries six histidine residues at its C-terminus; this polyhistidine has high affinity for Ni–NTA resin permitting single-step purification of cell-free synthesized peptides. The peptides were detected as single bands in 18 % native gel electrophoresis indicating the efficiency of purification. The four peptides showed significant antibacterial potency against E. coli, S. typhi, P. aeruginosa, S. aureus, and MRSA at MIC values between 1.25 and 10 μg/mL.
In this work, a new chimeric artificial peptide was engineered; ConLFcin that is anticipated to display stronger bioactivity than natural peptides as concluded from results of ProtParam tool – ExPASy and Helical Wheel tool analysis which revealed that the designed ConLFcin has the highest cationicity followed by cLFcin then bLFcin while hLFcin showed the lowest cationicity. This high cationic characteristic of ConLFcin and cLFcin is expected to allow these peptides to interact more readily and vigorously than other peptides with anionic bacterial membranes LPS and peptidoglycans leading to enhanced disruption of the bacterial cell membrane integrity and improved antibacterial activities [58]. ConLFcin and cLFcin achieved twice the inhibitory activity of bLFcin and hLFcin against test bacterial strains and even 4 times higher than that of hLFcin against S. typhi. Meanwhile, the high cationic characteristic of ConLFcin and cLFcin has significantly minor effect on normal mammalian PBMCs compared to its effect on bacterial cells (LC50 values of these peptides against PBMCs presented in Table 7 were significantly (P < 0.01) higher than their MIC values against test bacterial strains presented in Table 6). This makes ConLFcin a suitable candidate for further development into therapeutic use.
The use of cell-free protein expression as an alternative to cell-based protein production offers unique advantages and applications. CFPS systems have shown their utility for protein production at high titers, time saving, producing proteins that require posttranslational modifications, expressing proteins which are sensitive to proteolytic degradation or cytotoxic, establishing genetic regulatory element libraries (e.g., promoters, ribosome binding sites) in nonmodel organisms, sensing biomarkers for diagnostic applications, and biosynthetic pathways optimization before cells implementation [[28], [29], [30], [31], [32], [33], [34], [35], [36]]. Cell-free (in vitro) protein synthesis format avoids transformation, clone selection, expression induction, and cell lysis steps typically essential for cell-based (in vivo) protein synthesis. Cell lysates used for cell-free protein expression continue to be optimized, starting with the traditional E. coli, wheat germ, and rabbit reticulocyte lysates (batch, semi-batch, and continuous exchange formats) and extended with insect and human cell-free systems [30,31]. Recently, novel systems have been developed based on reconstituted highly purified components named PURE cell-free expression systems, which have considerably extended the range of applications of cell-free systems [32]. However, several problems have limited the use of cell-free systems as a protein production technology including expensive reagent costs, small reaction scales, lower recombinant protein yields, deleterious changes in the chemical environment associated with supplying needed energy and substrates for protein synthesis, and limited abilities of some source organisms from which the cell-free lysates are prepared to support posttranslational modifications. Nevertheless, new technical advances have dealt with these problems to increase the potential of cell-free systems to meet the increasing demands for recombinant protein synthesis at industrial scale [35].
Unfortunately, most previous efforts have focused on cell-free protein production rather than cell-free peptide production. Therapeutic proteins such as interferons, peptide hormones, vaccines, and diagnostic enzymes were produced using cell-free systems [[28], [29], [30],33,35]. In contrast, cell-free peptide production is certainly not widespread published method. Only two recent studies have described the production of short antimicrobial peptides using CFPS [37,38]. Using lyophilized E. coli CFPS system, ten different antimicrobial peptides have been synthesized successfully, three of which (BP100, Cecropin B, and Cecropin P1) were proved to be functional by E. coli inhibition assay [37]. Des Soye et al. [38] established Vibrio natriegens CFPS system and demonstrated its ability to be lyophilized and produce antimicrobial peptides. However, contrary to the former study, the bioactivity of the produced peptides was not demonstrated.
After our previous achievement of cell-free expression of consensus interferon-alpha [33], we conducted the first cell-free synthesis of lactoferricin and produced four recombinant peptides; three natural lactoferricin peptides and synthetic one. Cell-free peptide synthesis of ConLFcin enables us to avoid any problematic host-peptide interaction as well as laborious and lengthy in vivo expression of this peptide and subsequent purification for rapid analysis of its biological activity and comparing its antibacterial efficiency with that of its natural counterparts. We also could rapidly confirm that cLFcin is the most active among natural lactoferricin peptides comparable to its parent lactoferrin protein we proved previously [[8], [9], [10]]. The addition of histidine to the cell-free synthesized LFcin peptides was to facilitate their purification and does not affect the peptides 3D-functional structures, their net charge values calculated by Helical Wheel tool and ProtParam tool – ExPASy, and most importantly their antibacterial activity as documented previously [59,60]. Remarkably, the cell-free system was an ideal system for rapid expression and activity analysis of recombinant short (about 6 kDa) bioactive peptides as can be seen from the antibacterial activity assay.
5. Conclusion
Every day the pressure of antibiotic resistance increases. New and effective antimicrobial agents are important and urgent target. The study successfully produced four functional lactoferricin peptides in vitro for first time. Although, the synthesized bLFcin, hLFcin, cLFcin, and ConLFcin revealed a comparable antimicrobial activity, but cLFcin and ConLFcin antimicrobial activities were better. We suggest applicability of cell-free protein synthesis to provide quick access to the target peptides by rapid expression of the bioactive valuable peptides and their function analysis.
Authors' contributions
NAE-B, MAE, ESA and EMR conceived and designed research. HRK, NAE-B, and MMS conducted experiments. EMR contributed new reagents or analytical tools. HRK, NAE-B, EMR, MAE, MMS, and ESA analyzed data. HRK, and NAE-B wrote the manuscript and EMR revives the manuscript. All authors read and approved the manuscript.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding information
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We gratefully acknowledge the support from colleagues in Genetic Engineering and Biotechnology Research Institute, City of Scientific Research and Technological Applications during conducting this research.
References
- 1.Hancock R.E., Diamond G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Tomita M., Bellamy W., Takase M., Yamauchi K., Wakabayashi H., Kawase K. Potent antibacterial peptides generated by pepsin digestion of bovine lactoferrin. J. Dairy Sci. 1991;74:4137–4142. doi: 10.3168/jds.S0022-0302(91)78608-6. [DOI] [PubMed] [Google Scholar]
- 4.Harmsen M.C., Swart P.J., de Bethune M.P., Pauwels R., De Clercq E., The T.H., Meijer D.K. Antiviral effects of plasma and milk proteins: lactoferrin shows potent activity against both human immunodeficiency virus and human cytomegalovirus replication in vitro. J. Infect. Dis. 1995;172:380–388. doi: 10.1093/infdis/172.2.380. [DOI] [PubMed] [Google Scholar]
- 5.Valenti P., Antonini G. Lactoferrin: an important host defense against microbial and viral attack. Cell. Mol. Life Sci. 2005;62:2576–2587. doi: 10.1007/s00018-005-5372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redwan E.M., Tabll A. Camel lactoferrin markedly inhibits hepatitis C virus genotype 4 infection of human peripheral blood leukocytes. J. Immunoassay Immunochem. 2007;28:267–277. doi: 10.1080/15321810701454839. [DOI] [PubMed] [Google Scholar]
- 7.Conesa C., Sanchez L., Rota C., Perez M.D., Calvo M., Farnaud, Evans R.W. Isolation of lactoferrin from milk of different species: calorimetric and antimicrobial studies. Comp. Biochem. Physiol. - Part B: Biochem. Mol. Biol. 2008;150:131–139. doi: 10.1016/j.cbpb.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Redwan E.M., El-Baky N.A., Al-Hejin A.M., Baeshen M.N., Almehdar H.A., Elsaway A., Gomaa A.B., Al-Masaudi S.B., Al-Fassi F.A., AbuZeid I.E., Uversky V.N. Significant antibacterial activity and synergistic effects of camel lactoferrin with antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) Res. Microbiol. 2016;167:480–491. doi: 10.1016/j.resmic.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Almehdar H.A., El-Baky N.A., Alhaider A.A., Almuhaideb S.A., Alhaider A.A., Albiheyri R.S., Uversky V.N., Redwan E.M. Synergistic killing of pathogenic Escherichia coli using camel lactoferrin from different Saudi camel clans and various antibiotics. Protein J. 2019;38:479–496. doi: 10.1007/s10930-019-09828-5. [DOI] [PubMed] [Google Scholar]
- 10.Almehdar H.A., El-Baky N.A., Alhaider A.A., Almuhaideb S.A., Alhaider A.A., Albiheyri R.S., Uversky V.N., Redwan E.M. Bacteriostatic and bactericidal activities of camel lactoferrins against Salmonella enterica serovar typhi. Probiotics Antimicrob. Proteins. 2020;12:18–31. doi: 10.1007/s12602-019-9520-5. [DOI] [PubMed] [Google Scholar]
- 11.Liao Y., El-Fakharany E., Lonnerdal B., Redwan E.M. Inhibitory effects of native and recombinant full-length camel lactoferrin and its N and C lobes on hepatitis C virus infection of Huh7.5 cells. J. Med. Microbiol. 2012;61:375–383. doi: 10.1099/jmm.0.033894-0. [DOI] [PubMed] [Google Scholar]
- 12.El-Fakharany E.M., Sanchez L., Al-Mehdar H.A., Redwan E.M. Effectiveness of human, camel, bovine and sheep lactoferrin on the hepatitis C virus cellular infectivity: comparison study. Virol. J. 2013;10:199. doi: 10.1186/1743-422X-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redwan E.M., Uversky V.N., El-Fakharany E.M., Al-Mehdar H. Potential lactoferrin activity against pathogenic viruses. C. R. Biol. 2014;337:581–595. doi: 10.1016/j.crvi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Redwan E.M., El-Fakharany E.M., Uversky V.N., Linjawi M.H. Screening the anti-infectivity potentials of native N- and C-lobes derived from the camel lactoferrin against hepatitis C virus. BMC Complement. Altern. Med. 2014;14:219. doi: 10.1186/1472-6882-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Albar A.H., Almehdar H.A., Uversky V.N., Redwan E.M. Structural heterogeneity and multifunctionality of lactoferrin. Curr. Protein Pept. Sci. 2014;15:778–797. doi: 10.2174/1389203715666140919124530. [DOI] [PubMed] [Google Scholar]
- 16.Moore S.A., Anderson B.F., Groom C.R., Haridas M., Baker E.N. Three-dimensional structure of diferric bovine lactoferrin at 2.8 Å resolution. J. Mol. Biol. 1997;274:222–236. doi: 10.1006/jmbi.1997.1386. [DOI] [PubMed] [Google Scholar]
- 17.Khan J.A., Kumar P., Paramasivam M., Yadav R.S., Sahani M.S., Sharma S., Srinivasan A., Singh T.P., Lactoferrin Camel, Transferrin-cum-Lactoferrin a. Crystal structure of camel apolactoferrin at 2.6 Å resolution and structural basis of its dual role. J. Mol. Biol. 2001;309:751–761. doi: 10.1006/jmbi.2001.4692. [DOI] [PubMed] [Google Scholar]
- 18.Bellamy W., Takase M., Yamauchi K., Wakabayashi H., Kawase K., Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim. Biophys. Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 19.Bellamy W., Takase M., Wakabayashi H., Kawase K., Tomita M. Antibacterial spectrum of lactoferricin B, a potent bactericidal peptide derived from the N-terminal region of bovine lactoferrin. J. Appl. Bacteriol. 1992;73:472–479. doi: 10.1111/j.1365-2672.1992.tb05007.x. [DOI] [PubMed] [Google Scholar]
- 20.Mader J.S., Salsman J., Conrad D.M., Hoskin D.W. Bovine lactoferricin selectively induces apoptosis in human leukemia and carcinoma cell lines. Mol. Cancer Ther. 2005;4:612–624. doi: 10.1158/1535-7163.MCT-04-0077. [DOI] [PubMed] [Google Scholar]
- 21.Eliassen L.T., Berge G., Leknessund A., Wikman M., Lindin I., Lokke C., Ponthan F., Johnsen J.I., Sveinbjornsson B., Kogner P., Flaegstad T., Rekdal O. The antimicrobial peptide, lactoferricin B, is cytotoxic to neuroblastoma cells in vitro and inhibits xenograft growth in vivo. Int. J. Cancer. 2006;119:493–500. doi: 10.1002/ijc.21886. [DOI] [PubMed] [Google Scholar]
- 22.Andersen J.H., Jenssen H., Gutteberg T.J. Lactoferrin and lactoferricin inhibit Herpes simplex 1 and 2 infection and exhibit synergy when combined with acyclovir. Antiviral Res. 2003;58:209–215. doi: 10.1016/s0166-3542(02)00214-0. [DOI] [PubMed] [Google Scholar]
- 23.Jenssen H. Anti-herpes simplex virus activity of lactoferrin/lactoferricin-an example of antiviral activity of antimicrobial protein/peptide. Cell. Mol. Life Sci. CMLS. 2005;62:3002–3013. doi: 10.1007/s00018-005-5228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nibbering P.H., Ravensbergen E., Welling M.M., van Berkel L.A., van Berkel P.H., Pauwels E.K., Nuijens J.H. Human lactoferrin and peptides derived from its N terminus are highly effective against infections with antibiotic-resistant bacteria. Infect. Immun. 2001;69:1469–1476. doi: 10.1128/IAI.69.3.1469-1476.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chahardooli M., Niazi A., Aram F., Sohrabi S.M. Expression of recombinant Arabian camel lactoferricin-related peptide in Pichia pastoris and its antimicrobial identification. J. Sci. Food Agric. 2016;96:569–575. doi: 10.1002/jsfa.7125. [DOI] [PubMed] [Google Scholar]
- 26.Urtasun N., Baieli M.F., Romasanta P.N., Fernández M.M., Malchiodi E.L., Cascone O., Wolman F.J., Miranda M.V. Triazinic dye ligand selection by surface plasmon resonance for recombinant lactoferricin purification. Process. Biochem. 2013;48:1972–1979. [Google Scholar]
- 27.Feng X.J., Wang J.H., Shan A.S., Teng D., Yang Y.L., Yao Y., Yang G.P., Shao Y.C., Liu S., Zhang F. Fusion expression of bovine lactoferricin in Escherichia coli. Protein Expr. Purif. 2006;47:110–117. doi: 10.1016/j.pep.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 28.Jackson A.M., Boutell J., Cooley N., He M. Cell-free protein synthesis for proteomics. Brief. Funct. Genomic. Proteomic. 2004;2:308–319. doi: 10.1093/bfgp/2.4.308. [DOI] [PubMed] [Google Scholar]
- 29.Spirin A.S. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004;22:538–545. doi: 10.1016/j.tibtech.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Mikami S., Kobayashi T., Masutani M., Yokoyama S., Imataka H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr. Purif. 2008;62:190–198. doi: 10.1016/j.pep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Hino M., Kataoka M., Kajimoto K., Yamamoto T., Kido J., Shinohara Y., Baba Y. Efficiency of cell-free protein synthesis based on a crude cell extract from Escherichia coli, wheat germ, and rabbit reticulocytes. J. Biotechnol. 2008;133:183–189. doi: 10.1016/j.jbiotec.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu Y., Ueda T. PURE technology. Methods Mol. Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- 33.El-Baky N.A., Omar S.H., Redwan E.M. The anti-cancer activity of human consensus interferon-alpha synthesized in cell-free system. Protein Expr. Purif. 2011;80:61–67. doi: 10.1016/j.pep.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Orth J.H., Schorch B., Boundy S., Ffrench-Constant R., Kubick S., Aktories K. Cell-free synthesis and characterization of a novel cytotoxic pierisin-like protein from the cabbage butterfly Pieris rapae. Toxicon. 2011;57:199–207. doi: 10.1016/j.toxicon.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Zawada J.F., Yin G., Steiner A.R., Yang J., Naresh A., Roy S.M., Gold D.S., Heinsohn H.G., Murray C.J. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bechlars S., Wustenhagen D.A., Dragert K., Dieckmann R., Strauch E., Kubick S. Cell-free synthesis of functional thermostable direct hemolysins of Vibrio parahaemolyticus. Toxicon. 2013;76:132–142. doi: 10.1016/j.toxicon.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D. Portable, on-demand biomolecular manufacturing. Cell. 2016;167:248–259. doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 2018;7:2245–2255. doi: 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- 39.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwede T., Kopp J., Guex N., Peitsch M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.Mohammed Y., El-Baky N.A., Redwan E.M. Expression, purification, and characterization of recombinant human consensus interferon-alpha in Escherichia coli under λPL promoter. Prep. Biochem. Biotechnol. 2012;42:426–447. doi: 10.1080/10826068.2011.637600. [DOI] [PubMed] [Google Scholar]
- 43.Murray P.R., Zeitinger J.R. Evaluation of Muller-Hinton agar for disk diffusion susceptibility tests. J. Clin. Microbiol. 1983;18:1269–1271. doi: 10.1128/jcm.18.5.1269-1271.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 45.Andrews J.M. Determination of minimum inhibitory concentrations, Journal of Antimicrobial Chemotherapy. 48 (2001) 5-16. Erratum J. Antimicrobial Chemotherapy. 2002;49:1049. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 46.Redwan E.M. Simple, sensitive and quick protocol to detect less than 1 ng of bacterial lipopolysaccharide. Prep. Biochem. Biotechnol. 2012;42:171–182. doi: 10.1080/10826068.2011.586081. [DOI] [PubMed] [Google Scholar]
- 47.Lohr H.F., Goergen B., Meyer zum Buschenfelde K.H., Gerken G. HCV replication in mononuclear cells stimulates anti-HCV-secreting B cells and reflects nonresponsiveness to interferon-alpha. J. Med. Virol. 1995;46:314–320. doi: 10.1002/jmv.1890460405. [DOI] [PubMed] [Google Scholar]
- 48.Arnold R.R., Cole M.F., McGhee J.R. A bactericidal effect for human lactoferrin. Science (New York, N.Y.) 1977;197:263–265. doi: 10.1126/science.327545. [DOI] [PubMed] [Google Scholar]
- 49.Yamauchi K., Tomita M., Giehl T.J., Ellison R.T., 3rd Antibacterial activity of lactoferrin and a pepsin-derived lactoferrin peptide fragment. Infect. Immun. 1993;61:719–728. doi: 10.1128/iai.61.2.719-728.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellamy W., Yamauchi K., Wakabayashi H., Takase M., Takakura N., Shimamura S., Tomita M. Antifungal properties of lactoferricin B, a peptide derived from the N‐terminal region of bovine lactoferrin. Lett. Appl. Microbiol. 1994;18:230–233. [Google Scholar]
- 51.Turchany J.M., Aley S.B., Gillin F.D. Giardicidal activity of lactoferrin and N-terminal peptides. Infect. Immun. 1995;63:4550–4552. doi: 10.1128/iai.63.11.4550-4552.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Omata Y., Satake M., Maeda R., Saito A., Shimazaki K., Yamauchi K., Uzuka Y., Tanabe S., Sarashina T., Mikami T. Reduction of the infectivity of Toxoplasma gondii and Eimeria stiedai sporozoites by treatment with bovine lactoferricin. J. Vet. Med. Sci. 2001;63:187–190. doi: 10.1292/jvms.63.187. [DOI] [PubMed] [Google Scholar]
- 53.Andersen J.H., Osbakk S.A., Vorland L.H., Traavik T., Gutteberg T.J. Lactoferrin and cyclic lactoferricin inhibit the entry of human cytomegalovirus into human fibroblasts. Antiviral Res. 2001;51:141–149. doi: 10.1016/s0166-3542(01)00146-2. [DOI] [PubMed] [Google Scholar]
- 54.McCann K.B., Lee A., Wan J., Roginski H., Coventry M.J. The effect of bovine lactoferrin and lactoferricin B on the ability of feline calicivirus (a norovirus surrogate) and poliovirus to infect cell cultures. J. Appl. Microbiol. 2003;95:1026–1033. doi: 10.1046/j.1365-2672.2003.02071.x. [DOI] [PubMed] [Google Scholar]
- 55.Yoo Y.C., Watanabe S., Watanabe R., Hata K., Shimazaki K., Azuma I. Bovine lactoferrin and lactoferricin, a peptide derived from bovine lactoferrin, inhibit tumor metastasis in mice. Jpn. J. Cancer Res. 1997;88:184–190. doi: 10.1111/j.1349-7006.1997.tb00364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eliassen L.T., Berge G., Sveinbjornsson B., Svendsen J.S., Vorland L.H., Rekdal O. Evidence for a direct antitumor mechanism of action of bovine lactoferricin. Anticancer Res. 2002;22:2703–2710. [PubMed] [Google Scholar]
- 57.Tanhaeian A., Ahmadi F.S., Sekhavati M.H., Mamarabadi M. Expression and purification of the main component contained in camel milk and its antimicrobial activities against bacterial plant pathogens. Probiotics Antimicrob. Proteins. 2018;10:787–793. doi: 10.1007/s12602-018-9416-9. [DOI] [PubMed] [Google Scholar]
- 58.El-Fakharany E.M., Redwan E.M. Protein-lipid complexes: molecular structure, current scenarios and mechanisms of cytotoxicity. RSC Adv. 2019;9:36890–36906. doi: 10.1039/c9ra07127j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen G.H., Yin L.J., Chiang I.H., Jiang S.T. Expression and purification of goat lactoferrin from Pichia pastoris expression system. J. Food Sci. 2007;72:67–71. doi: 10.1111/j.1750-3841.2007.00281.x. [DOI] [PubMed] [Google Scholar]
- 60.Chen G.H., Chen W.M., Huang G.T., Chen Y.W., Jiang S.T. Expression of recombinant antibacterial lactoferricin-related peptides from Pichia pastoris expression system. J. Agric. Food Chem. 2009;57:9509–9515. doi: 10.1021/jf902611h. [DOI] [PubMed] [Google Scholar]