Abstract
Enterocytozoon bieneusi, a fungus-like protist parasite, causes symptomatic and asymptomatic intestinal infections in terrestrial animals and is also abundant in the environment. This parasite has been isolated from a variety of host types including humans, livestock, companion animals, birds, and wildlife, as well as the natural and urban environments including drinking source water, coastal water, recreational water, wastewater, vegetables in retail markets, and raw milk on farms. E. bieneusi exhibits high genetic diversity among host species and environmental sources and at least 500 genotypes have been identified thus far. Since its discovery in AIDS patients in 1985, scientists across the world have worked to demonstrate the natural history and public health potential of this pathogen. Here we review molecular typing studies on E. bieneusi and summarize relevant data to identify the potential sources of human and nonhuman infections and environmental contamination. This review also discusses the possible transmission routes of E. bieneusi and the associated risk factors, and advocates the importance of the One Health approach to tackle E. bieneusi infections.
Keywords: Enterocytozoon bieneusi, Ecology, Public health, Host, Environment, Transmission
1. Introduction
In the broadest sense, the science of ecology can be defined as the study of the interactions between organisms and their environment. Microbial ecology examines the interactions between microorganisms, their hosts, and the environment. Publications on the abundance, distribution, and genetic variation of Enterocytozoon bieneusi among hosts in the context of host specificity and zoonotic potential have been reviewed [[1], [2], [3], [4], [5], [6]], whereas there has been no review interpreting the ecological aspects relating to the environmental transport of this pathogen in nature and the associated public health implications.
Microsporidia comprises a highly diverse group of obligate intracellular parasites colonizing an extremely wide range of eukaryotes [[7], [8], [9]]. Over 200 microsporidian genera and nearly 1500 species have now been described [3], of which the most frequently reported in human microsporidiosis is E. bieneusi [10]. The life cycle of E. bieneusi involves two major stages, an infective or environmental spore stage and a disease-causing vegetative stage. Spores are ingested by the susceptible hosts, invade the epithelial cells lining the gut lumen, and replicate intracellularly. Fresh spores are released into the environment via feces [3]. The rigid three-layered spore wall may play a vital role in the survival of the parasite in hostile environments [11,12]. In line with the importance of environmental contamination in its transmission, E. bieneusi has caused food- and waterborne and hospital-related outbreaks [[13], [14], [15], [16]]. Most cases of microsporidian infections in HIV-infected persons are attributable to E. bieneusi and the infections manifest as varying clinical symptoms, typically chronic diarrhea and wasting [7]. While oral fumagillin has been considered as a possible method for eradicating this opportunistic pathogen from the intestinal tract of AIDS patients, it is highly toxic and not commercially available in most countries [17,18]. As a consequence, a promising therapy for E. bieneusi-associated microsporidiosis remains to be explored. In addition, it is very difficult to expand anti-E. bieneusi treatment options due to the limitation of in vitro propagation of the parasite [19]. As there are repeated reports of the parasite in asymptomatic immunocompetent individuals, notably children and the elderly, it may be argued that the association between E. bieneusi infection and diarrhea is not as tight as it was initially believed to be [10].
Specific diagnosis of E. bieneusi by conventional staining methods is difficult to achieve due to the extremely small size of the spore (around 1 μm) and the interference from food particles, bacteria, fungi, and other microsporidian species like Encephalitozoon spp.; transmission electron microscopy is a powerful means for diagnosis, but is not available in most settings [7,12,20]. Immunodetection with monoclonal antibodies is readily applicable to large-scale epidemiological surveys, but no genotyping information can be provided [[21], [22], [23]]. With the advent of molecular techniques, such as PCR and DNA sequencing of the ribosomal internal transcribed spacer (ITS), E. bieneusi has been characterized worldwide from humans, a great variety of domestic and wild animals (pigs, monkeys, cattle, deer, sheep, goats, dogs, cats, horses, carnivores, rodents, birds, etc.), and water environment [3,4], arousing our curiosity about the sources of infection and modes of transmission. Current literature lacks consensus on the definition of the natural reservoir hosts of E. bieneusi since the parasite can infect multiple animal species, rather than specific target populations. Asymptomatic E. bieneusi infections are quite common in animals [4]. There is seemingly no consistent correction between E. bieneusi occurrence and animal diarrhea [[24], [25], [26]]. Human E. bieneusi infections may be acquired by contact-associated zoonotic transmission or by ingestion of spores from environmental sources [10,27]. Many cases are likely resulted from human to human transmission [28,29].
Water contamination with pathogenic microorganisms is one of the serious threats to human and animal health, and is also an important issue of environmental and ecological concerns all over the world. Major sources of microbial contamination in water include feces from humans, livestock, poultry, and wild animals through surface water runoff, wastewater discharge, and agricultural effluents [30]. Intestinal carriage of E. bieneusi is ubiquitous in mammalian and avian hosts [3], and via fecal matter from those hosts, environments (mainly water) can readily become contaminated [[31], [32], [33], [34], [35]]. ITS genotyping potentially allows the discrimination of environmental E. bieneusi isolates, helping to identify the host source of environmental contamination as well as improving our knowledge of the routes of cross-species or zoonotic E. bieneusi transmission [[36], [37], [38]]. In addition, concerns have been raised regarding the importance of fresh produce contaminated with E. bieneusi spores in foodborne microsporidiosis [39].
Despite the abundance, richness, diversity, and zoonotic potential of E. bieneusi, the ecological aspects of this parasite remain largely unknown. This creates serious difficulties in elucidating the natural history and mode of transmission of E. bieneusi and catching the inherent link between the environment and the health of humans and animals. All these strongly urge on us the need for applying a One Health approach to tackle E. bieneusi infections.
2. Molecular typing and phylogenetic groups of E. bieneusi
Polymorphism analysis of the ITS region of the rRNA gene and phylogenetic assessment allowed the identification of at least 500 E. bieneusi genotypes distributed in 11 phylogenetic groups [2,3]. Three sets of nested PCR primers that cover the entire ITS region have been broadly applied in genotyping studies [2,[40], [41], [42], [43]]. E. bieneusi genotypes in different groups display varying degree of host specificity and zoonotic potential [3]. Group 1 is the largest group that comprises over 300 genotypes isolated from almost all current known host types, some (e.g., D, EbpC, and Type IV) of which exhibit remarkable adaption to life within a diverse array of host and natural environments [2,3]. By contrast, some Group 1 genotypes show strong host specificity, for instance, genotypes B and C are thus far confined to human infections in multiple geographic areas, and EbpB to pig infections likewise [2,3]. Only minor ITS sequence divergence (about 20 single nucleotide substitutions) is present between E. bieneusi genotypes from Group 1 and Group 2. Group 2 is the second largest cluster previously considered adapted to ruminants; most of its members are the most frequent contributors to E. bieneusi infections in cattle, deer, sheep, and goats, including BEB4, BEB6, I, and J [3,5]. These four successful genotypes probably have experienced host range expansion, leading to their human infectivity [3]. In recent years, there has been a rapid increase in the number of Group 1 and Group 2 genotypes and the host range of some of them has been extended [3]. E. bieneusi genotypes in Groups 3 to 11 are genetically divergent from those in Groups 1 and 2; the former mostly have a restricted host range and thus represent a minor or unknown public health threat [3]. Currently, there are many reports of genetic variants of the existing E. bieneusi genotypes as new ones; some of these could be due to sequencing errors, lack of proofreading of nucleotide differences in the ITS region of the PCR products, and improper consideration of polymorphisms in the flanking regions (small and large subunits) of the ITS [3,4,6,44]. In addition, caution should be taken when discussing cross-species transmission potential of E. bieneusi genotypes among humans, animals, and birds, as clinical, genetic, epidemiological, or ecological factors could also affect the outcome. After all, genetic identity or relatedness in the ~243-bp ITS region might not reflect the overall genetic characteristics of the E. bieneusi genome (~6 megabases) [45,46].
Multilocus sequence typing (MLST) has been shown to be more discriminatory since it considers genetic polymorphisms of four mini- and microsatellites in addition to the ITS [47]. MLST-based population genetic analysis by evaluating the intragenic and intergenic linkage disequilibrium (LD), standardized index of association, neutrality, and recombination events has revealed the widespread occurrence of clonality among E. bieneusi populations from various host species and geographical regions [[48], [49], [50], [51], [52]]. Population subdivision was determined by both multilocus phylogenetic analysis and substructural analysis; the estimation of Wright's fixation index and gene flow was performed to assess the level of genetic differentiation between subpopulations [[48], [49], [50], [51], [52]]. Further genetic analysis and host range analysis have indicated that E. bieneusi subpopulations might have different genetic structures (clonal/epidemic), levels of host specificity (low/strong), and transmission pathways (interspecies/intraspecies) [1,2]. Genetic network analysis has demonstrated that E. bieneusi isolates more often evolve from low to strong host specificity; the LD decay and relatively high genetic diversity in the subpopulations with a clonal structure might be consequences of broad host range and adaptation to new host environments [48]. Such findings have implications for the application of the current MLST scheme as a One Health typing tool for ecological surveillance of E. bieneusi. One limitation of the current MLST tool is its poor amplification efficiency in the analysis of the isolates from Groups 2 to 11 [2]. Based on the existing MLST evidence, whilst limited, it can be speculated that there might be other species in the genus Enterocytozoon [1]. Additional genetic markers are desirable for delimiting Enterocytozoon species [6].
3. Reservoir hosts of human-pathogenic E. bieneusi
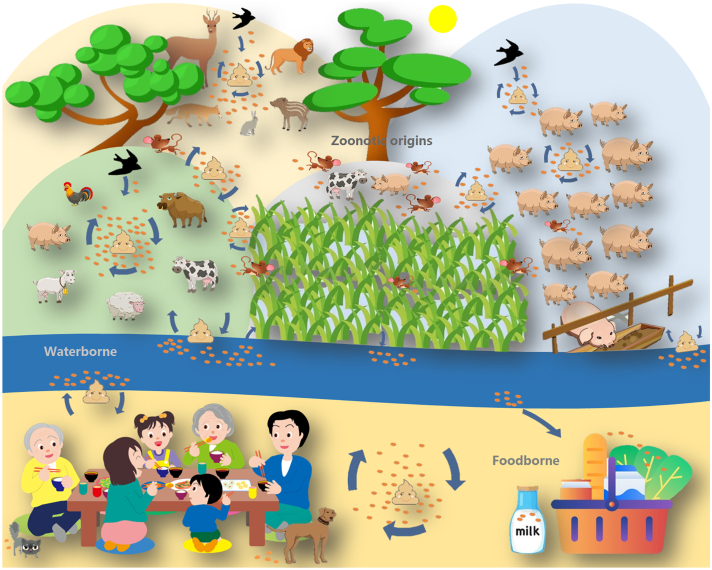
The usual vehicles of E. bieneusi dispersal include both domestic and wild animals (Fig. 1), and among host species the pathogen displays genetic and phenotypic diversity [4,5]. Humans are infected predominately by Group 1 genotypes (e.g., D, EbpC, Type IV, A, and Peru8), and very occasionally by the genotypes from other genetic groups (e.g., Group 2 genotypes I, J, BEB4, and BEB6, Group 5 genotype KIN-3, and Group 6 genotypes MAY1, Nig3, and Nig4) [3]. There seems no geographic segregation among human E. bieneusi isolates as inferred from either ITS or MLST phylogeny [50,53]. Nonhuman primates (NHPs) are well-known carriers of human-pathogenic E. bieneusi genotypes, such as D, Type IV, EbpC, EbpA, Peru8, and Peru11, and they can also harbor some potentially NHP-specific genotypes like KB-5, KB-6, and Gorilla 3 [3]. There appears to be a high possibility of cross-species transmission of E. bieneusi between humans and laboratory NHPs since the latter can be the host of some common human-pathogenic genotypes such as D, Type IV, Peru8, and Peru11 [[54], [55], [56]]. There is seemingly no barrier to E. bieneusi transmission among free-ranging park/zoo NHPs and other animals nearby, because they are often in close contact with each other and the NHPs can spread generalist genotypes D, EbpC, and Type IV [[57], [58], [59], [60], [61], [62]]. Wild NHPs that often carry genotype D are also of environmental and public health interest, since in many cases they serve as habitual frequenters of rural or suburban areas and interact directly with villagers and grazing domestic animals, potentially initiating cross-species transmission of E. bieneusi [[63], [64], [65], [66], [67]].
Fig. 1.
Schematic diagram showing the ecological and public health significance of Enterocytozoon bieneusi and the major routes of transmission.
Pigs. Pigs have a high carriage rate of E. bieneusi and the great majority of genotypes identified belong to Group 1, with zoonotic genotypes EbpC, EbpA, O, H, and D being the most frequent and abundant. Therefore, pigs are considered major reservoir hosts of human-pathogenic E. bieneusi [3]. The zoonotic nature of some genotype D/EbpC isolates was already supported by MLST analysis [1]. The first documented case of pig E. bieneusi infection occurred on a farm in 1996 in Switzerland and the disease persisted for months [68]. Experimental infection of gnotobiotic piglets with E. bieneusi spores derived from humans, macaques, and infected pigs provided convincing evidence for cross-species transmission potential [42,69]. Substantial molecular epidemiological studies on a global scale highlighted the role of farmed pigs in the dispersal of E. bieneusi genotypes of public health importance, notably EbpC [24,26,42,[70], [71], [72], [73], [74], [75]], probably through animal waste slurry from intensive piggeries, which facilitates contamination of surface water and vegetables, thereby promoting waterborne and foodborne transmission of the pathogen (Fig. 1). Indeed, it has been suggested that persons who come into contact with farmed and household-raised pigs may face high risk of E. bieneusi infection [[75], [76], [77]]. Free-roaming feral pigs may also play a role in E. bieneusi dispersal since they usually shuttle between human and animal hosts and between home and natural environments (Fig. 1). Wild boars and farmed pigs have similar distribution pattern of E. bieneusi genotypes [78,79]. It has become evident that genotypes EbpA and PigSpEb1 might be transmitted between sympatric extensively farmed pigs and free-ranging wild boars [80].
Ruminants. Cattle, sheep, goats, and deer are common reservoir hosts of E. bieneusi [3]. Phenotypic and genotypic profiles of E. bieneusi isolates from ruminants show marked heterogeneity comparable with those from pigs; ruminants are major hosts of Group 2 genotypes, such as BEB4, BEB6, I, and J, and they can also carry some Group 1 genotypes such as EbpC, D, Type IV, and Peru6, and several divergent genotypes (CM4 and S7) in other genetic groups [3]. Frequent interactions among farmed ruminants (cattle, sheep, and goats) may facilitate the transmission of some E. bieneusi genotypes; animal wastes from feedlots can conceivably spread some genotypes (e.g., BEB4, BEB6, I, J, EbpC, D, and Type IV) with broad host ranges to the residents and uninfected animals, and contaminate vegetable field and water supplies [[81], [82], [83], [84], [85], [86], [87], [88]]. Milk contaminated with genotypes D, Type IV, I, and J can also act as an infection source of human microsporidiosis [89]. Due caution should be exercised to the potential cross-species transmission of generalist genotypes EbpC, D, Type IV, BEB4, BEB6, I, and J between free-ranging domestic ruminants and other domestic and wild animals that share the same outdoor or grazing areas (Fig. 1). In addition, we should not discount domestic and wild deer as potential reservoirs for some human-infective genotypes such as D, EbpA, EbpC, Type IV, Peru6, BEB6, I, and J [[90], [91], [92], [93]], as well as farmed takins and zoo-bred alpacas for genotypes D, BEB6, I, and J [61,83,94].
Companion animals. Cats and dogs living as human companions and free-roaming animals can potentiate zoonotic or cross-species transmission of E. bieneusi. Cats are the common carriers of the most common human-pathogenic genotypes D and Type IV. In contrast, dogs around the world mostly harbor a host-adapted genotype PtEb IX (a Group 11 genotype), although they can also host some human-pathogenic genotypes such as D, EbpC, O, and A [[95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105]]. There appears to be no significant differences in genotype distribution between household and stray cats/dogs [3]. Companion animals might be the most direct source of E. bieneusi contamination in the home environment, and in turn can acquire infections through contacts with fecal matters of humans and other household animals (Fig. 1). Stray cats and dogs are at increased risk of exposure to E. bieneusi infections from various sources including spore-containing human and animal wastes, food, and water, thus facilitating the dispersal of E. bieneusi in the environment.
Birds. Close contacts with birds can also transmit E. bieneusi infections [3]. Similar to livestock, fecal materials from farmed chickens, geese, ducks, pigeons, falcons, and ostriches and the organic wastes from these farms can also be important contributors to E. bieneusi in the environment (Fig. 1), due to the common occurrence of zoonotic genotypes D, Type IV, BEB6, J, Peru6, Peru11, and Henan-IV in those birds [72,[106], [107], [108], [109], [110], [111]]. Humans and household animals in theory can acquire E. bieneusi infections by genotypes D, EbpC, A, EbpA, and Peru6 from exotic birds, although direct evidence for this type of cross-species transmission is lacking [83,[112], [113], [114], [115]]. Urban pigeons and rooks and wild migratory cranes can act as reservoirs of genotypes D, J, EbpA, and Peru6 [109,112,114,[116], [117], [118], [119], [120], [121]]; through fecal droppings of those free-flying birds, human settlement environments, livestock farms and grazing areas, wildlife habitats, livestock/wildlife interface areas, farmlands, and surface water may become contaminated with E. bieneusi spores (Fig. 1).
Other domestic, captive/exotic, and wild animals. Those animals such as equines, carnivores, rodents, and lagomorphs are also commonly infected with E. bieneusi, thus have the potential for disseminating E. bieneusi spores. The role of such reservoir hosts, particularly free-roaming wild rodents, in the environmental transport and ecology of human-pathogenic E. bieneusi, cannot be ignored because of their carriages of genotypes D, EbpC, Type IV, Peru6, I, and J [3]. A number of epidemiologic investigations have assessed the public health potential of E. bieneusi infections from farmed/grazing horses and donkeys, farmed/household-raised rabbits, and farmed/zoo-bred foxes, raccoon dogs, raccoons, lions, tigers, bears, pandas, meerkats, and kangaroos [61,72,83,[122], [123], [124], [125], [126], [127], [128], [129]]. A previous report showed the possible transmission of a unique genotype Peru16 between children and guinea pigs that live in the same household [27]. As shown in Fig. 1, E. bieneusi may be maintained in a natural transmission cycle involving wild free-living mammals, such as foxes, raccoons, bears, otters, beavers, squirrels, chipmunks, woodchucks, muskrats, voles, mice, and cottontails, since they can carry zoonotic genotypes D, EbpC, Peru11, or Type IV [37,43,72,130,131]. E. bieneusi spores in feces of wild animal reservoirs can contaminate source water via storm runoff [37,43]. The free-ranging wildlife, notably small rodents, may maintain the cycling of E. bieneusi among humans, livestock, wildlife, and the environment (Fig. 1) [130,131].
In spite of widespread presence of E. bieneusi in terrestrial hosts and the important ecological roles of those hosts in dispersing spores, the parasite was also identified in some aquatic mollusks like mussels [132,133] and oysters [134], which have been used as biomonitors for ecological risk assessment; however, the potential role of mollusks as vectors in transmission of E. bieneusi remains elusive due to the lack of molecular typing data. While E. bieneusi has been identified in bottlenose dolphins [135], further detailed evidence is needed to determine their infectivity to other hosts.
4. E. bieneusi in the environment
E. bieneusi spores can be transported into water bodies and become waterborne pathogens (Fig. 1). The discovery of E. bieneusi in surface water can be traced back to the late 1990s [31,136]. A waterborne outbreak of microsporidiosis by E. bieneusi was documented in 1999 [14]. Humans and domestic animals are possible sources of E. bieneusi contamination in drinking source water, as judged by identification of both host-specific genotypes C (human), EbpB (pig), and PtEb IX (dog) and zoonotic genotypes EbpC, D, EbpA, Peru8, and Peru11 in the rivers impacted by human and livestock activities [36,137]. The negative impacts of E. bieneusi-infected wild mammals may extend beyond natural ecosystems to human and livestock communities during storm events; genotypes D and Type IV are commonly found in wildlife within drinking source watershed [37]. Urban storm overflow that contains human-origin genotypes such as D, Type IV, Peru8, and Peru11 can potentially act as a contamination source for drinking source water and recreational water [138]. Beaches, lakes, and rivers used for bathing or yachting can harbor viable E. bieneusi spores; bathing in those public waters or direct contact with contaminated water constitutes a potential infection risk [32,[139], [140], [141]]. The contamination of E. bieneusi spores in lakes, rivers, and coastal areas may be attributed to urban sewage discharge or agricultural runoff with human and animal wastes [133]. Potentially host-specific (e.g., C and PtEb IX) and zoonotic (e.g., D, EbpC, Type IV, and BEB6) E. bieneusi genotypes have been frequently detected in untreated sewages or raw wastewater, which might come from humans and domestic and wild animals; despite sewage treatment, E. bieneusi spores persist in the wastewater treatment effluents, thereby posing a threat for the contamination of surface water used for recreation and drinking water abstraction [33,34,38,137,138,[142], [143], [144], [145], [146], [147], [148]].
The contamination of retail fresh food produce such as berries, sprouts, and green-leafed vegetables with potentially viable E. bieneusi spores could result from the application of dirty water in the production process, untreated sewage or animal feces to fertilize ready-to-eat crops, and runoff-influenced surface water for crop irrigation (Fig. 1) [39,149,150]. In reality, E. bieneusi has caused an outbreak of gastrointestinal illness in a group of apparently immunocompetent persons who visited a hotel in Sweden and consumed cheese sandwiches and salad. Cucumber slices were identified as the most probable vehicle of transmission. The identification of the human-adapted genotype C implicated the potential contamination source to human waste [13].
5. One Health perspectives
Evidence accumulated thus far has shown a common occurrence of E. bieneusi in the ecosystem and high genetic diversity of the pathogen among isolates from different hosts and the environment. This has allowed the evolution of diverse genotypes with different host ranges and adaptation to various ecological niches, presenting substantial challenges in the assessment of the sources and public health importance of the small-sized pathogens found in the ecosystem. Molecular ecological studies of E. bieneusi have been greatly hampered due to multiple factors, including 1) low discriminatory power or effectiveness in the currently used typing loci, 2) poor understanding of the relationship between genetic variation and phenotypic traits, 3) absence of standardized methods used for differentiating E. bieneusi isolates and tracing the routes of infection in epidemiological investigations, and 4) lack of genotype data from humans in many geographical areas of the world. A novel MLST strategy with significantly expanded typing range is urgently needed for E. bieneusi to open up new opportunities for solving a central question regarding how genetic variations drive phenotypic traits and their effects on the host and natural environment. In addition, there is a critical need for molecular epidemiological studies conducted on diverse reservoir hosts including humans in the same areas to fully elucidate the cross-species or zoonotic transmissibility of E. bieneusi. Some recent studies concerning several other important protozoan parasites such as Cryptosporidium parvum, Giardia duodenalis, Toxoplasma gondii, and Leishmania spp. have already highlighted the value of a One Health approach in helping control the diseases they cause [[151], [152], [153], [154], [155]].
The reservoir hosts of E. bieneusi are comprised of a number of epidemiologically connected populations of the same or different species, potentially including vectors. E. bieneusi can be maintained by those hosts and transmitted to each other. Domestic and wild animals both represent the well-documented reservoir hosts of E. bieneusi and many animal species are infected with both potentially zoonotic and host-specific E. bieneusi genotypes. In this context, effort should be directed to search on genetic predictors of E. bieneusi adaptation to specific hosts or host switching. E. bieneusi infections are frequently asymptomatic in the host species investigated, although clinical disease characterized by diarrhea does occur. These subclinical infections in reservoir hosts may be beneficial to the spread of E. bieneusi to other reservoir or susceptible hosts, thus could be a long-term strategy in the coevolution of host-parasite interactions. In light of the absence of safe and effective therapeutic agents and vaccines for E. bieneusi, One Health preventive measures should be taken to reduce the close contacts between infected and susceptible hosts and contamination of food and water with E. bieneusi spores commonly found in animal feces and human sewage.
Although there is increasing recognition of the major threat of E. bieneusi to public health, the ecological complexity of its transmission continues to pose challenges in combating zoonotic infections. Animals are believed to be the major sources of environmental contamination with E. bieneusi; spores released from animal stools can be dispersed into water sources, especially via wastewater discharges and farm runoff. Surface water for irrigation contaminated with animal fecal matters represents a latent source for foodborne E. bieneusi infection. It may be worthwhile to investigate how surface water and ground water contamination is facilitated by the small size of E. bieneusi spores, as there have been no studies on the transport of E. bieneusi spore in the environment. The cognition of infection sources and transmission routes, particularly the importance of waterborne or foodborne transmission, could be helpful in the formulation of preventive measures to limit the dissemination of E. bieneusi to urban environment. All these call for adopting a One Health approach to the control and prevention of microsporidiosis by E. bieneusi, via collaborative and interdisciplinary efforts that span the medical, veterinary, and ecological realms.
Acknowledgments
Acknowledgements
This work is funded by the Natural Science Fund of Heilongjiang Province for Excellent Young Scholars (YQ2020C010) and National Natural Science Foundation of China (32030109).
Declaration of Competing Interest
The authors Wei Li and Lihua Xiao declare that no competing interests exist.
Contributor Information
Wei Li, Email: neaulw@gmail.com.
Lihua Xiao, Email: lxiao@scau.edu.cn.
References
- 1.Li W., Xiao L. Multilocus sequence typing and population genetic analysis of Enterocytozoon bieneusi: host specificity and its impacts on public health. Front. Genet. 2019;10:307. doi: 10.3389/fgene.2019.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Feng Y., Zhang L., Xiao L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.104033. [DOI] [PubMed] [Google Scholar]
- 3.Li W., Feng Y., Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019;35(6):436–451. doi: 10.1016/j.pt.2019.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Santin M., Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011;90(3):363–371. doi: 10.1016/j.rvsc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 5.Thellier M., Breton J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite. 2008;15(3):349–358. doi: 10.1051/parasite/2008153349. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Feng Y., Xiao L. Diagnosis and molecular typing of Enterocytozoon bieneusi: the significant role of domestic animals in transmission of human microsporidiosis. Res. Vet. Sci. 2020;133:251–261. doi: 10.1016/j.rvsc.2020.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Weber R., Bryan R.T., Schwartz D.A., Owen R.L. Human microsporidial infections. Clin. Microbiol. Rev. 1994;7(4):426–461. doi: 10.1128/cmr.7.4.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mathis A., Weber R., Deplazes P. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 2005;18(3):423–445. doi: 10.1128/CMR.18.3.423-445.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vavra J., Lukes J. Microsporidia and ‘the art of living together’. Adv. Parasitol. 2013;82:253–319. doi: 10.1016/B978-0-12-407706-5.00004-6. [DOI] [PubMed] [Google Scholar]
- 10.Matos O., Lobo M.L., Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012;2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigliardi E., Sacchi L. Cell biology and invasion of the microsporidia. Microbes Infect. 2001;3(5):373–379. doi: 10.1016/s1286-4579(01)01393-4. [DOI] [PubMed] [Google Scholar]
- 12.Wasson K., Peper R.L. Mammalian microsporidiosis. Vet. Pathol. 2000;37(2):113–128. doi: 10.1354/vp.37-2-113. [DOI] [PubMed] [Google Scholar]
- 13.Decraene V., Lebbad M., Botero-Kleiven S., Gustavsson A.M., Lofdahl M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 2012;140(3):519–527. doi: 10.1017/S095026881100077X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cotte L., Rabodonirina M., Chapuis F., Bailly F., Bissuel F., Raynal C. Waterborne outbreak of intestinal microsporidiosis in persons with and without human immunodeficiency virus infection. J. Infect. Dis. 1999;180(6):2003–2008. doi: 10.1086/315112. [DOI] [PubMed] [Google Scholar]
- 15.Desoubeaux G., Nourrisson C., Moniot M., De Kyvon M.A., Bonnin V., De La Bretonniere M.E. Genotyping approach for potential common source of Enterocytozoon bieneusi infection in hematology unit. Emerg. Infect. Dis. 2019;25(9):1625–1631. doi: 10.3201/eid2509.190311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L., Xiao L., Duan L., Ye J., Guo Y., Guo M. Concurrent infections of Giardia duodenalis, Enterocytozoon bieneusi, and Clostridium difficile in children during a cryptosporidiosis outbreak in a pediatric hospital in China. PLoS Negl. Trop. Dis. 2013;7(9) doi: 10.1371/journal.pntd.0002437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Didier E.S. Effects of albendazole, fumagillin, and TNP-470 on microsporidial replication in vitro. Antimicrob. Agents Chemother. 1997;41(7):1541–1546. doi: 10.1128/aac.41.7.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molina J.M., Goguel J., Sarfati C., Chastang C., Desportes-Livage I., Michiels J.F. Potential efficacy of fumagillin in intestinal microsporidiosis due to Enterocytozoon bieneusi in patients with HIV infection: results of a drug screening study. AIDS. 1997;11(13):1603–1610. doi: 10.1097/00002030-199713000-00009. [DOI] [PubMed] [Google Scholar]
- 19.Visvesvara G.S. In vitro cultivation of microsporidia of clinical importance. Clin. Microbiol. Rev. 2002;15(3):401–413. doi: 10.1128/CMR.15.3.401-413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia L.S. Laboratory identification of the microsporidia. J. Clin. Microbiol. 2002;40(6):1892–1901. doi: 10.1128/JCM.40.6.1892-1901.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Accoceberry I., Thellier M., Desportes-Livage I., Achbarou A., Biligui S., Danis M. Production of monoclonal antibodies directed against the microsporidium Enterocytozoon bieneusi. J. Clin. Microbiol. 1999;37(12):4107–4112. doi: 10.1128/jcm.37.12.4107-4112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q., Singh I., Sheoran A., Feng X., Nunnari J., Carville A. Production and characterization of monoclonal antibodies against Enterocytozoon bieneusi purified from rhesus macaques. Infect. Immun. 2005;73(8):5166–5172. doi: 10.1128/IAI.73.8.5166-5172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheoran A.S., Feng X., Singh I., Chapman-Bonofiglio S., Kitaka S., Hanawalt J. Monoclonal antibodies against Enterocytozoon bieneusi of human origin. Clin. Diagn. Lab. Immunol. 2005;12(9):1109–1113. doi: 10.1128/CDLI.12.9.1109-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan Q., Lin Y., Mao Y., Yang Y., Li Q., Zhang S. High Prevalence and Widespread Distribution of zoonotic Enterocytozoon bieneusi genotypes in swine in Northeast China: implications for public health. J. Eukaryot. Microbiol. 2016;63(2):162–170. doi: 10.1111/jeu.12264. [DOI] [PubMed] [Google Scholar]
- 25.da Silva Fiuza V.R., Lopes C.W., de Oliveira F.C., Fayer R., Santin M. New findings of Enterocytozoon bieneusi in beef and dairy cattle in Brazil. Vet. Parasitol. 2016;216:46–51. doi: 10.1016/j.vetpar.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Jeong D.K., Won G.Y., Park B.K., Hur J., You J.Y., Kang S.J. Occurrence and genotypic characteristics of Enterocytozoon bieneusi in pigs with diarrhea. Parasitol. Res. 2007;102(1):123–128. doi: 10.1007/s00436-007-0740-3. [DOI] [PubMed] [Google Scholar]
- 27.Cama V.A., Pearson J., Cabrera L., Pacheco L., Gilman R., Meyer S. Transmission of Enterocytozoon bieneusi between a child and Guinea pigs. J. Clin. Microbiol. 2007;45(8):2708–2710. doi: 10.1128/JCM.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leelayoova S., Subrungruang I., Rangsin R., Chavalitshewinkoon-Petmitr P., Worapong J., Naaglor T. Transmission of Enterocytozoon bieneusi genotype a in a Thai orphanage. Am J. Trop. Med. Hygiene. 2005;73(1):104–107. [PubMed] [Google Scholar]
- 29.Pagornrat W., Leelayoova S., Rangsin R., Tan-Ariya P., Naaglor T., Mungthin M. Carriage rate of Enterocytozoon bieneusi in an orphanage in Bangkok, Thailand. J. Clin. Microbiol. 2009;47(11):3739–3741. doi: 10.1128/JCM.01606-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harwood V.J., Staley C., Badgley B.D., Borges K., Korajkic A. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol. Rev. 2014;38(1):1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 31.Dowd S.E., Gerba C.P., Pepper I.L. Confirmation of the human-pathogenic microsporidia Enterocytozoon bieneusi, Encephalitozoon intestinalis, and Vittaforma corneae in water. Appl. Environ. Microbiol. 1998;64(9):3332–3335. doi: 10.1128/aem.64.9.3332-3335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coupe S., Delabre K., Pouillot R., Houdart S., Santillana-Hayat M., Derouin F. Detection of Cryptosporidium, Giardia and Enterocytozoon bieneusi in surface water, including recreational areas: a one-year prospective study. FEMS Immunol. Med. Microbiol. 2006;47(3):351–359. doi: 10.1111/j.1574-695X.2006.00098.x. [DOI] [PubMed] [Google Scholar]
- 33.Graczyk T.K., Lucy F.E., Mashinsky Y., Andrew Thompson R.C., Koru O., Dasilva A.J. Human zoonotic enteropathogens in a constructed free-surface flow wetland. Parasitol. Res. 2009;105(2):423–428. doi: 10.1007/s00436-009-1400-6. [DOI] [PubMed] [Google Scholar]
- 34.Cheng H.W., Lucy F.E., Graczyk T.K., Broaders M.A., Mastitsky S.E. Municipal wastewater treatment plants as removal systems and environmental sources of human-virulent microsporidian spores. Parasitol. Res. 2011;109(3):595–603. doi: 10.1007/s00436-011-2291-x. [DOI] [PubMed] [Google Scholar]
- 35.Izquierdo F., Castro Hermida J.A., Fenoy S., Mezo M., Gonzalez-Warleta M., del Aguila C. Detection of microsporidia in drinking water, wastewater and recreational rivers. Water Res. 2011;45(16):4837–4843. doi: 10.1016/j.watres.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 36.Hu Y., Feng Y., Huang C., Xiao L. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ. Sci. Technol. 2014;48(24):14219–14227. doi: 10.1021/es504464t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y., Alderisio K.A., Yang W., Cama V., Feng Y., Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl. Environ. Microbiol. 2014;80(1):218–225. doi: 10.1128/AEM.02997-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li N., Xiao L., Wang L., Zhao S., Zhao X., Duan L. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl. Trop. Dis. 2012;6(9) doi: 10.1371/journal.pntd.0001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jedrzejewski S., Graczyk T.K., Slodkowicz-Kowalska A., Tamang L., Majewska A.C. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl. Environ. Microbiol. 2007;73(12):4071–4073. doi: 10.1128/AEM.00477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katzwinkel-Wladarsch S., Lieb M., Helse W., Loscher T., Rinder H. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop. Med. Int. Health. 1996;1(3):373–378. doi: 10.1046/j.1365-3156.1996.d01-51.x. [DOI] [PubMed] [Google Scholar]
- 41.Rinder H., Katzwinkel-Wladarsch S., Loscher T. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol. Res. 1997;83(7):670–672. doi: 10.1007/s004360050317. [DOI] [PubMed] [Google Scholar]
- 42.Buckholt M.A., Lee J.H., Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002;68(5):2595–2599. doi: 10.1128/AEM.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulaiman I.M., Fayer R., Lal A.A., Trout J.M., Schaefer F.W., 3rd, Xiao L. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003;69(8):4495–4501. doi: 10.1128/AEM.69.8.4495-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santin M., Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J. Eukaryot. Microbiol. 2009;56(1):34–38. doi: 10.1111/j.1550-7408.2008.00380.x. [DOI] [PubMed] [Google Scholar]
- 45.Akiyoshi D.E., Morrison H.G., Lei S., Feng X., Zhang Q., Corradi N. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 2009;5(1) doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keeling P.J., Corradi N., Morrison H.G., Haag K.L., Ebert D., Weiss L.M. The reduced genome of the parasitic microsporidian Enterocytozoon bieneusi lacks genes for core carbon metabolism. Genome Biol. Evol. 2010;2:304–309. doi: 10.1093/gbe/evq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng Y., Li N., Dearen T., Lobo M.L., Matos O., Cama V. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2011;77(14):4822–4828. doi: 10.1128/AEM.02803-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wan Q., Xiao L., Zhang X., Li Y., Lu Y., Song M. Clonal evolution of Enterocytozoon bieneusi populations in swine and genetic differentiation in subpopulations between isolates from swine and humans. PLoS Negl. Trop. Dis. 2016;10(8) doi: 10.1371/journal.pntd.0004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W., Wan Q., Yu Q., Yang Y., Tao W., Jiang Y. Genetic variation of mini- and microsatellites and a clonal structure in Enterocytozoon bieneusi population in foxes and raccoon dogs and population differentiation of the parasite between fur animals and humans. Parasitol. Res. 2016;115(7):2899–2904. doi: 10.1007/s00436-016-5069-3. [DOI] [PubMed] [Google Scholar]
- 50.Li W., Cama V., Akinbo F.O., Ganguly S., Kiulia N.M., Zhang X. Multilocus sequence typing of Enterocytozoon bieneusi: lack of geographic segregation and existence of genetically isolated sub-populations. Infect. Genet. Evol. 2013;14:111–119. doi: 10.1016/j.meegid.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 51.Li W., Cama V., Feng Y., Gilman R.H., Bern C., Zhang X. Population genetic analysis of Enterocytozoon bieneusi in humans. Int. J. Parasitol. 2012;42(3):287–293. doi: 10.1016/j.ijpara.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Karim M.R., Wang R., He X., Zhang L., Li J., Rume F.I. Multilocus sequence typing of Enterocytozoon bieneusi in nonhuman primates in China. Vet. Parasitol. 2014;200(1–2):13–23. doi: 10.1016/j.vetpar.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Widmer G., Akiyoshi D.E. Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi. Infect. Genet. Evol. 2010;10(1):122–128. doi: 10.1016/j.meegid.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drosten C., Laabs J., Kuhn E.M., Schottelius J. Interspecies transmission of Enterozytozoon bieneusi supported by observations in laboratory animals and phylogeny. Med. Microbiol. Immunol. 2005;194(4):207–209. doi: 10.1007/s00430-005-0240-y. [DOI] [PubMed] [Google Scholar]
- 55.Ye J., Xiao L., Li J., Huang W., Amer S.E., Guo Y. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol. Int. 2014;63(1):132–137. doi: 10.1016/j.parint.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Yang H., Lin Y., Li Y., Song M., Lu Y., Li W. Molecular characterization of Enterocytozoon bieneusi isolates in laboratory macaques in North China: zoonotic concerns. Parasitol. Res. 2017;116(10):2877–2882. doi: 10.1007/s00436-017-5603-y. [DOI] [PubMed] [Google Scholar]
- 57.Ye J., Xiao L., Ma J., Guo M., Liu L., Feng Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg. Infect. Dis. 2012;18(10):1640–1643. doi: 10.3201/eid1810.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karim M.R., Wang R., Dong H., Zhang L., Li J., Zhang S. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl. Environ. Microbiol. 2014;80(6):1893–1898. doi: 10.1128/AEM.03845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sak B., Kvac M., Petrzelkova K., Kvetonova D., Pomajbikova K., Mulama M. Diversity of microsporidia (fungi: microsporidia) among captive great apes in European zoos and African sanctuaries: evidence for zoonotic transmission? Folia Parasitol. 2011;58(2):81–86. doi: 10.14411/fp.2011.008. [DOI] [PubMed] [Google Scholar]
- 60.Karim M.R., Dong H., Li T., Yu F., Li D., Zhang L. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W., Deng L., Yu X., Zhong Z., Wang Q., Liu X. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasit. Vectors. 2016;9(1):395. doi: 10.1186/s13071-016-1668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu F., Wu Y., Li T., Cao J., Wang J., Hu S. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet. Res. 2017;13(1):158. doi: 10.1186/s12917-017-1084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W., Kiulia N.M., Mwenda J.M., Nyachieo A., Taylor M.B., Zhang X. Cyclospora papionis, Cryptosporidium hominis, and human-pathogenic Enterocytozoon bieneusi in captive baboons in Kenya. J. Clin. Microbiol. 2011;49(12):4326–4329. doi: 10.1128/JCM.05051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du S.Z., Zhao G.H., Shao J.F., Fang Y.Q., Tian G.R., Zhang L.X. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling mountains. Korean J. Parasitol. 2015;53(4):395–402. doi: 10.3347/kjp.2015.53.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mynarova A., Foitova I., Kvac M., Kvetonova D., Rost M., Morrogh-Bernard H. Prevalence of Cryptosporidium spp., Enterocytozoon bieneusi, Encephalitozoon spp. and Giardia intestinalis in wild, semi-wild and captive orangutans (Pongo abelii and Pongo pygmaeus) on Sumatra and Borneo, Indonesia. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0152771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sak B., Petrzelkova K.J., Kvetonova D., Mynarova A., Shutt K.A., Pomajbikova K. Long-term monitoring of microsporidia, Cryptosporidium and Giardia infections in western Lowland Gorillas (Gorilla gorilla gorilla) at different stages of habituation in Dzanga Sangha Protected Areas, Central African Republic. PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0071840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sak B., Petrzelkova K.J., Kvetonova D., Mynarova A., Pomajbikova K., Modry D. Diversity of microsporidia, Cryptosporidium and Giardia in mountain gorillas (Gorilla beringei beringei) in Volcanoes National Park, Rwanda. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0109751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deplazes P., Mathis A., Muller C., Weber R. Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J. Eukaryot. Microbiol. 1996;43(5):93S. doi: 10.1111/j.1550-7408.1996.tb05018.x. [DOI] [PubMed] [Google Scholar]
- 69.Kondova I., Mansfield K., Buckholt M.A., Stein B., Widmer G., Carville A. Transmission and serial propagation of Enterocytozoon bieneusi from humans and rhesus macaques in gnotobiotic piglets. Infect. Immun. 1998;66(11):5515–5519. doi: 10.1128/iai.66.11.5515-5519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sak B., Kvac M., Hanzlikova D., Cama V. First report of Enterocytozoon bieneusi infection on a pig farm in the Czech Republic. Vet. Parasitol. 2008;153(3–4):220–224. doi: 10.1016/j.vetpar.2008.01.043. [DOI] [PubMed] [Google Scholar]
- 71.Reetz J., Nockler K., Reckinger S., Vargas M.M., Weiske W., Broglia A. Identification of Encephalitozoon cuniculi genotype III and two novel genotypes of Enterocytozoon bieneusi in swine. Parasitol. Int. 2009;58(3):285–292. doi: 10.1016/j.parint.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 72.Galvan-Diaz A.L., Magnet A., Fenoy S., Henriques-Gil N., Haro M., Gordo F.P. Microsporidia detection and genotyping study of human pathogenic E. bieneusi in animals from Spain. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0092289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W., Diao R., Yang J., Xiao L., Lu Y., Li Y. High diversity of human-pathogenic Enterocytozoon bieneusi genotypes in swine in Northeast China. Parasitol. Res. 2014;113(3):1147–1153. doi: 10.1007/s00436-014-3752-9. [DOI] [PubMed] [Google Scholar]
- 74.Zhao W., Zhang W., Yang F., Cao J., Liu H., Yang D. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl. Environ. Microbiol. 2014;80(12):3699–3707. doi: 10.1128/AEM.00807-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prasertbun R., Mori H., Pintong A.R., Sanyanusin S., Popruk S., Komalamisra C. Zoonotic potential of Enterocytozoon genotypes in humans and pigs in Thailand. Vet. Parasitol. 2017;233:73–79. doi: 10.1016/j.vetpar.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 76.Wang L., Zhang H., Zhao X., Zhang L., Zhang G., Guo M. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013;51(2):557–563. doi: 10.1128/JCM.02758-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang J., Song M., Wan Q., Li Y., Lu Y., Jiang Y. Enterocytozoon bieneusi genotypes in children in Northeast China and assessment of risk of zoonotic transmission. J. Clin. Microbiol. 2014;52(12):4363–4367. doi: 10.1128/JCM.02295-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nemejc K., Sak B., Kvetonova D., Hanzal V., Janiszewski P., Forejtek P. Prevalence and diversity of Encephalitozoon spp. and Enterocytozoon bieneusi in wild boars (Sus scrofa) in Central Europe. Parasitol. Res. 2014;113(2):761–767. doi: 10.1007/s00436-013-3707-6. [DOI] [PubMed] [Google Scholar]
- 79.Li W., Deng L., Wu K., Huang X., Song Y., Su H. Presence of zoonotic Cryptosporidium scrofarum, Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotypes in captive Eurasian wild boars (Sus scrofa) in China: potential for zoonotic transmission. Parasit. Vectors. 2017;10(1):10. doi: 10.1186/s13071-016-1942-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dashti A., Rivero-Juarez A., Santin M., Lopez-Lopez P., Caballero-Gomez J., Frias-Casas M. Enterocytozoon bieneusi (microsporidia): identification of novel genotypes and evidence of transmission between sympatric wild boars (Sus scrofa ferus) and Iberian pigs (Sus scrofa domesticus) in Southern Spain. Transbound. Emerg. Dis. 2020;67(6):2869–2880. doi: 10.1111/tbed.13658. [DOI] [PubMed] [Google Scholar]
- 81.Santin M., Dargatz D., Fayer R. Prevalence and genotypes of Enterocytozoon bieneusi in weaned beef calves on cow-calf operations in the USA. Parasitol. Res. 2012;110(5):2033–2041. doi: 10.1007/s00436-011-2732-6. [DOI] [PubMed] [Google Scholar]
- 82.Jiang Y., Tao W., Wan Q., Li Q., Yang Y., Lin Y. Zoonotic and potentially Host-Adapted Enterocytozoon bieneusi genotypes in sheep and cattle in Northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl. Environ. Microbiol. 2015;81(10):3326–3335. doi: 10.1128/AEM.00328-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li J., Qi M., Chang Y., Wang R., Li T., Dong H. Molecular characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in captive wildlife at Zhengzhou zoo, China. J. Eukaryot. Microbiol. 2015;62(6):833–839. doi: 10.1111/jeu.12269. [DOI] [PubMed] [Google Scholar]
- 84.Ma J., Li P., Zhao X., Xu H., Wu W., Wang Y. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015;207(3–4):220–227. doi: 10.1016/j.vetpar.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 85.Zhao W., Zhang W., Yang D., Zhang L., Wang R., Liu A. Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infect. Genet. Evol. 2015;32:265–270. doi: 10.1016/j.meegid.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 86.Ye J., Xiao L., Wang Y., Guo Y., Roellig D.M., Feng Y. Dominance of Giardia duodenalis assemblage A and Enterocytozoon bieneusi genotype BEB6 in sheep in Inner Mongolia, China. Vet. Parasitol. 2015;210(3–4):235–239. doi: 10.1016/j.vetpar.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 87.Fiuza V.R., Lopes C.W., Cosendey R.I., de Oliveira F.C., Fayer R., Santin M. Zoonotic Enterocytozoon bieneusi genotypes found in brazilian sheep. Res. Vet. Sci. 2016;107:196–201. doi: 10.1016/j.rvsc.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Y., Koehler A.V., Wang T., Haydon S.R., Gasser R.B. Enterocytozoon bieneusi genotypes in cattle on farms located within a water catchment area. J. Eukaryot. Microbiol. 2019;66(4):553–559. doi: 10.1111/jeu.12696. [DOI] [PubMed] [Google Scholar]
- 89.Lee J.H. Molecular detection of Enterocytozoon bieneusi and identification of a potentially human-pathogenic genotype in milk. Appl. Environ. Microbiol. 2008;74(5):1664–1666. doi: 10.1128/AEM.02110-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y., Koehler A.V., Wang T., Haydon S.R., Gasser R.B. First detection and genetic characterisation of Enterocytozoon bieneusi in wild deer in Melbourne’s water catchments in Australia. Parasit. Vectors. 2018;11(1):2. doi: 10.1186/s13071-017-2577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Santin M., Fayer R. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J. Eukaryot. Microbiol. 2015;62(1):34–43. doi: 10.1111/jeu.12155. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Z., Huang J., Karim M.R., Zhao J., Dong H., Ai W. Zoonotic Enterocytozoon bieneusi genotypes in Pere David’s deer (Elaphurus davidianus) in Henan, China. Exp. Parasitol. 2015;155:46–48. doi: 10.1016/j.exppara.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 93.Liu W., Nie C., Zhang L., Wang R., Liu A., Zhao W. First detection and genotyping of Enterocytozoon bieneusi in reindeers (Rangifer tarandus): a zoonotic potential of ITS genotypes. Parasit. Vectors. 2015;8:526. doi: 10.1186/s13071-015-1155-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao G.H., Du SZ Wang H.B., Hu X.F., Deng M.J., Yu S.K. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi) Infect. Genet. Evol. 2015;34:394–401. doi: 10.1016/j.meegid.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 95.Mathis A., Breitenmoser A.C., Deplazes P. Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite. 1999;6(2):189–193. doi: 10.1051/parasite/1999062189. [DOI] [PubMed] [Google Scholar]
- 96.Dengjel B., Zahler M., Hermanns W., Heinritzi K., Spillmann T., Thomschke A. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 2001;39(12):4495–4499. doi: 10.1128/JCM.39.12.4495-4499.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lobo M.L., Xiao L., Cama V., Stevens T., Antunes F., Matos O. Genotypes of Enterocytozoon bieneusi in mammals in Portugal. J. Eukaryot. Microbiol. 2006;53:S61–S64. doi: 10.1111/j.1550-7408.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 98.Santin M., Cortes Vecino J.A., Fayer R. Enterocytozoon bieneusi genotypes in dogs in Bogota, Colombia. Am J. Trop. Med. Hygiene. 2008;79(2):215–217. [PubMed] [Google Scholar]
- 99.Santin M., Trout J.M., Vecino J.A., Dubey J.P., Fayer R. Cryptosporidium, Giardia and Enterocytozoon bieneusi in cats from Bogota (Colombia) and genotyping of isolates. Vet. Parasitol. 2006;141(3–4):334–339. doi: 10.1016/j.vetpar.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 100.Abe N., Kimata I., Iseki M. Molecular evidence of Enterocytozoon bieneusi in Japan. J. Vet. Med. Sci. 2009;71(2):217–219. doi: 10.1292/jvms.71.217. [DOI] [PubMed] [Google Scholar]
- 101.Zhang X., Wang Z., Su Y., Liang X., Sun X., Peng S. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 2011;49(5):2006–2008. doi: 10.1128/JCM.00372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Karim M.R., Dong H., Yu F., Jian F., Zhang L., Wang R. Genetic diversity in Enterocytozoon bieneusi isolates from dogs and cats in China: host specificity and public health implications. J. Clin. Microbiol. 2014;52(9):3297–3302. doi: 10.1128/JCM.01352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li W., Li Y., Song M., Lu Y., Yang J., Tao W. Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet. Parasitol. 2015;208(3–4):125–134. doi: 10.1016/j.vetpar.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 104.Xu H., Jin Y., Wu W., Li P., Wang L., Li N. Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai, China. Parasit. Vectors. 2016;9:121. doi: 10.1186/s13071-016-1409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Piekarska J., Kicia M., Wesolowska M., Kopacz Z., Gorczykowski M., Szczepankiewicz B. Zoonotic microsporidia in dogs and cats in Poland. Vet. Parasitol. 2017;246:108–111. doi: 10.1016/j.vetpar.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 106.Reetz J., Rinder H., Thomschke A., Manke H., Schwebs M., Bruderek A. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens) Int. J. Parasitol. 2002;32(7):785–787. doi: 10.1016/s0020-7519(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 107.Li W., Tao W., Jiang Y., Diao R., Yang J., Xiao L. Genotypic distribution and phylogenetic characterization of Enterocytozoon bieneusi in diarrheic chickens and pigs in multiple cities, China: potential zoonotic transmission. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0108279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.da Cunha M.J., Cury M.C., Santin M. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotypes in chickens. Vet. Parasitol. 2016;217:108–112. doi: 10.1016/j.vetpar.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 109.Zhao W., Yu S., Yang Z., Zhang Y., Zhang L., Wang R. Genotyping of Enterocytozoon bieneusi (microsporidia) isolated from various birds in China. Infect. Genet. Evol. 2016;40:151–154. doi: 10.1016/j.meegid.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 110.da Cunha M.J.R., Cury M.C., Santin M. Molecular identification of Enterocytozoon bieneusi, Cryptosporidium, and Giardia in Brazilian captive birds. Parasitol. Res. 2017;116(2):487–493. doi: 10.1007/s00436-016-5309-6. [DOI] [PubMed] [Google Scholar]
- 111.Muller M.G., Kinne J., Schuster R.K., Walochnik J. Outbreak of microsporidiosis caused by Enterocytozoon bieneusi in falcons. Vet. Parasitol. 2008;152(1–2):67–78. doi: 10.1016/j.vetpar.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 112.Lobo M.L., Xiao L., Cama V., Magalhaes N., Antunes F., Matos O. Identification of potentially human-pathogenic Enterocytozoon bieneusi genotypes in various birds. Appl. Environ. Microbiol. 2006;72(11):7380–7382. doi: 10.1128/AEM.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kasickova D., Sak B., Kvac M., Ditrich O. Sources of potentially infectious human microsporidia: molecular characterisation of microsporidia isolates from exotic birds in the Czech Republic, prevalence study and importance of birds in epidemiology of the human microsporidial infections. Vet. Parasitol. 2009;165(1–2):125–130. doi: 10.1016/j.vetpar.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 114.Lallo M.A., Calabria P., Milanelo L. Encephalitozoon and Enterocytozoon (microsporidia) spores in stool from pigeons and exotic birds: microsporidia spores in birds. Vet. Parasitol. 2012;190(3–4):418–422. doi: 10.1016/j.vetpar.2012.06.030. [DOI] [PubMed] [Google Scholar]
- 115.Tavalla M., Mardani-Kateki M., Abdizadeh R., Soltani S., Saki J. Molecular diagnosis of potentially human pathogenic Enterocytozoon bieneusi and Encephalitozoon species in exotic birds in southwestern Iran. J. Infec. Public Health. 2018;11(2):192–196. doi: 10.1016/j.jiph.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 116.Haro M., Izquierdo F., Henriques-Gil N., Andres I., Alonso F., Fenoy S. First detection and genotyping of human-associated microsporidia in pigeons from urban parks. Appl. Environ. Microbiol. 2005;71(6):3153–3157. doi: 10.1128/AEM.71.6.3153-3157.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Graczyk T.K., Sunderland D., Rule A.M., da Silva A.J., Moura I.N., Tamang L. Urban feral pigeons (Columba livia) as a source for air- and waterborne contamination with Enterocytozoon bieneusi spores. Appl. Environ. Microbiol. 2007;73(13):4357–4358. doi: 10.1128/AEM.00202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bart A., Wentink-Bonnema E.M., Heddema E.R., Buijs J., van Gool T. Frequent occurrence of human-associated microsporidia in fecal droppings of urban pigeons in Amsterdam, the Netherlands. Appl. Environ. Microbiol. 2008;74(22):7056–7058. doi: 10.1128/AEM.01429-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pirestani M., Sadraei J., Forouzandeh M. Molecular characterization and genotyping of human related microsporidia in free-ranging and captive pigeons of Tehran, Iran. Infect. Genet. Evol. 2013;20:495–499. [PubMed] [Google Scholar]
- 120.Perec-Matysiak A., Wesolowska M., Lesnianska K., Bunkowska-Gawlik K., Hildebrand J., Kicia M. Survey for zoonotic microsporidian pathogens in wild living urban rooks (Corvus frugilegus) J. Eukaryot. Microbiol. 2017;64(5):721–724. doi: 10.1111/jeu.12402. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y., Zhang K., Zhang Y., Wang K., Gazizova A., Wang L. First detection of Enterocytozoon bieneusi in whooper swans (Cygnus cygnus) in China. Parasit. Vectors. 2020;13(1):5. doi: 10.1186/s13071-020-3884-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Santin M., Vecino J.A., Fayer R. A zoonotic genotype of Enterocytozoon bieneusi in horses. J. Parasitol. 2010;96(1):157–161. doi: 10.1645/GE-2184.1. [DOI] [PubMed] [Google Scholar]
- 123.Wagnerova P., Sak B., Kvetonova D., Bunatova Z., Civisova H., Marsalek M. Enterocytozoon bieneusi and Encephalitozoon cuniculi in horses kept under different management systems in the Czech Republic. Vet. Parasitol. 2012;190(3–4):573–577. doi: 10.1016/j.vetpar.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 124.Laatamna A.E., Wagnerova P., Sak B., Kvetonova D., Xiao L., Rost M. Microsporidia and Cryptosporidium in horses and donkeys in Algeria: detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol. 2015;208(3–4):135–142. doi: 10.1016/j.vetpar.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 125.Qi M., Wang R., Wang H., Jian F., Li J., Zhao J. Enterocytozoon bieneusi genotypes in grazing horses in China and their zoonotic transmission potential. J. Eukaryot. Microbiol. 2016;63(5):591–597. doi: 10.1111/jeu.12308. [DOI] [PubMed] [Google Scholar]
- 126.Yue D.M., Ma J.G., Li F.C., Hou J.L., Zheng W.B., Zhao Q. Occurrence of Enterocytozoon bieneusi in donkeys (Equus asinus) in China: a public health concern. Front. Microbiol. 2017;8:565. doi: 10.3389/fmicb.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang Y., Lin Y., Li Q., Zhang S., Tao W., Wan Q. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotype D in farmed foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in China: first identification and zoonotic concern. Parasitol. Res. 2015;114(11):4341–4348. doi: 10.1007/s00436-015-4714-6. [DOI] [PubMed] [Google Scholar]
- 128.Tian G.R., Zhao G.H., Du S.Z., Hu X.F., Wang H.B., Zhang L.X. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect. Genet. Evol. 2015;34:32–35. doi: 10.1016/j.meegid.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 129.Zhang X.X., Jiang J., Cai Y.N., Wang C.F., Xu P., Yang G.L. Molecular characterization of Enterocytozoon bieneusi in domestic rabbits (Oryctolagus cuniculus) in Northeastern China. Korean J. Parasitol. 2016;54(1):81–85. doi: 10.3347/kjp.2016.54.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sak B., Kvac M., Kvetonova D., Albrecht T., Pialek J. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European House Mice (Mus musculus musculus) and West-European House Mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Vet. Parasitol. 2011;178(3–4):246–250. doi: 10.1016/j.vetpar.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 131.Perec-Matysiak A., Bunkowska-Gawlik K., Kvac M., Sak B., Hildebrand J., Lesnianska K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet. Parasitol. 2015;214(3–4):242–246. doi: 10.1016/j.vetpar.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 132.Graczyk T.K., Conn D.B., Lucy F., Minchin D., Tamang L., Moura L.N. Human waterborne parasites in zebra mussels (Dreissena polymorpha) from the Shannon River drainage area, Ireland. Parasitol. Res. 2004;93(5):385–391. doi: 10.1007/s00436-004-1142-4. [DOI] [PubMed] [Google Scholar]
- 133.Lucy F.E., Graczyk T.K., Tamang L., Miraflor A., Minchin D. Biomonitoring of surface and coastal water for Cryptosporidium, Giardia, and human-virulent microsporidia using molluscan shellfish. Parasitol. Res. 2008;103(6):1369–1375. doi: 10.1007/s00436-008-1143-9. [DOI] [PubMed] [Google Scholar]
- 134.Graczyk T.K., Girouard A.S., Tamang L., Nappier S.P., Schwab K.J. Recovery, bioaccumulation, and inactivation of human waterborne pathogens by the Chesapeake Bay nonnative oyster, Crassostrea ariakensis. Appl. Environ. Microbiol. 2006;72(5):3390–3395. doi: 10.1128/AEM.72.5.3390-3395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fayer R., Fair P.A., Bossart G.D., Santin M. Examination of naturally exposed bottlenose dolphins (Tursiops truncatus) for microsporidia, Cryptosporidium, and Giardia. J. Parasitol. 2008;94(1):143–147. doi: 10.1645/GE-1262.1. [DOI] [PubMed] [Google Scholar]
- 136.Sparfel J.M., Sarfati C., Liguory O., Caroff B., Dumoutier N., Gueglio B. Detection of microsporidia and identification of Enterocytozoon bieneusi in surface water by filtration followed by specific PCR. J. Eukaryot. Microbiol. 1997;44(6):78S. doi: 10.1111/j.1550-7408.1997.tb05792.x. [DOI] [PubMed] [Google Scholar]
- 137.Galvan A.L., Magnet A., Izquierdo F., Fenoy S., Rueda C., Fernandez Vadillo C. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: a year-long longitudinal study. Appl. Environ. Microbiol. 2013;79(2):449–459. doi: 10.1128/AEM.02737-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Huang C., Hu Y., Wang L., Wang Y., Li N., Guo Y. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl. Environ. Microbiol. 2017;83(16):e00682–17. doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Graczyk T.K., Sunderland D., Tamang L., Lucy F.E., Breysse P.N. Bather density and levels of Cryptosporidium, Giardia, and pathogenic microsporidian spores in recreational bathing water. Parasitol. Res. 2007;101(6):1729–1731. doi: 10.1007/s00436-007-0734-1. [DOI] [PubMed] [Google Scholar]
- 140.Graczyk T.K., Sunderland D., Tamang L., Shields T.M., Lucy F.E., Breysse P.N. Quantitative evaluation of the impact of bather density on levels of human-virulent microsporidian spores in recreational water. Appl. Environ. Microbiol. 2007;73(13):4095–4099. doi: 10.1128/AEM.00365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Graczyk T.K., Sunderland D., Awantang G.N., Mashinski Y., Lucy F.E., Graczyk Z. Relationships among bather density, levels of human waterborne pathogens, and fecal coliform counts in marine recreational beach water. Parasitol. Res. 2010;106(5):1103–1108. doi: 10.1007/s00436-010-1769-2. [DOI] [PubMed] [Google Scholar]
- 142.Graczyk T.K., Lucy F.E., Tamang L., Mashinski Y., Broaders M.A., Connolly M. Propagation of human enteropathogens in constructed horizontal wetlands used for tertiary wastewater treatment. Appl. Environ. Microbiol. 2009;75(13):4531–4538. doi: 10.1128/AEM.02873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ben Ayed L., Yang W., Widmer G., Cama V., Ortega Y., Xiao L. Survey and genetic characterization of wastewater in Tunisia for Cryptosporidium spp., Giardia duodenalis, Enterocytozoon bieneusi, Cyclospora cayetanensis and Eimeria spp. J. Water Health. 2012;10(3):431–444. doi: 10.2166/wh.2012.204. [DOI] [PubMed] [Google Scholar]
- 144.Cheng H.W., Broaders M.A., Lucy F.E., Mastitsky S.E., Graczyk T.K. Determining potential indicators of Cryptosporidium oocysts throughout the wastewater treatment process. Water Sci. Technol. 2012;65(5):875–882. doi: 10.2166/wst.2012.918. [DOI] [PubMed] [Google Scholar]
- 145.Ma J., Feng Y., Hu Y., Villegas E.N., Xiao L. Human infective potential of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in urban wastewater treatment plant effluents. J. Water Health. 2016;14(3):411–423. doi: 10.2166/wh.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yamashiro S., Fiuza V., Teixeira A., Branco N., Levy C.E., Castro I. Enterocytozoon bieneusi detected by molecular methods in raw sewage and treated effluent from a combined system in Brazil. Mem. Inst. Oswaldo Cruz. 2017;112(6):403–410. doi: 10.1590/0074-02760160435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ye J., Ji Y., Xu J., Ma K., Yang X. Zoonotic Enterocytozoon bieneusi in raw wastewater in Zhengzhou, China. Folia Parasitol. 2017;64 doi: 10.14411/fp.2017.002. [DOI] [PubMed] [Google Scholar]
- 148.Graczyk T.K., Kacprzak M., Neczaj E., Tamang L., Graczyk H., Lucy F.E. Human-virulent microsporidian spores in solid waste landfill leachate and sewage sludge, and effects of sanitization treatments on their inactivation. Parasitol. Res. 2007;101(3):569–575. doi: 10.1007/s00436-007-0515-x. [DOI] [PubMed] [Google Scholar]
- 149.Graczyk T.K., Lucy F.E., Tamang L., Miraflor A. Human enteropathogen load in activated sewage sludge and corresponding sewage sludge end products. Appl. Environ. Microbiol. 2007;73(6):2013–2015. doi: 10.1128/AEM.02412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thurston-Enriquez J.A., Watt P., Dowd S.E., Enriquez R., Pepper I.L., Gerba C.P. Detection of protozoan parasites and microsporidia in irrigation waters used for crop production. J. Food Prot. 2002;65(2):378–382. doi: 10.4315/0362-028x-65.2.378. [DOI] [PubMed] [Google Scholar]
- 151.Innes E.A., Chalmers R.M., Wells B., Pawlowic M.C. A One Health approach to tackle cryptosporidiosis. Trends Parasitol. 2020;36(3):290–303. doi: 10.1016/j.pt.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hong A., Zampieri R.A., Shaw J.J., Floeter-Winter L.M., Laranjeira-Silva M.F. One Health approach to leishmaniases: understanding the disease dynamics through diagnostic tools. Pathogens. 2020;9(10):809. doi: 10.3390/pathogens9100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krecek R.C., Rabinowitz P.M., Conrad P.A. Demystifying and demonstrating the value of a One Health approach to parasitological challenges. Vet. Parasitol. 2020;287:109202. doi: 10.1016/j.vetpar.2020.109202. [DOI] [PubMed] [Google Scholar]
- 154.Aguirre A.A., Longcore T., Barbieri M., Dabritz H., Hill D., Klein P.N. The One Health approach to toxoplasmosis: epidemiology, control, and prevention strategies. EcoHealth. 2019;16(2):378–390. doi: 10.1007/s10393-019-01405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Suijkerbuijk A.W.M., van Gils P.F., Bonacic Marinovic A.A., Feenstra T.L., Kortbeek L.M., Mangen M.J. The design of a social cost-benefit analysis of preventive interventions for toxoplasmosis: an example of the One Health approach. Zoonoses Public Health. 2018;65(1):185–194. doi: 10.1111/zph.12417. [DOI] [PubMed] [Google Scholar]