Abstract
Mixed strain infection (MSI) refers to the concurrent infection of a susceptible host with multiple strains of a single pathogenic species. Known to occur in humans and animals, MSIs deserve special consideration when studying transmission dynamics, evolution, and treatment of mycobacterial diseases, notably tuberculosis in humans and paratuberculosis (or Johne's disease) in ruminants. Therefore, a systematic review was conducted to examine how MSIs are defined in the literature, how widespread the phenomenon is across the host species spectrum, and to document common methods used to detect such infections. Our search strategy identified 121 articles reporting MSIs in both humans and animals, the majority (78.5%) of which involved members of the Mycobacterium tuberculosis complex, while only a few (21.5%) examined non-tuberculous mycobacteria (NTM). In addition, MSIs exist across various host species, but most reports focused on humans due to the extensive amount of work done on tuberculosis. We reviewed the strain typing methods that allowed for MSI detection and found a few that were commonly employed but were associated with specific challenges. Our review notes the need for standardization, as some highly discriminatory methods are not adapted to distinguish between microevolution of one strain and concurrent infection with multiple strains. Further research is also warranted to examine the prevalence of NTM MSIs in both humans and animals. In addition, it is envisioned that the accurate identification and a better understanding of the distribution of MSIs in the future will lead to important information on the epidemiology and pathophysiology of mycobacterial diseases.
Keywords: Mycobacterium, tuberculosis, non-tuberculous, complex, mixed strain, infections, detection, methods
Introduction
The genus Mycobacterium includes 192 different species with diverse growth characteristics and (Schulze-Röbbecke, 1993; Primm et al., 2004; Parte, 2018) and host tropism (Ahmed et al., 2013). Mycobacteria can be categorized into three major groups: those that cause tuberculosis (TB) and are part of the Mycobacterium tuberculosis complex (MTBC), those that cause leprosy (including Mycobacterium leprae and Mycobacterium lepromatosis), and the remaining members, commonly referred to as atypical mycobacteria (Siddiqi, 1978), non-tuberculous mycobacteria (NTM) or mycobacteria other than M. tuberculosis (MOTT) (Ahmed et al., 2013). In addition, members from this genus can be further categorized based on their growth rates into rapid and slow growers, where the latter have prolonged doubling times, making it challenging to cultivate them (Wayne and Kubica, 1986).
Tuberculosis is caused by M. tuberculosis infecting the lungs of the host, though the pathogen can also spread to other parts of the body (Sia and Wieland, 2011). Members of the MTBC such as Mycobacterium africanum also cause TB in humans (De Jong et al., 2010), while non-human host tropism is reported for other bacteria from the group. For example, Mycobacterium bovis causes bovine TB (Morris et al., 1994; Cosivi et al., 1998; Grange, 2001), Mycobacterium caprae can infect a variety of wild and domesticated animals and Mycobacterium pinnipedii causes TB in pinniped species (Roe et al., 2019). Tuberculosis is an ancient disease afflicting humans and M. tuberculosis has been studied for over a century, but the disease remains a significant cause of global morbidity and mortality (WHO, 2019). One reason why TB remains problematic is due to the complex interaction between MTBC members and their hosts, many aspects of which are still not fully understood. In addition, the emergence and spread of drug resistant forms of M. tuberculosis further exacerbates the situation, leaving few effective treatment options in some cases (WHO, 2019).
The NTM group comprises over 150 different species, including several pathogens from the Mycobacterium avium and Mycobacterium abscessus complexes (Tortoli, 2014). Members of the M. avium complex (MAC) are commonly found in the environment and cause opportunistic infections (Ichiyama et al., 1988; Von Reyn et al., 1993; Yajko et al., 1995; Reed et al., 2006), especially in immunocompromised individuals such as those suffering from acquired immunodeficiency syndrome (AIDS) (Jacobson et al., 1991; Havlik et al., 1992; Griffith et al., 2007). M. avium and Mycobacterium intracellulare (also a MAC member) cause pulmonary infections (Guthertz et al., 1989; Hocqueloux et al., 1998), where the latter can also infect immunocompetent individuals (Han et al., 2005; Koh et al., 2012). Furthermore, M. avium includes several subspecies (avium, paratuberculosis, hominissuis and silvaticum), which can infect organs other than the lungs (Thorel et al., 1990; Mijs et al., 2002). For example, M. avium subspecies paratuberculosis (MAP) causes Johne's disease in ruminants and has been linked to Crohn's disease in humans, conditions that afflict the small intestine (Behr and Kapur, 2008; Sweeney et al., 2012).
The M. abscessus complex includes three fast-growing subspecies (abscessus, massiliense and bolletii), which are highly resistant to many antibiotics and cause a wide range of human infections (Cho et al., 2013; Sassi and Drancourt, 2014; Lee et al., 2015; Adekambi et al., 2017). Another NTM of significance is M. genavense, an opportunistic pathogen that often causes disease in immunocompromised patients and has also been found to infect various domestic companion animals (Hoop et al., 1993; Böttger, 1994; Kiehn et al., 1996; Hughes et al., 1999; Krebs et al., 2000; Lucas et al., 2000; Hoefsloot et al., 2013). The NTM discussed above are just a few of many that are of concern to human and animal health (Piersimoni and Scarparo, 2008; Cook, 2010; Griffith, 2010; Atkins and Gottlieb, 2014; Biet and Boschiroli, 2014), demonstrating the propensity of members from this group to cause diverse diseases if given the opportunity.
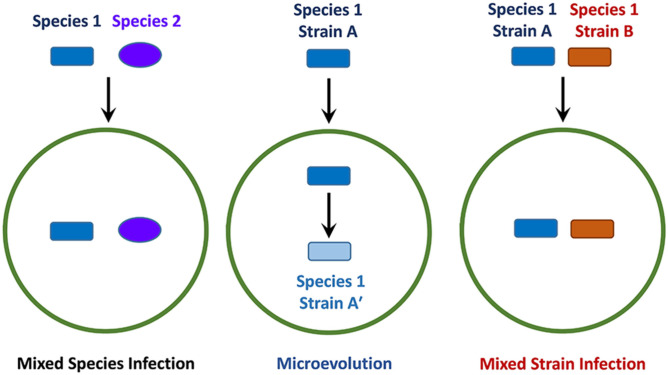
The progression and outcome of an infection is dependent on many factors, which include the resident host-microbiome and the presence of other pathogens, sometimes from the same genus (Figure 1) (Adami and Cervantes, 2015; Namasivayam et al., 2019, 2020). Mixed-species infections refer to the phenomenon where different species belonging to the same genus concurrently infect a single host (Figure 1). Another important factor to consider is the potential for genetically distinct strains (or isolates) of the same pathogenic species to infect a single host at any given time, which is sometimes referred to as an polyclonal infection (Taylor et al., 1997; Cohen et al., 2012; McNaughton et al., 2018). This situation can potentially arise if an isolate undergoes intra-host evolution (also referred to as microevolution) following infection (Figure 1), leading to minor genetic differences in the resulting progeny (Jordan et al., 2002; Feil, 2004; Ley et al., 2019). Another mechanism leading to polyclonal infections involves concomitant or sequential infection by genetically distinct strains (Figure 1), a process that is referred to as mixed strain infection (MSI). The presence of multiple strains with varying genotypes can result in altered physiological characteristics or pathogenicity, which in turn can affect transmission dynamics (Taylor et al., 1997), or lead to treatment complications due to varying antibiotic resistance profiles (also known as heteroresistance) (Van Rie et al., 2005; Shin et al., 2018). It is possible that a single treatment regimen will not be optimal against all strains in an individual, causing the infection to persist or return after briefly subsiding. MSIs are particularly relevant in slow-growing pathogens such as M. tuberculosis, MAC members and other related mycobacteria, as these organisms can remain undetectable for long periods of time (Whitlock and Buergelt, 1996; Gengenbacher and Kaufmann, 2012). If an MSI exists and the initial treatment is unsuccessful, the persistence of these infections may result in the development of more severe disease over time (Baldeviano-Vidalón et al., 2005). Additionally, MSIs have the potential to interfere with host immune responses due to antigenic variations that might exist between different strains (Huang et al., 2010; Cohen et al., 2012; Yoshida et al., 2018). Therefore, by examining MSIs and their transmission, successful treatment methods can be devised, and essential information might also be gained for use in future vaccine development endeavors.
Figure 1.
Schematic of different infection events involving pathogens from the same genus. The green circle represents a permissive host whereas the dark blue rectangle and purple oval indicate different species of pathogens belonging to the same genus. The rectangles of different colors indicate distinct strains derived from the same species, where blue indicates microevolution and red, mixed strain infection.
The purpose of this study was to conduct a systematic review to gain a better understanding of MSIs across the genus Mycobacterium and the methods used to detect them. Typically, the detection of such infections is challenging due to the lack of distinct intraspecies markers that allow for discrimination between isolates/strains. Despite this, MSIs in mycobacteria were found using a variety of strain typing methods, each with a different level of discriminatory ability and ease of use, with different methods focusing on specific aspects of the Mycobacterium genome. Mycobacterial strain discrimination is made possible by the analysis of restriction fragment length polymorphisms (RFLP) in species specific insertion sequences (IS) such as IS6110 (associated with the MTBC, with some exceptions) (Coros et al., 2008; Gonzalo-Asensio et al., 2018), and IS1245 or IS1311 (both associated with the MAC) (Guerrero et al., 1995; Johansen et al., 2005; Coll et al., 2014). Another general method used to discriminate between strains exploits the nucleotide sequences present in variable number tandem repeats (VNTR), which are dispersed throughout mycobacterial genomes. By examining differences in the number of nucleotide repeats present at distinct loci, individual strains can be typed. Different mycobacteria harbor a variety of VNTR loci, though depending on the species and loci examined, they may instead be referred to as multi-locus variable-number tandem repeats (MLV) (Overduin et al., 2004; Hill et al., 2012; Biffa et al., 2014), mycobacterial interspersed repetitive unit–variable number tandem repeats (MIRU-VNTR) (Supply et al., 2001, 2006) or short sequence repeats (SSRs) (Amonsin et al., 2004; Podder et al., 2015). Analysis of the entire genome at the individual nucleotide level using methods based on whole-genome sequencing (WGS) also allows for examination of strain diversity, but at a resolution unmatched by RFLP or VNTR based methods. By using WGS, strains can be typed and compared without focusing on a given set of loci allowing for more accurate detection of MSIs, re-infections, and relapses (Homolka et al., 2012; Coll et al., 2014; Witney et al., 2017; Lipworth et al., 2019). Heterogeneous (also referred to as heterozygous) single nucleotide polymorphisms (SNPs) are predominantly used in strain comparisons, and the presence of many different SNPs in isolates from a single sample is suggestive of MSIs (Sobkowiak et al., 2018).
It was also our intention to help clarify true MSIs as compared to similar events such as re-infection, relapses, and microevolution. While polyclonal infection may refer to microevolution, some studies have also used the term to describe infections that fit the criteria of an MSI (Adams et al., 2012; Fujita et al., 2014; Farmanfarmaei et al., 2017; Kamakoli et al., 2017, 2020b; Nathavitharana et al., 2017). Due to this lack of consensus regarding the terminology used in the literature and to be consistent in our study, we have selected definitions to describe the different events (Table 1). For our purposes, MSIs refer to an infection where multiple unrelated strains, which did not evolve from an initial infecting strain, are present within a single host at the same time.
Table 1.
Glossary of terms.
| Terma | Descriptionb |
|---|---|
| Sample/specimen | A sputum, blood, feces or otherwise uncultured biological sample from a host/patient, which is directly used for strain-typing analysis or is subsequently used for culturing. |
| Culture | Refers to the amplification of bacteria following growth on solid or in liquid media using a sample/specimen as an inoculant. Does not explicitly imply a pure or axenic culture. |
| Isolate (strain, if characterized) | Refers to a single colony from an agar plate or an axenic bacterial culture derived from a sample/specimen. |
| Microevolution | A co-infection that involves multiple strains/sub-strains in a single host that evolved from a single strain that caused the initial infection. |
| Mixed genotype infection (MGI)/polyclonal infection | A co-infection caused by two or more strains of the same species in a single host. Encompasses both microevolution and mixed strain infections. |
| Mixed strain infection (MSI) | A co-infection caused by phylogenetically distinct strains of the same species in a single host (contrary to microevolution). Sometimes also referred to simply as mixed infection in the review. |
| Re-infection | Recurrent disease caused by a strain that is unrelated to the one that caused the initial infection. |
| Relapse | Recurrent disease due to the same strain that caused the initial infection. |
Only non-standard terms or those that have been modified for use in the review are listed.
Descriptions for the purposes as used in the current review.
Materials and Methods
A systematic review of the literature was performed to identify methods used for detecting MSIs caused by a single species of mycobacteria. This process comprised four stages, where the first one entailed utilizing a comprehensive search strategy to locate published studies. An initial limited search was conducted in Ovid MEDLINE to compile a list of keywords and index terms from relevant articles. A full search strategy was then developed by a librarian by testing search terms in MEDLINE and only those terms yielding unique results were retained for further use. The full search strategy was externally peer-reviewed by a second librarian using the Peer Review of Electronic Search Strategies (PRESS) guidelines (McGowan et al., 2016). The search strategy was then adapted for EMBASE (Elsevier), following which both the MEDLINE and EMBASE were initially searched on June 11, 2020, with no prerequisite limits. An updated search was performed on August 25th, 2020 to include the term “polyclonal” (Supplementary Table 1). Search results were collated and uploaded into Endnote version X8.2 (Clarivate Analytics, PA, USA) for organization followed by Covidence™ (Veritas Health Innovation Ltd, Melbourne, Australia), a screening and extraction tool for systematic reviews. After removal of duplicates, reference lists of all selected studies were screened for additional articles of interest using Google Scholar (Figure 2). The second stage involved the screening of article titles and abstracts by two reviewers. The inclusion criteria for articles at this stage were as follows: (i) presence of both a title and abstract; (ii) abstracts in English or French; (iii) primary article or review; (iv) explicit mention of mixed infection or other synonymous terms like double infection, multiple infection, simultaneous infection or polyclonal infection; (v) described molecular or phenotypic methods; and (vi) mention of M. tuberculosis (MTBC), non-tuberculous mycobacteria (NTM), mycobacteria other than tuberculosis (MOTT), atypical mycobacteria, environmental mycobacteria or M. leprae. The presence of additional terms including “mixed, simultaneous, multiple, mycobacteria, concomitant, concurrent, co-infection, polyclonal, Mycobacterium, heterogeneity, subtype, sub-type, double, more than one, dual, and superinfection” were searched for within abstracts to assist in study identification. Articles were excluded if they were: (i) opinion pieces; (ii) commentaries; or (iii) not written in English or French. Conflicts on inclusion/exclusion of specific articles were resolved after analysis by a third reviewer or by mutual agreement by the team.
Figure 2.
Flow diagram of search strategy. Modified preferred reporting items for systematic reviews and meta-analyses (PRISMA) results for search on mycobacterial MSI detection. Details are provided in material and methods section and Supplementary Table 1.
Full texts of articles were uploaded to Covidence™ and subsequently assessed for eligibility at the third screening stage using the following inclusion criteria: (i) full-text availability; (ii) article in English or French; and (iii) reported mixed infection, double infection, multiple infection, simultaneous infection or polyclonal infections involving one Mycobacterium species using molecular or phenotypic methods. Articles were excluded from the main list if: (i) they reported clonal variants suggestive of microevolution of a single strain; (ii) reported detecting mixed-species infections, e.g., M. tuberculosis and M. bovis together; (iii) article was unavailable; (iv) they were dissertations, conference or poster presentation abstracts; (v) did not contain sufficient information; or (v) were not in English or French. In the last stage of the analysis, initial findings from the full-text screening were analyzed to identify data for extraction and use in the review. All articles citing the identified reports for use in the review were also examined for relevant information, which was included in the final results.
Results and Discussion
An initial screen of the literature yielded 14,952 records, and after the removal of duplicate and non-relevant entries based on abstracts and content, 253 articles were retained for full-text review (Figure 2). Examination of these articles resulted in the further exclusion of 156 entries for various reasons as described in Figure 2, leaving 97 reports for inclusion in the review. Additionally, 24 other relevant studies cited in the 97 reports were also included and are presented in the modified Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA), to adhere to the systematic review format (Figure 2).
Data was extracted from all 121 selected articles, and general information, including the publication year, primary author, study location, bacterial and host species involved, was recorded (Supplementary Table 2). The number of samples/isolates, if they were derived from clinical specimens or cultures, the prevalence of MSIs reported in each study and the human immunodeficiency viruses (HIV) status of human subjects was also noted when possible. The studies were allocated into two separate groups based on MSI reports in humans and animals, respectively (Table 2). Reports were further classified based on the location of study by continent, publication year (grouped by decade), the analysis method used, and whether the study examined MSIs involving members of the MTBC or NTM. If a study examined multiple types of hosts or used different methods that successfully identified MSIs, then it was counted in more than one category. A brief overview of the different methods that successfully detected mycobacterial MSIs is presented in Table 3. It was noted that reports involving M. tuberculosis MSIs from humans were significantly overrepresented than those involving NTM or animals (Table 2). Thus, results from the analysis on MTBC and NTM are discussed separately to highlight methods and significant findings.
Table 2.
General characteristics of reviewed mycobacterial studies involving MSIs (n = 121).
| Attribute | Human (%)a | Animal (%)a |
|---|---|---|
| Mycobacterial group responsible for MSIs | ||
| M. tuberculosis Complex (MTBC) | 87 (71.9%) | 8 (6.6%) |
| Non-tuberculous Mycobacteria (NTM)b | 18 (14.9%) | 9 (7.4%) |
| Geographic locationc | ||
| Africab | 28 (23.1%) | 4 (3.3%) |
| Asiab | 31 (25.6%) | 0 (0.0%) |
| Europeb | 20 (16.5%) | 8 (6.6%) |
| North America | 15 (12.4%) | 3 (2.5%) |
| Oceania | 1 (0.8%) | 0 (0.0%) |
| South Americab | 11 (9.1%) | 3 (2.5%) |
| Publication year (decade) | ||
| Pre-2000 | 12 (9.9%) | 0 (0.0%) |
| 2000-2009b | 32 (26.4%) | 4 (3.3%) |
| 2010-2019 | 54 (44.6%) | 13 (10.7%) |
| 2020 | 7 (5.8%) | 1 (0.8%) |
| MSI detection methodd | ||
| Insertion Sequence Based RFLPb | 34 (28.1%) | 3 (2.5%) |
| Variable Number Tandem Repeat | 44 (36.4%) | 12 (9.9%) |
| Whole Genome Sequencing | 12 (9.9%) | 3 (2.5%) |
| Spoligotyping | 14 (11.6%) | 6 (5.0%) |
| Region Specific PCRb | 16 (13.2%) | 0 (0.0%) |
| Othere | 11 (9.1%) | 0 (0.0%) |
Studies were grouped based on multiple criteria (host, geographical location, publication date and methods used). Totals for some sections may not add up to 100% as certain studies met multiple criteria and were therefore counted more than once.
The subsection contains studies counted under more than one criterion.
Excluding one study (Gan et al., 2016), which used information on isolates from a global database.
Not counting methods that were unable to identify MSIs in the respective studies. Methods are briefly described in Table 3 and details are included in Supplementary Table 2.
Includes methods such as phage typing, PFGE, etc. Details are provided in Supplementary Table 2.
Table 3.
Overview of methods that were used to detect mycobacterial MSIs.
| Methoda | Targetb | Output or readoutc | Sample typed |
|---|---|---|---|
| Insertion sequence based RFLP | Species specific insertion sequences | Differences in restriction enzyme band hybridization patterns | Culture |
| Variable number tandem repeat | Species specific tandem repeat loci | Differences in copy numbers based on PCR | Specimen or culture |
| Whole genome sequencing | Entire genome | Heterogeneous SNPs | Specimen or culture |
| Spoligotyping | Direct repeat spacers (CRISPR) | Differences in spacer sequences based on hybridization | Specimen or culture |
| Region-specific PCR | Chromosomal targets depending on the specific method and species | Differences in PCR band patterns | Specimen or culture |
| Phage typinge | Lysis of colonies by mycobacteriophages | Susceptibility to a select mycobacteriophages | Culture |
| Pulse field gel electrophoresise | Enzymatic digestion of chromosomal DNA | Differences in restriction enzyme band patterns | Culture |
| pTBN12 digestione | Species specific plasmid fingerprinting | Differences in restriction enzyme band patterns | Culture |
| hsp65 PCR-RFLPe | hsp65 gene | Differences in restriction enzyme band patterns | Culture |
| Random amplified polymorphic DNAe | Species specific chromosomal DNA | Differences in PCR band patterns | Culture |
| Other RFLPe | Species specific DNA sequences | Differences in restriction enzyme band hybridization patterns | Specimen or culture |
References for methods are present in the main text and Supplementary Table 2.
The component or property of the bacterial cell analyzed by the respective methods.
The result obtained for interpretation using the respective methods.
The type of material that can be used for the assay. Note that all methods except for phage typing use extracted DNA for analysis.
Methods listed come under the “other” category in Table 2.
M. tuberculosis Complex
One of the earliest methods developed to discriminate between strains of M. tuberculosis involved the use of mycobacteriophages (Table 3), which specifically lyse certain strains, leading to the formation of plaques on solid agar plates (Jones and Greenberg, 1978). Phage typing has also been used to identify cross-contamination, transmission dynamics and MSIs based on the sensitivities of M. tuberculosis isolates to a panel of selective phages (Mankiewicz and Liivak, 1975; Bates et al., 1976; Snider et al., 1984; Jones, 1988), but more modern methods are faster and offer better discriminatory power (Snider et al., 1984). Our primary workflow did not find any reports on the identification of MSIs using phage typing, but secondary searches found two such studies. Mankiewicz and Liivak (1975) sampled 233 patients, of which 33 (14.2%) showed evidence of MSIs due to the presence of multiple M. tuberculosis phage types in a single culture. In the other study, Bates et al. (1976) analyzed samples from 87 different patients and identified three (3.4%) as having mixed phage-typing profiles. While the possibility that the presence of multiple M. tuberculosis phage types within the same patient could indicate an MSI, the limited discriminatory power of the method prevents definitive confirmation and cannot completely rule out intra-host microevolution.
Until recently, RFLP based on the insertion sequence IS6110 was the standard method used for comparing the genetic relatedness of M. tuberculosis isolates (Van Soolingen et al., 1993, 1995). IS6110 (sometimes referred to as IS986) belongs to the IS3 family, members of which are only present in the MTBC (Hermans et al., 1990; Thierry et al., 1990a,b). M. tuberculosis and M. bovis strains can contain 0–25 and 0–3 copies each of IS6110, respectively (Cave et al., 1991, 1994; Van Soolingen et al., 1991; Yuen et al., 1993; Fomukong et al., 1994; Brosch et al., 2000; Lok et al., 2002; Singh et al., 2004; Steensels et al., 2013). Variations in the copy number of IS6110 elements within different M. tuberculosis strains make it an attractive target for epidemio-typing isolates containing multiple copies of the insertion sequence, but not in low copy number strains. Therefore, IS6110-typing has led to the development of extensively used standardized protocols (Table 3) (Van Embden et al., 1993).
In total, 26 (21.5%) of the publications reported herein included the use of IS6110-typing methods for detecting MSIs involving MTBC members (Chaves et al., 1999; Pavlic et al., 1999; Yeh et al., 1999; Braden et al., 2001; Du Plessis et al., 2001; Richardson et al., 2002; García De Viedma et al., 2003, 2005; Allix et al., 2004; Das et al., 2004; Shamputa et al., 2004, 2006; Baldeviano-Vidalón et al., 2005; Cox et al., 2005, 2007; Van Rie et al., 2005; Van Der Zanden et al., 2007; Andrade et al., 2009; Mokrousov et al., 2009; Gardy et al., 2011; Navarro et al., 2011; Von Reyn et al., 2011; Adams et al., 2012; Cerezo et al., 2012; Huyen et al., 2012; De Almeida et al., 2019), 11 of which exclusively used IS6110-RFLP (Supplementary Table 2) and one study used IS6110-inverse polymerase chain reaction (PCR) (Mokrousov et al., 2009). All the studies employed cultures in their analysis, and IS6110-inverse PCR was primarily used as a tool to identify strains belonging to the Beijing evolutionary lineage (Mokrousov et al., 2003, 2009). The first report on MSI detection using IS6110-RFLP was by Yeh et al. (1999), who noticed multiple bands of varying intensities in a sample from a patient due to the presence of two separate strains. Another study found an MSI with two drug-susceptible isolates in an immunosuppressed patient (Pavlic et al., 1999), and similar infections with drug-susceptible and drug-resistant M. tuberculosis have also been detected by IS6110-RFLP in separate reports (Braden et al., 2001; Hofmann-Thiel et al., 2009). Overall, MSIs were identified in 0.4–100% of cases using IS6110-RFLP in these studies (Supplementary Table 2), where most reports with 100% MSI detection rates involved a single patient (Pavlic et al., 1999; Yeh et al., 1999; Andrade et al., 2009). In addition, a recent study detected MSIs in 3 out of 17 samples from patients using two probes for IS6110-RFLP instead of a single probe used conventionally (De Almeida et al., 2019).
Spacer oligotyping (spoligotyping) is a commonly used method for the simultaneous detection and identification of MTBC members (Table 3) (Kamerbeek et al., 1997). Spoligotyping exploits the nucleotide sequence diversity of clustered regularly interspaced palindromic repeats (CRISPRs) (Haft et al., 2005), which are present in many bacteria and archaea (Barrangou et al., 2007). The chromosomal locus specifically used in this assay is known as direct repeat (DR) in mycobacteria (Hermans et al., 1991). Spoligotyping is traditionally performed by amplifying the entire DR region using PCR with a pair of oligonucleotide primers, one of which is labeled with biotin to aid in the detection of products by hybridization. Membranes containing a unique set of 43 covalently bound synthetic oligonucleotide spacer sequences derived from M. tuberculosis and M. bovis Bacille Calmette-Guérin (BCG) are used in the hybridization (Kamerbeek et al., 1997), and can differentiate between MTBC isolates based on the presence or absence of spacers.
Spoligotyping has been widely used in epidemiological studies to investigate the cause of recurrent TB (defined as endogenous reactivation of an initially infecting strain or exogenous reinfection with a different strain) (Small et al., 1994; Warren et al., 2002; Van Rie et al., 2005; Andrews et al., 2008), tracking epidemics (Goyal et al., 1999; Källenius et al., 1999; Caminero et al., 2001), and investigating laboratory cross-contamination (Nivin et al., 2000). Twenty (16.5%) publications reported using spoligotyping as one of the methods for detecting MSIs, with a majority employing one (Pavlic et al., 1999; Cox et al., 2005; García De Viedma et al., 2005; Umubyeyi et al., 2007; Furphy et al., 2012; Biffa et al., 2014; Chaoui et al., 2014; Lamine-Khemiri et al., 2014; Ssengooba et al., 2015; Wang et al., 2015; Egbe et al., 2017; Baffoe-Bonnie et al., 2019) or two (García De Viedma et al., 2003; Mokrousov et al., 2009; Huyen et al., 2012; Navarro et al., 2015; Silva-Pereira et al., 2019) additional methods (Supplementary Table 2). Only three reports used spoligotyping as the sole genotyping method and reported MSIs at frequencies ranging from 11.8 to 57.1% (Andrews et al., 2008; Kamakoli et al., 2018; Guernier-Cambert et al., 2019).
One significant limitation of spoligotyping is that it can underestimate MSIs as hybridization signals from multiple strains in a sample can overlap and appear as a single pattern (Kamakoli et al., 2018). For this reason, when spoligotyping is used to investigate MSIs, subculturing is usually performed to obtain single isolated colonies for testing (García De Viedma et al., 2003; Shamputa et al., 2004; Huyen et al., 2012). The ability to detect MSIs in the latter case is dependent upon the proportion of different strains in the original sample and the number of colonies picked for analysis. To help resolve this problem, Lazzarini et al. (2012) developed a computational method that can predict if individual spoligotypes contained signatures from more than one of four major global lineages, which would indicate an MSI. In most cases, a secondary typing method like MIRU-VNTR, IS6110-RFLP or WGS may be required to verify results that may appear to contain a single spoligotyping pattern. It is worth noting that although spoligotyping can be applied directly to clinical specimens, all the studies reported herein used cultures, possibly due to the requirement of purified DNA for other methods employed by the authors (Shamputa et al., 2004, 2010; Hofmann-Thiel et al., 2009). Warren et al. (2004) used a combination of a lineage-specific PCR and spoligotyping to distinguish between M. tuberculosis strains belonging to the Beijing- and non-Beijing evolutionary lineages. They detected MSIs in 19% of samples from patients associated with retreatment in their study. In addition, WGS was used as second technique in three spoligotyping based studies (Witney et al., 2017; Baffoe-Bonnie et al., 2019; Silva-Pereira et al., 2019), one of which focused on animals. In their work, Silva-Pereira et al. (2019) detected an M. pinnipedii MSI in a South American sea lion (Otaria flavescens) using WGS, which was not suggested by in silico spoligotyping or MIRU-VNTR initially. The above-mentioned studies demonstrate the importance of using more discriminatory methods along with traditional screening techniques to ensure the detection of different strains that might be present in a single sample.
Variable number tandem repeats (VNTR) are short DNA sequences (Nakamura et al., 1987), which are dispersed throughout the genomes of many bacteria and eukaryotes (Cox and Mirkin, 1997). They vary in repeat unit length and repeat number depending on the specific organism and locus being analyzed (Supply et al., 1997). Since the repeat unit length at specific VNTR loci is known for each species, determining the numbers of repeats present at the respective loci can be used to discriminate between strains (Table 3). The use of VNTRs for typing M. tuberculosis strains was first reported in 1998 (Frothingham and Meeker-O'Connell, 1998), and since then the discrimination power has been improved by using combinations of mycobacteria-interspersed repetitive units (MIRUs) located at different loci throughout the genome (Smittipat and Palittapongarnpim, 2000; Le Flèche et al., 2002; Roring et al., 2002; Skuce et al., 2002). Initially, 12-locus MIRU-VNTR was used widely (Supply et al., 2001), but the method had some limitations in its ability to discriminate between unrelated isolates (Supply et al., 2006). To overcome this problem, the stability and resolution power of 29 MIRU-VNTR loci was evaluated using predominant global M. tuberculosis lineages, resulting in the standardization of 24-MIRU-VNTR loci for high-resolution epidemiological studies (Supply et al., 2006). In addition, the 15 most discriminatory loci of the 24 were selected for use in routine epidemiological investigations involving M. tuberculosis (Supply et al., 2006).
To generate a MIRU-VNTR profile, several genomic regions known to contain VNTRs are amplified by PCR using specific primer pairs either individually (simplex) or in multiples (multiplex). In this way, the number of repeats at each VNTR locus can be determined using different DNA sizing techniques for comparing isolates. It was found that 50 studies used VNTR-typing to detect M. tuberculosis MSIs, where 28 studies used it as the sole discriminatory method for this purpose (Allix et al., 2004; Shamputa et al., 2004; Umubyeyi et al., 2007; Fang et al., 2008; Hofmann-Thiel et al., 2009; Mokrousov et al., 2009; Stavrum et al., 2009; Dickman et al., 2010; Mulenga et al., 2010; Cohen et al., 2011; Gardy et al., 2011; Navarro et al., 2011, 2015; Cerezo et al., 2012; Furphy et al., 2012; Huyen et al., 2012; Hingley-Wilson et al., 2013; Muwonge et al., 2013; Peng et al., 2013; Biffa et al., 2014; Chaoui et al., 2014; Lamine-Khemiri et al., 2014; Zetola et al., 2014; Barletta et al., 2015; Mei et al., 2015; Pang et al., 2015; Shin et al., 2015, 2018; Ssengooba et al., 2015; Streit et al., 2015; Wang et al., 2015; Zheng et al., 2015; Antusheva et al., 2016; Hajimiri et al., 2016; Hu et al., 2016; Egbe et al., 2017; Farmanfarmaei et al., 2017; Ghielmetti et al., 2017; Kamakoli et al., 2017, 2019, 2020a,b; Khosravi et al., 2017; Kontsevaya et al., 2017; Nathavitharana et al., 2017; Sadegh et al., 2017; Silva-Pereira et al., 2019; Abascal et al., 2020; Baik et al., 2020; Sichewo et al., 2020). In addition, 24-locus MIRU-VNTR was the most commonly used method for detecting M. tuberculosis MSIs, and nearly all (47/50, 94.0%) of the reports used some form of culturing for analysis. Comparatively, MIRU-VNTR detected more MSIs than any other method based in the current review (Table 2). One reason for this could be the use of PCR amplification during MIRU-VNTR, which increases the sensitivity and detection power of the method (Shamputa et al., 2006), especially in instances where different strains are not proportionally present in a single sample.
Among other PCR-based methods used to detect MSIs, the majority were focused on differentiating between M. tuberculosis strains belonging to the Beijing and non-Beijing evolutionary lineages (Warren et al., 2004; Van Rie et al., 2005; Huang et al., 2010; Wang et al., 2011; Mustafa et al., 2016). Of the 121 studies we examined, 13 (10.7%) used region-specific PCR methods and detected MSIs in 3.2–100% of samples (Supplementary Table 2) (Theisen et al., 1995; Lourenço et al., 2000; García De Viedma et al., 2003; Warren et al., 2004; Baldeviano-Vidalón et al., 2005; Van Rie et al., 2005; Huang et al., 2010; Mallard et al., 2010; Wang et al., 2011; Hanekom et al., 2013; Peng et al., 2013; Müller et al., 2014; Mustafa et al., 2016). For example, Warren et al. (2004) detected MSIs in 35 (18.8%) of the 186 sputum cultures from TB patients tested during an epidemiologic study in South Africa. They reported that MSIs were more often associated with retreatment (23%) vs. new cases (17%), and the sensitivity and specificity of their method was comparable to IS6110-RFLP and spoligotyping (Warren et al., 2004). Using the same method, Van Rie et al. (2005) also detected one case of MSI with drug-susceptible and drug-resistant M. tuberculosis isolates. While the method developed by Warren et al. (2004) used simplex PCR, Huang et al. (2010) utilized multiplex PCR to detect MSIs caused by Beijing and non-Beijing lineage strains in 11.3% of the 185 sputum samples from patients without any prior history of TB treatment. Another group used quantitative PCR to detect MSIs based on the presence of both Beijing and non-Beijing lineages in 3% of TB patients <25 years in age using clinical specimens and cultures (Wang et al., 2011), whereas M. tuberculosis isolates belonging to the two lineages were also detected together in 14.7% cases by Mustafa et al. (2016). In contrast, a Latin American and Mediterranean (LAM) and non-LAM lineage-based PCR found MSIs in 4 out of 160 (2.5%) culture-positive sputa analyzed (Mallard et al., 2010), suggesting that such methods are useful in identifying MSIs under settings where the occurrence of M. tuberculosis isolates from mixed lineages is high. Therefore, by using an in-house PCR to identify isolates from the Beijing, Haarlem, S-family, and LAM evolutionary lineages, Hanekom et al. (2013) were able to detect MSIs in 31 (15%) of the 206 samples analyzed in their study.
M. tuberculosis MSIs have also been reported in patients with discordant drug susceptibility profiles on more than one occasion. Isolates from 10 such TB patients out of 89 (11.2%) were confirmed to have MSIs using Beijing lineage-specific PCR (Chen et al., 2007) and 16-locus MIRU-VNTR (Peng et al., 2013). Other techniques such as linker PCR (Theisen et al., 1995) and gyrA PCR/sequencing (Sreevatsan et al., 1997; Müller et al., 2014) have also detected M. tuberculosis MSIs in cases involving discordant drug susceptibility profiles and also in archaeological samples. In addition, two studies employed double repetitive element PCR (Lourenço et al., 2000; Baldeviano-Vidalón et al., 2005) based on the IS6110 and a GC-rich repetitive sequence described by Friedman et al. (1995). Baldeviano-Vidalón et al. (2005) also observed multiple discrepancies in drug susceptibility testing results among follow up samples from patients, which were attributed to MSIs based on IS6110-RFLP analysis. Such studies emphasize the importance of considering MSIs during TB drug susceptibility testing and while devising appropriate treatment regimens.
Methods based on WGS provide the ability to examine strain diversity at very high resolution, which cannot be achieved by other techniques such as RFLP or MIRU-VNTR. Heterogeneous SNPs are predominantly used for strain discrimination, and the presence of many different SNPs in isolates from a single sample is suggestive of MSIs (Sobkowiak et al., 2018). While the concept is simple–a SNP is confirmed within several sequencing reads used to assemble the locus being examined (Table 3), the technical criteria for identifying bona fide SNPs varies between studies. Factors that can affect SNP detection include the quality and depth of sequencing, experimental design, sample preparation and pathogen species, to name a few (Hatherell et al., 2016). Some reports require that the frequency of the alternative base at a specific locus be found from anywhere between 5 and 30% or more of the reads for SNP calling (Bryant et al., 2013a; Guerra-Assuncaõ et al., 2015; Dippenaar et al., 2019), or even just two reads in some deep sequencing studies (Gan et al., 2016). Some studies also include threshold nucleotide base quality scores to minimize artifacts (Bryant et al., 2013a; Gan et al., 2016; Nimmo et al., 2020). Additionally, several loci with heterogeneous bases must be identified between isolates to qualify them as MSIs. However, the minimum number of SNPs used to qualify the presence of an MSI using isolates also varies, as anywhere over 16 to 80 have been used for the purpose depending on the sequencing technology (Bryant et al., 2013a; Gan et al., 2016). In addition, WGS analysis cannot be performed directly on clinical samples in most cases because the genomic complexity of the sample limits the confidence at which SNPs are called. Therefore, WGS often requires axenic cultures for strain typing and MSI identification (Van Den Berg et al., 2005; Döpfer et al., 2008; Davidson et al., 2016).
Of the 97 studies identified in Covidence, eight (8.2%) primarily used WGS to detect MSIs in humans (Bryant et al., 2013a; Gan et al., 2016; Witney et al., 2017; Baffoe-Bonnie et al., 2019; Dippenaar et al., 2019; O'Donnell et al., 2019; Anyansi et al., 2020; Wollenberg et al., 2020), while three others (3.1%) used the method to detect MSIs in various animal species (Davidson et al., 2016; Pfeiffer et al., 2017; Silva-Pereira et al., 2019), only one of which focused on a MTBC bacteria (Silva-Pereira et al., 2019). Secondary searches found four more human studies on WGS and mycobacterial MSIs (Chan et al., 2013; Kay et al., 2015; Dheda et al., 2017; Nimmo et al., 2020), in addition to reports where the method was used to confirm such infection that were initially identified using other means (Supplementary Table 2). For example, six isolates from 47 paired patient samples taken before and after treatment during the REMoxTB clinical trial were initially classified as relapses or re-infections by MIRU-VNTR but were later determined to be MSIs by WGS analysis (Bryant et al., 2013a). A follow-up study re-examined the same data using QuantTB (Anyansi et al., 2020), a tool developed to identify MSIs through the iterative comparison of SNPs and suggested that only four of the six MSI cases could be classified as such. O'Donnell et al. (2019) used isolates from a patient where drug susceptibility testing alluded toward an MSI involving susceptible and resistant M. tuberculosis, which was confirmed using WGS. They showed that the patient was initially infected with a drug-susceptible strain followed by an extensively drug-resistant M. tuberculosis isolate, which was selected for during antibiotic therapy. The high resolution of WGS underscores its importance in strain typing for devising individualized treatments for TB infections, although its widespread use may be limited in many high TB burden settings due to insufficient resources or technical capabilities. The ability to use traditional culture based drug susceptibility testing methods have limitations for many slow growing pathogenic mycobacteria, but genomics based technologies are more rapid and allow for the detection of resistance based on the presence of conferring mutations (Nicol and Cox, 2019). While PCR methods targeting specific genes can detect important drug resistance mutations for early diagnosis, WGS can additionally infer potential resistance, allowing for individualized treatment regimens (Nicol and Cox, 2019).
The use of WGS also enhances the detection of potential MSIs. For example, while examining pre-and post-treatment isolates from a TB patient, Witney et al. (2017) noticed 57 SNP differences between two of them, indicating a re-infection by a second strain. More detailed analysis of the WGS data indicated a potential MSI in the pre-treatment sample at a 3:1 genotypic ratio by two strains, where the minor genotype was closely related to the post-treatment isolate. This suggested that recurrent disease was caused by a relapse, where one of the two strains from the MSI was eliminated during the initial course of therapy. Interestingly, 24-locus MIRU-VNTR typing did not detect the genotype of the post-treatment isolate in the pre-treatment sample, which is intriguing, as the method was previously shown to detect MSIs in proportions as low as 1:99 (García De Viedma et al., 2005). Such reports suggest that many MSIs might have gone undetected due to technical limitations and could have potentially affected disease outcomes.
Although more TB MSIs are now being reported, evidence from archaeological studies indicate that the phenomenon has been around for a long time. Through metagenomics analysis of ancient DNA, one study identified an MSI (difference of 398 SNPs) within an eighteenth-century Hungarian mummy (Chan et al., 2013). A follow up study by a related group reported five MSIs in eight mummified bodies examined from the same archaeological site using similar metagenomics-based methods (Kay et al., 2015). In addition, a separate study employed gyrA PCR to successfully identify an MSI using ancient DNA from a variety of archaeological samples across Britain and France (Müller et al., 2014). The detection of MSIs using ancient DNA and complex samples is intriguing. Such reports also provide precedence for using metagenomics and other technologies to examine the prevalence and impact of such infections in future prospective and retrospective studies.
Non-tuberculous and Other Mycobacteria
Our review showed that MSIs involving NTM have not been investigated to the same extent as compared to those caused by M. tuberculosis (Table 2). Amongst the 121 mycobacterial studies identified, 26 (21.5%) examined NTM, of which 17 (14.0%) and eight (6.6%) found MSIs in humans and animals, respectively, whereas one report identified MSIs in both human (Arbeit et al., 1993; Slutsky et al., 1994; Von Reyn et al., 1995; Devallois and Rastogi, 1997; Picardeau et al., 1997; Wallace et al., 1998; Legrand et al., 2000a,b; Oliveira et al., 2000; Saad et al., 2000; Dvorska et al., 2002; Panunto et al., 2003; Ohkusu et al., 2004; De Sequeira et al., 2005; Fujita et al., 2014; García-Pedrazuela et al., 2015; Kimizuka et al., 2019) and animal (Dvorska et al., 2007; Shitaye et al., 2008; Furphy et al., 2012; Gerritsmann et al., 2014; Johansen et al., 2014; Gioffré et al., 2015; Podder et al., 2015; Davidson et al., 2016; Pfeiffer et al., 2017) populations simultaneously (Pate et al., 2008). Many early studies used pulsed field gel electrophoresis (PFGE) to identify MSIs in 14.3–100% of patients infected with MAC bacteria (Arbeit et al., 1993; Slutsky et al., 1994; Von Reyn et al., 1995; Wallace et al., 1998; Ohkusu et al., 2004). PFGE was also used as the sole method to identify MSIs associated with other NTMs. For example, Legrand et al. (2000a) found 33.3% M. simiae MSI prevalence (3 of 9 hosts) in their study involving AIDS patients. In addition, PFGE was used in combination with other methods in studies on NTM MSIs. A report by Picardeau et al. (1997), detected 3 M. avium MSIs while examining 93 samples from AIDS patients using simple double repetitive element PCR (MaDRE-PCR), which amplifies a region of the M. avium chromosome between IS1245 and IS1311 (Picardeau and Véronique, 1996). They confirmed these results using IS1245-RFLP, which showed the presence of multiple low-intensity bands in the same sample. Pulsed field gel electrophoresis was also able to pick up multiple banding patterns, including in some samples from other patients (Picardeau et al., 1997), but the criteria for attributing them to different strains (≥3 differences) was not surpassed (Tenover et al., 1995). A similar study on 31 AIDS patients from the Caribbean islands initially identified three potential polyclonal M. avium infections based on IS1245-RFLP, but PFGE analysis showed that two isolates had identical banding patterns (Legrand et al., 2000b). Therefore, results from PFGE analysis do not corroborate those obtained by other methods on multiple occasions, which might allude to differences in their discriminatory powers.
IS1245 and IS1311 have also been used as general fingerprinting targets to identify an MSI (25%) involving MAC members from 25 patients (Devallois and Rastogi, 1997). In a separate study, Oliveira et al. (2000) used RFLP analysis of hsp65 PCR products, IS1245 and IS1311, respectively, to detect an M. avium MSI. Though the presence of IS1245-RFLP may be supplemented using additional methods, studies have shown it alone is capable of identifying 2.6–100% of NTM MSIs in samples from human subjects, some with HIV-AIDS (Supplementary Table 2) (Picardeau et al., 1997; Saad et al., 2000; Dvorska et al., 2002; Panunto et al., 2003; Pate et al., 2008). In addition, a study examined 41 samples from 14 AIDS patients using MaDRE-PCR, which found four cases with multiple banding patterns, though re-evaluation using IS1245-RFLP only confirmed two as M. avium MSIs (De Sequeira et al., 2005). Based on the reports mentioned above, the feasibility of using IS1245-RFLP as a screening method for detecting NTM MSIs warrants further evaluation.
Kimizuka et al. (2019) have also used a variety of VNTR loci, including 16 M. avium tandem repeats (MATR) and 5 Higashi Nagoya tandem repeats (HNTR) to identify M. avium MSIs in samples from nine patients (13.8%) out of 65 examined. Another study examined samples from 120 patients with pulmonary MAC infections (94 M. avium and 26 M. intracellulare) (Fujita et al., 2014). MIRU-VNTR analysis using 15 loci for M. avium and 16 loci for M. intracellulare successfully identified 20 and 7 cases of M. avium and M. intracellulare MSIs, respectively. Other methods that have detected NTM MSIs in humans include random amplified polymorphic DNA (RAPD) analysis (Zhang et al., 1997). García-Pedrazuela et al. (2015) used the method and found MSIs involving several species including M. abscessus, Mycobacterium chelonae, Mycobacterium fortuitum and Mycobacterium mucogenicum while examining 64 isolates from Spanish patients.
Systematic screening using the Covidence platform also identified some reports on NTM MSIs in animals, including a few involving M. avium subsp. paratuberculosis (MAP). Gerritsmann et al. (2014) examined 39 MAP containing samples from a variety wild and domestic ruminant species, five of which were classified as MSIs using 8-locus MIRU-VNTR by them (Supplementary Table 2). In another study, Gioffré et al. (2015) used 8-locus MIRU-VNTR to identify a MAP MSI from a group to 97 cattle, sheep and goats based on differences in two loci. MAP MSIs have also been identified using multi-locus short sequence repeat (MLSSR or SSR) typing (Podder et al., 2015), which examines sequences that vary between isolates and allows for their discrimination (Amonsin et al., 2004). In addition, the use of DNA detection and sizing techniques such as fragment analysis of labeled PCR products further improves the resolution or SSR typing (Oakey et al., 2014; Podder et al., 2015). Using this strategy, Podder et al. (2015) identified MAP MSIs in all 18 animals from their study, which was subsequently confirmed by WGS analysis using some of their isolates (Davidson et al., 2016). MAP strains from the same animal had significantly different SNPs at high frequencies, which ruled out microevolution based evolutionary rates (Bryant et al., 2016).
In addition to MAP, there were reports on MSIs due to other NTM. M. avium subsp. avium MSIs were identified in domestic chickens using IS901-RFLP, where multiple banding patterns were detected in 7 of the 16 (43.8%) tissue samples tested (Shitaye et al., 2008). Eight-locus VNTR was used to detect M. avium subsp. hominissuis MSIs in pigs using isolates from multiple organs of a single animal (Johansen et al., 2014). Another study that previously reported a single M. avium subsp. hominissuis MSI in a human (out of 26 patients, 3.8%) using IS1245-RFLP also detected similar infections in 33 pigs (13.5%) (Pate et al., 2008). WGS was also used to detect non-MAP MSIs from 113 birds by Pfeiffer et al. (2017). They reported 12 cases of MSIs (2 involving M. avium and 10 involving M. genavense) based on differences in at least 12 SNPs as suggested by Walker et al. (2013). Therefore, NTM MSIs seem to occur in a variety of animal species, but studies examining their prevalence and impact are few and far between.
Our review did not reveal the occurrence of MSIs within leprosy causing mycobacteria. However, using artificially co-infected armadillos, Shin et al. (2015) demonstrated that distinct strains of M. leprae could simultaneously exist in the same host. They noted that the in vivo growth rate of the non-armadillo strain was significantly higher in the absence of competing strains, suggesting that pathological variations exist between the different strain types. Due to the challenges associated with culturing M. leprae (Scollard et al., 2006), the same experimental approaches used to study MTBC and NTM members might not be feasible, thereby limiting the potential for detecting M. leprae MSIs. Therefore, with the advent of more sensitive and powerful discriminatory methods, it is possible that the use of culture-independent techniques such as metagenomics will help to shed light on MSIs involving M. leprae in the future.
Challenges Faced in Defining MSIs
Many questions are raised regarding the use of standard strain typing methods for delineating true MSIs from microevolution. When using IS6110-RFLP and MIRU-VNTR on M. tuberculosis isolates derived from the same host, MSIs are identified based on differences in specific DNA/PCR fragment profiles (Table 4). IS6110-RFLP profiles that differ by at least 2–3 bands indicate the occurrence of concomitant infection by distinct strains (Shamputa et al., 2004, 2006; Marx et al., 2014). Conversely, minor variations, i.e., anything <2 discriminatory bands, is considered as microevolution. In the case of MIRU-VNTR, MSIs are predicted based on heterogeneity at two or more loci (Shamputa et al., 2004, 2006; Cohen et al., 2011; Maghradze et al., 2019), whereas differences in a single locus suggests microevolution (Shamputa et al., 2004, 2006; Maghradze et al., 2019). Therefore, allelic diversity at more than one locus is a criterion for differentiating microevolution from true MSI when using MIRU-VNTR.
Table 4.
Thresholds for distinguishing MSIs from microevolution events as per reviewed studies.
| Yeara | Methodb | Microevolutionc | MSIc | References |
|---|---|---|---|---|
| 2004 | IS6110 RFLP | 1-3 different bands | >3 different bands | Shamputa et al., 2004 |
| MIRU-VNTR | Double allele at 1 locus | Double alleles at ≥2 Loci | ||
| 2011 | MIRU-VNTR | Double allele at 1 locus | Double alleles at ≥2 Loci | Cohen et al., 2011 |
| 2013 | WGS | <80 heterogeneous SNPs | >80 heterogeneous SNPs | Bryant et al., 2013b |
| 2014 | IS6110 RFLP | 1-2 different bands | >2 different bands | Marx et al., 2014 |
| 2015 | WGS | ≤140 heterogeneous SNPs | >140 heterogeneous SNPs | Guerra-Assuncaõ et al., 2015 |
| 2019 | MIRU-VNTR | Double allele at 1 locus | Double alleles at ≥2 Loci | Maghradze et al., 2019 |
| 2019 | WGS | 0-5 heterogeneous SNPs | 757-833 heterogeneous SNPs | Dippenaar et al., 2019 |
Year of publication in chronological order.
Details regarding specific methods are included in the main text and Supplementary Table 2.
Thresholds or criteria used by the respective methods to differentiate between the two forms of infection.
Defining MSIs using WGS based methods also has inherent challenges. However, the high discriminatory power of WGS allows for more definitive explanations for TB recurrence post-treatment (Bryant et al., 2013a; Guerra-Assuncaõ et al., 2015; Dippenaar et al., 2019). The classification of infections based on SNP differences between isolates from a single patient varies, where 5–10 suggest the occurrence of a relapse and anything over 100, a re-infection (Bryant et al., 2013a; Guerra-Assuncaõ et al., 2015; Dippenaar et al., 2019) (Table 4). These differences in SNP numbers are apparently set by taking into account the calculated evolutionary rate of M. tuberculosis, which ranges from 0.3 to 0.5 SNPs per genome per year, and limits the number of SNPs that can accumulate within strains in a defined period of time (Bryant et al., 2013b; Ford et al., 2013; Roetzer et al., 2013; Walker et al., 2013). Varying SNP thresholds have been used to define MSIs, where Bryant et al. (2013a) called them as such if isolates derived from the same patient had 80 or more different SNPs based on manual inspection. In comparison, Guerra-Assuncaõ et al. (2015) used a threshold of 140 heterogeneous SNPs to define MSIs based on an empirical cutoff generated during their data analysis. Furthermore, Dippenaar et al. (2019) did not define a specific threshold, as the numbers of heterogeneous SNPs in their study were either limited (1–2, classified as microevolution), or rather numerous (757–883, classified as MSI). These differences in SNP numbers used to identify M. tuberculosis MSIs shows that the field requires some form of standardization so that results can be compared between studies. It is possible that the lack of a clear definition for MSIs using WGS could hinder the identification of some reports for inclusion in this review. Although this is unlikely as only a few studies have used WGS technology for such purposes to date, and most of them were manually screened for their relevance.
Considering that the criteria necessary for identifying different types of M. tuberculosis polyclonal infections using IS6110-RFLP and MIRU-VNTR are well-defined, several studies were also found in the current review that reported on microevolution (Martín et al., 2007; Streit et al., 2015; Cohen et al., 2016; Jajou et al., 2018). The classification was not evident in some other reports that used IS6110-RFLP or MIRU-VNTR, due to results potentially indicating either an MSI or a microevolution event depending on the applied criteria (Al-Hajoj et al., 2010; Alves et al., 2011), or due to WGS not having a well-defined SNP threshold (Liu et al., 2015). While microevolution may initially appear to have little significance when compared to MSIs in terms of virulence and pathogenicity, one study found that the presence of highly evolvable repeats near genes (VNTR52, QUB26, or MIRU10/27) can influence gene expression in different M. tuberculosis isolates (Pérez-Lago et al., 2013). In addition, microevolution has obvious implication on the emergence of drug resistance due to selective pressures applied during treatment regimens (Fonseca et al., 2015).
Microevolution derived infections may not be as distinct as MSIs, but different clonal variants can still spread in unique patterns. While a strain undergoes a genetic drift, some of the progeny might spread both within and between hosts. For example, WGS identified separate M. tuberculosis clonal variants derived from within host microevolution at both respiratory and extrapulmonary sites in a patient (Pérez-Lago et al., 2014; Ssengooba et al., 2016). A study by Buff et al. (2009) examined several TB cases where IS6110-RFLP and spoligotyping of isolates from community transmission events showed identical patterns but exhibited variations at a single locus during 12-locus MIRU-VNTR analysis. It was shown that M. tuberculosis isolates from the source patient also displayed varying MIRU-VNTR profiles that matched the secondary patients, suggesting that different clonal variants were transmitted individually.
In addition to the MSIs described in our review, there is also evidence for the microevolution of mycobacterial pathogens in animals. For example, microevolution events and MSIs involving M. avium subsp. hominissuis in bongo antelopes were identified using 8-locus MIRU-VNTR and IS1245-RFLP (Moravkova et al., 2013). Another study used 8-locus MIRU-VNTR to examine M. caprae from 55 different animal hosts (including goats, cattle, sheep and wild boar) in Portugal and identified microevolution derived infections in 12 of them (Reis et al., 2020). Therefore, mycobacterial microevolution also occurs in animals, which is expected given the current state of knowledge regarding such infections in humans.
Conclusion
As summarized in this systematic review, a variety of terminologies have been used for describing MSIs, some of which overlap with microevolution. Findings show that MSIs exist among many different mycobacteria, although the majority of studies have been conducted in humans and predominantly focus on M. tuberculosis. In addition, most studies used VNTR based methods, though more recent reports involved WGS. This change in methodology might represent an overall shift as newer technologies are developed and used more widely for strain typing. With methods suitable for large-scale screening of genetic variations by the massively-parallel sequencing approaches, the potential for the accurate identification of MSIs is now accessible at lower cost. Methods based on WGS offer unprecedented resolution, but appear to lack uniform SNP thresholds (Bryant et al., 2013a; Guerra-Assuncaõ et al., 2015; Dippenaar et al., 2019), sometimes making it ambiguous to differentiate between a true MSI or microevolution event. Therefore, there is scope for further standardizing WGS criteria for discriminating MSIs from genomic drift or technical aberrations, but as of now a universal definition for calling mycobacterial MSIs using the technology is lacking. Consideration of both microevolution and MSI has potential implications for developing personalized medical treatments for diseases such as TB. A select number of publications examined the historical context of M. tuberculosis using ancient DNA (Chan et al., 2013; Müller et al., 2014; Kay et al., 2015), suggesting that MSIs have likely been around for a long time. Furthermore, reports on discrepant drug susceptibilities between one or more M. tuberculosis isolates from the same host during a single disease episode underscores the importance of considering MSIs when managing mycobacterial infections. Reports have shown that TB patients with MSIs associated with heteroresistance are at a higher risk of poor treatment outcomes (Zetola et al., 2014; Cohen et al., 2016; Kamakoli et al., 2017; Shin et al., 2018). Therefore, additional strain typing is recommended under certain incidences where heteroresistance is detected, to determine if it is caused due to MSIs or microevolution. It was also noted that studies on NTM MSIs in humans and animals are limited, but such infections are found across many species. Given the importance of NTM in causing opportunistic and nosocomial infections in humans, and diseases in farmed animal, the prevalence and impact of MSIs caused by this large and important group of mycobacteria warrants further investigation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
IS, AB, and KT developed the framework for the review. AG formulated the search strategy used to identify publications, with some input on key terms from AB, IS, and KT. AB and IS performed primary screening in Covidence, with KT resolving any conflicts on paper inclusion. Data extraction and analysis was conducted by AB and IS. AB, IS, and KT performed writing and primary editing with technical and editorial input from NB. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank John Reynolds, Librarian Assistant Professor, University of Miami Miller School of Medicine for peer reviewing the search using the Peer Review of Electronic Search Strategies (PRESS) guidelines.
Footnotes
Funding. Work on mycobacterial MSIs at Memorial University of Newfoundland (MUN) was supported by a grant from the Dairy Farmers of Canada (DFC: ASC-07) to KT and NB. The Natural Sciences and Engineering Research Council of Canada (NSERC: 2018–05949) also funds research on Actinobacteria in KTs laboratory, whereas DFC and MUN provided a graduate student stipend to AB.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2020.600692/full#supplementary-material
References
- Abascal E., Herranz M., Acosta F., Agapito J., Cabibbe A. M., Monteserin J., et al. (2020). Screening of inmates transferred to Spain reveals a Peruvian prison as a reservoir of persistent Mycobacterium tuberculosis MDR strains and mixed infections. Sci. Rep. 10, 1–8. 10.1038/s41598-020-59373-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adami A. J., Cervantes J. L. (2015). The microbiome at the pulmonary alveolar niche and its role in Mycobacterium tuberculosis infection. Tuberculosis 95, 651–658. 10.1016/j.tube.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams L. V., Kreiswirth B. N., Arbeit R. D., Soini H., Mtei L., Matee M., et al. (2012). Molecular epidemiology of HIV-associated tuberculosis in Dar es Salaam, Tanzania: strain predominance, clustering, and polyclonal disease. J. Clin. Microbiol. 50, 2645–2650. 10.1128/JCM.00624-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adekambi T., Sassi M., Van Ingen J., Drancourt M. (2017). Reinstating Mycobacterium massiliense and Mycobacterium bolletii as species of the Mycobacterium abscessus complex. Int. J. Syst. Evol. Microbiol. 67, 2726–2730. 10.1099/ijsem.0.002011 [DOI] [PubMed] [Google Scholar]
- Ahmed I., Jabeen K., Hasan R. (2013). Identification of non-tuberculous mycobacteria isolated from clinical specimens at a tertiary care hospital: a cross-sectional study. BMC Infect. Dis. 13:493. 10.1186/1471-2334-13-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajoj S. A. M., Akkerman O., Parwati I., Al-Gamdi S., Rahim Z., Van Soolingen D., et al. (2010). Microevolution of Mycobacterium tuberculosis in a tuberculosis patient. J. Clin. Microbiol. 48, 3813–3816. 10.1128/JCM.00556-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allix C., Supply P., Fauville-Dufaux M. (2004). Utility of fast mycobacterial interspersed repetitive unit-variable number tandem repeat genotyping in clinical mycobacteriological analysis. Clin. Infect. Dis. 39, 783–789. 10.1086/423383 [DOI] [PubMed] [Google Scholar]
- Alves S. L., Metzker F. S., de Araújo-Filho J. A., Junqueira-Kipnis P., Kipnis A. (2011). Clinical data and molecular analysis of Mycobacterium tuberculosis isolates from drug-resistant tuberculosis patients in Goiás, Brazil. Mem. Inst. Oswaldo Cruz 106, 655–661. 10.1590/S0074-02762011000600003 [DOI] [PubMed] [Google Scholar]
- Amonsin A., Li L. L., Zhang Q., Bannantine J. P., Motiwala A. S., Sreevatsan S., et al. (2004). Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42, 1694–1702. 10.1128/JCM.42.4.1694-1702.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade M. K. N., Machado S. M. A., Leite M. L., Saad M. H. F. (2009). Phenotypic and genotypic variant of MDR-Mycobacterium tuberculosis multiple isolates in the same tuberculosis episode, Rio de Janeiro, Brazil. Brazilian J. Med. Biol. Res. 42, 433–437. 10.1590/S0100-879X2009000500006 [DOI] [PubMed] [Google Scholar]
- Andrews J. R. R., Gandhi N. R. R., Moodley P., Shah N. S. S., Bohlken L., Moll A. P. P., et al. (2008). Exogenous reinfection as a cause of multidrug-resistant and extensively drug-resistant tuberculosis in rural South Africa. J. Infect. Dis. 198, 1582–1589. 10.1086/592991 [DOI] [PubMed] [Google Scholar]
- Antusheva E., Mironuk O., Tarasova I., Eliseev P., Plusnina G., Ridell M., et al. (2016). Outbreak of tuberculosis in a closed setting: views on transmission based on results from molecular and conventional methods. J. Hosp. Infect. 93, 187–190. 10.1016/j.jhin.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Anyansi C., Keo A., Walker B. J., Straub T. J., Manson A. L., Earl A. M., et al. (2020). QuantTB- A method to classify mixed Mycobacterium tuberculosis infections within whole genome sequencing data. BMC Genomics 21:80 10.1186/s12864-020-6486-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeit R. D., Slutsky A., Barber T. W., Maslow J. N., Niemczyk S., Falkinham J. O., et al. (1993). Genetic diversity among strains of Mycobacterium avium causing monoclonal and polyclonal bacteremia in patients with AIDS. J. Infect. Dis. 167, 1384–1390. 10.1093/infdis/167.6.1384 [DOI] [PubMed] [Google Scholar]
- Atkins B. L., Gottlieb T. (2014). Skin and soft tissue infections caused by nontuberculous mycobacteria. Curr. Opin. Infect. Dis. 27, 137–145. 10.1097/QCO.0000000000000041 [DOI] [PubMed] [Google Scholar]
- Baffoe-Bonnie A., Houpt E. R., Turner L., Dodge D., Heysell S. K. (2019). Drug-susceptible and multidrug-resistant Mycobacterium tuberculosis in a single patient. Emerg. Infect. Dis. 25, 2120–2121. 10.3201/eid2511.180638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik Y., Modongo C., Moonan P. K., Click E. S., Tobias J. L., Boyd R., et al. (2020). Possible transmission mechanisms of mixed mycobacterium tuberculosis infection in high HIV prevalence country, Botswana. Emerg. Infect. Dis. 26, 953–960. 10.3201/eid2605.191638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldeviano-Vidalón G. C., Quispe-Torres N., Bonilla-Asalde C., Gastiaburú-Rodriguez D., Pro-Cuba J. E., Llanos-Zavalaga F. (2005). Multiple infection with resistant and sensitive M. tuberculosis strains during treatment of pulmonary tuberculosis patients. Int. J. Tuberc. Lung Dis. 9, 1155–1160. [PubMed] [Google Scholar]
- Barletta F., Otero L., De Jong B. C., Iwamoto T., Arikawa K., Van Der Stuyft P., et al. (2015). Predominant Mycobacterium tuberculosis families and high rates of recent transmission among new cases are not associated with primary multidrug resistance in Lima, Peru. J. Clin. Microbiol. 53, 1854–1863. 10.1128/JCM.03585-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Bates J. H., Stead W. W., Rado T. A. (1976). Phage type of tubercle bacilli isolated from patients with two or more sites of organ involvement. Am. Rev. Respir. Dis. 114, 353–358. [DOI] [PubMed] [Google Scholar]
- Behr M. A., Kapur V. (2008). The evidence for Mycobacterium paratuberculosis in crohn's disease. Curr. Opin. Gastroenterol. 24, 17–21. 10.1097/MOG.0b013e3282f1dcc4 [DOI] [PubMed] [Google Scholar]
- Biet F., Boschiroli M. L. (2014). Non-tuberculous mycobacterial infections of veterinary relevance. Res. Vet. Sci. 97, S69–S77. 10.1016/j.rvsc.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Biffa D., Johansen T. B., Godfroid J., Muwonge A., Skjerve E., Djønne B. (2014). Multi-locus variable-number tandem repeat analysis (MLVA) reveals heterogeneity of Mycobacterium bovis strains and multiple genotype infections of cattle in Ethiopia. Infect. Genet. Evol. 23, 13–19. 10.1016/j.meegid.2014.01.021 [DOI] [PubMed] [Google Scholar]
- Böttger E. C. (1994). Mycobacterium genavense: an emerging pathogen. Eur. J. Clin. Microbiol. Infect. Dis. 13, 932–936. 10.1007/BF02111494 [DOI] [PubMed] [Google Scholar]
- Braden C. R. R., Morlock G. P. P., Woodley C. L. L., Johnson K. R. R., Colombel A. C. C., Cave M. D. D., et al. (2001). Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin. Infect. Dis. 33, e42–e47. 10.1086/322635 [DOI] [PubMed] [Google Scholar]
- Brosch R. G., Gordon S. V., Eiglmeier K., Garnier T., Tekaia F., Yeramian E., et al. (2000). Genomics, biology and evolution of the Mycobacterium tuberculosis complex, in Molecular Genetics of Mycobacteria, eds Hatful G. F., Jacobs W. R., Jr (Washington DC: American Society for Microbiology Press; ), 19–36. [Google Scholar]
- Bryant J. M., Harris S. R., Parkhill J., Dawson R., Diacon A. H., van Helden P., et al. (2013a). Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir. Med. 1, 786–792. 10.1016/S2213-2600(13)70231-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J. M., Schürch A. C., van Deutekom H., Harris S. R., de Beer J. L., de Jager V., et al. (2013b). Inferring patient to patient transmission of Mycobacterium tuberculosis from whole genome sequencing data. BMC Infect. Dis. 13:110. 10.1186/1471-2334-13-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant J. M., Thibault V. C., Smith D. G. E., McLuckie J., Heron I., Sevilla I. A., et al. (2016). Phylogenomic exploration of the relationships between strains of Mycobacterium avium subspecies paratuberculosis. BMC Genomics 17:79. 10.1186/s12864-015-2234-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buff A. M., Sosa L. E., Hoopes A. J., Buxton-Morris D., Condren T. B., Hadler J. L., et al. (2009). Two tuberculosis genotyping clusters, one preventable outbreak. Public Health Rep. 124, 490–494. 10.1177/003335490912400405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminero J. A., Pena M. J., Campos-Herrero M. I., Rodríguez J. C., García I., Cabrera P., et al. (2001). Epidemiological evidence of the spread of a Mycobacterium tuberculosis strain of the Beijing genotype on Gran Canaria Island. Am. J. Respir. Crit. Care Med. 164, 1165–1170. 10.1164/ajrccm.164.7.2101031 [DOI] [PubMed] [Google Scholar]
- Cave M. D., Eisenach K. D., McDermott P. F., Bates J. H., Crawford J. T. (1991). IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol. Cell. Probes 5, 73–80. 10.1016/0890-8508(91)90040-Q [DOI] [PubMed] [Google Scholar]
- Cave M. D., Eisenach K. D., Templeton G., Salfinger M., Mazurek G., Bates J. H., et al. (1994). Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 32, 262–266. 10.1128/JCM.32.1.262-266.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo I., Jiménez Y., Hernandez J., Zozio T., Murcia M. I., Rastogi N. (2012). A first insight on the population structure of Mycobacterium tuberculosis complex as studied by spoligotyping and MIRU-VNTRs in Bogotá, Colombia. Infect. Genet. Evol. 12, 657–663. 10.1016/j.meegid.2011.07.006 [DOI] [PubMed] [Google Scholar]
- Chan J. Z. M. Z.-M. Z. M., Sergeant M. J., Lee O. Y. C. Y.-C., Minnikin D. E., Besra G. S., Pap I., et al. (2013). Metagenomic analysis of tuberculosis in a mummy. N. Engl. J. Med. 369, 289–290. 10.1056/NEJMc1302295 [DOI] [PubMed] [Google Scholar]
- Chaoui I., Zozio T., Lahlou O., Sabouni R., Abid M., El Aouad R., et al. (2014). Contribution of spoligotyping and MIRU-VNTRs to characterize prevalent Mycobacterium tuberculosis genotypes infecting tuberculosis patients in Morocco. Infect. Genet. Evol. 21, 463–471. 10.1016/j.meegid.2013.05.023 [DOI] [PubMed] [Google Scholar]
- Chaves F., Dronda F., Alonso-Sanz M., Noriega A. R. (1999). Evidence of exogenous reinfection and mixed infection with more than one strain of Mycobacterium tuberculosis among Spanish HIV-infected inmates. AIDS 13, 615–620. 10.1097/00002030-199904010-00011 [DOI] [PubMed] [Google Scholar]
- Chen J., Tsolaki A. G., Shen X., Jiang X., Mei J., Gao Q. (2007). Deletion-targeted multiplex PCR (DTM-PCR) for identification of Beijing/W genotypes of Mycobacterium tuberculosis. Tuberculosis 87, 446–449. 10.1016/j.tube.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Cho Y. J., Yi H., Chun J., Cho S. N., Daley C. L., Koh W. J., et al. (2013). The genome sequence of “Mycobacterium massiliense” strain CIP 108297 suggests the independent taxonomic status of the Mycobacterium abscessus complex at the subspecies level. PLoS ONE 8:e81560. 10.1371/journal.pone.0081560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T., Chindelevitch L., Misra R., Kempner M. E., Galea J., Moodley P., et al. (2016). Within-host heterogeneity of Mycobacterium tuberculosis infection is associated with poor early treatment response: a prospective cohort study. J. Infect. Dis. 213, 1796–1799. 10.1093/infdis/jiw014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T., van Helden P. D., Wilson D., Colijn C., McLaughlin M. M., Abubakar I., et al. (2012). Mixed-strain Mycobacterium tuberculosis infections and the implications for tuberculosis treatment and control. Clin. Microbiol. Rev. 25, 708–719. 10.1128/CMR.00021-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen T., Wilson D., Wallengren K., Samuel E. Y., Murray M. (2011). Mixed-strain Mycobacterium tuberculosis infections among patients dying in a hospital in KwaZulu-Natal, South Africa. J. Clin. Microbiol. 49, 385–388. 10.1128/JCM.01378-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll F., McNerney R., Guerra-Assunção J. A., Glynn J. R., Perdigão J., Viveiros M., et al. (2014). A robust SNP barcode for typing Mycobacterium tuberculosis complex strains. Nat. Commun. 5, 1–5. 10.1038/ncomms5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. L. (2010). Nontuberculous mycobacteria: opportunistic environmental pathogens for predisposed hosts. Br. Med. Bull. 96, 45–59. 10.1093/bmb/ldq035 [DOI] [PubMed] [Google Scholar]
- Coros A., DeConno E., Derbyshire K. M. (2008). IS6110, a Mycobacterium tuberculosis complex-specific insertion sequence, is also present in the genome of Mycobacterium smegmatis, suggestive of lateral gene transfer among mycobacterial species. J. Bacteriol. 190, 3408–3410. 10.1128/JB.00009-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosivi O., Grange J. M., Daborn C. J., Raviglione M. C., Fujikura T., Cousins D., et al. (1998). Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4, 59–70. 10.3201/eid0401.980108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox H. S., Kubica T., Doshetov D., Kebede Y., Rüesch-Gerdes S., Niemann S. (2005). The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 6, 1–9. 10.1186/1465-9921-6-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox H. S., Niemann S., Ismailov G., Doshetov D., Orozco J. D., Blok L., et al. (2007). Risk of acquired drug resistance during short-course directly observed treatment of tuberculosis in an area with high levels of drug resistance. Clin. Infect. Dis. 44, 1421–1427. 10.1086/517536 [DOI] [PubMed] [Google Scholar]
- Cox R., Mirkin S. M. (1997). Characteristic enrichment of DNA repeats in different genomes. Proc. Natl. Acad. Sci. U.S.A. 94, 5237–5242. 10.1073/pnas.94.10.5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Narayanan S., Hari L., Mohan N. S., Somasundaram S., Selvakumar N., et al. (2004). Simultaneous infection with multiple strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 8, 267–270. [PubMed] [Google Scholar]
- Davidson F. W., Ahlstrom C., de Buck J., Whitney H. G., Tahlan K. (2016). Examination of Mycobacterium avium subspecies paratuberculosis mixed genotype infections in dairy animals using a whole genome sequencing approach. PeerJ. 4:e2793. 10.7717/peerj.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida S. M., Malaspina A. C., Fujimura Leite C. Q., Féres Saad M. H. (2019). Usefulness of 3′- 5′ IS6110-RFLP genotyping and spoligotyping of Mycobacterium tuberculosis isolated in a tertiary hospital: a retrospective study detecting unsuspected epidemiological events. Rev. Inst. Med. Trop. São Paulo 61:e51 10.1590/s1678-9946201961051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong B. C., Antonio M., Gagneux S. (2010). Mycobacterium africanum-review of an important cause of human tuberculosis in West Africa. PLoS Negl. Trop. Dis. 4:e744. 10.1371/journal.pntd.0000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sequeira P. C., De Souza Fonseca L., Da Silva M. G., Féres Saad M. H. (2005). Mycobacterium avium restriction fragment length polymorphism-IS IS1245 and the simple double repetitive element polymerase chain reaction typing method to screen genetic diversity in Brazilian strains. Mem. Inst. Oswaldo Cruz 100, 743–748. 10.1590/S0074-02762005000700012 [DOI] [PubMed] [Google Scholar]
- Devallois A., Rastogi N. (1997). Computer-assisted analysis of Mycobacterium avium fingerprints using insertion elements IS1245 and IS1311 in a caribbean setting. Res. Microbiol. 148, 703–713. 10.1016/S0923-2508(99)80069-2 [DOI] [PubMed] [Google Scholar]
- Dheda K., Limberis J. D., Pietersen E., Phelan J., Esmail A., Lesosky M., et al. (2017). Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study. Lancet Respir. Med. 5, 269–281. 10.1016/S2213-2600(16)30433-7 [DOI] [PubMed] [Google Scholar]
- Dickman K. R., Nabyonga L., Kateete D. P., Katabazi F. A., Asiimwe B. B., Mayanja H. K., et al. (2010). Detection of multiple strains of Mycobacterium tuberculosis using MIRU-VNTR in patients with pulmonary tuberculosis in Kampala, Uganda. BMC Infect. Dis. 10:349 10.1186/1471-2334-10-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippenaar A., De Vos M., Marx F. M., Adroub S. A., van Helden P. D., Pain A., et al. (2019). Whole genome sequencing provides additional insights into recurrent tuberculosis classified as endogenous reactivation by IS6110 DNA fingerprinting. Infect. Genet. Evol. 75:103948 10.1016/j.meegid.2019.103948 [DOI] [PubMed] [Google Scholar]
- Döpfer D., Buist W., Soyer Y., Munoz M. A., Zadoks R. N., Geue L., et al. (2008). Assessing genetic heterogeneity within bacterial species isolated from gastrointestinal and environmental samples: how many isolates does it take? Appl. Environ. Microbiol. 74, 3490–3496. 10.1128/AEM.02789-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis D. G., Warren R., Richardson M., Joubert J. J., Van Helden P. D. (2001). Demonstration of reinfection and reactivation in HIV-negative autopsied cases of secondary tuberculosis: multilesional genotyping of Mycobacterium tuberculosis utilizing IS6110 and other repetitive element-based DNA fingerprinting. Tuberculosis 81, 211–220. 10.1054/tube.2000.0278 [DOI] [PubMed] [Google Scholar]
- Dvorska L., Bartos M., Ostadal O., Kaustova J., Matlova L., Pavlik I. (2002). IS1311 and IS1245 restriction fragment length polymorphism analyses, serotypes, and drug susceptibilities of Mycobacterium avium complex isolates obtained from a human immunodeficiency virus-negative patient. J. Clin. Microbiol. 40, 3712–3719. 10.1128/JCM.40.10.3712-3719.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorska L., Matlova L., Ayele W. Y., Fischer O. A., Amemori T., Weston R. T., et al. (2007). Avian tuberculosis in naturally infected captive water birds of the Ardeideae and Threskiornithidae families studied by serotyping, IS901 RFLP typing, and virulence for poultry. Vet. Microbiol. 119, 366–374. 10.1016/j.vetmic.2006.09.010 [DOI] [PubMed] [Google Scholar]
- Egbe N. F., Muwonge A., Ndip L., Kelly R. F., Sander M., Tanya V., et al. (2017). Molecular epidemiology of Mycobacterium bovis in Cameroon. Sci. Rep. 7:4652. 10.1038/s41598-017-04230-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Li X., Li J., Wu J., Shen X., Gui X., et al. (2008). Mixed infections of Mycobacterium tuberculosis in tuberculosis patients in Shanghai, China. Tuberculosis 88, 469–473. 10.1016/j.tube.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmanfarmaei G., Kargarpour Kamakoli M., Sadegh H. R., Masoumi M., Abdolrahimi F., Fateh A., et al. (2017). Bias in detection of Mycobacterium tuberculosis polyclonal infection: use clinical samples or cultures? Mol. Cell. Probes 33, 1–3. 10.1016/j.mcp.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Feil E. J. (2004). Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2, 483–495. 10.1038/nrmicro904 [DOI] [PubMed] [Google Scholar]
- Fomukong N. G., Tang T. H., Al-Maamary S., Ibrahim W. A., Ramayah S., Yates M., et al. (1994). Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuber. Lung Dis. 75, 435–440. 10.1016/0962-8479(94)90117-1 [DOI] [PubMed] [Google Scholar]
- Fonseca J. D., Knight G. M., McHugh T. D. (2015). The complex evolution of antibiotic resistance in Mycobacterium tuberculosis. Int. J. Infect. Dis. 32, 94–100. 10.1016/j.ijid.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Ford C. B., Shah R. R., Maeda M. K., Gagneux S., Murray M. B., Cohen T., et al. (2013). Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat. Genet. 45, 784–790. 10.1038/ng.2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman C. R., Stoeckle M. Y., Johnson W. D., Riley L. W. (1995). Double-repetitive-element PCR method for subtyping Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 33, 1383–1384. 10.1128/JCM.33.5.1383-1384.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frothingham R., Meeker-O'Connell W. A. (1998). Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144, 1189–1196. 10.1099/00221287-144-5-1189 [DOI] [PubMed] [Google Scholar]
- Fujita K., Ito Y., Hirai T., Kubo T., Maekawa K., Togashi K., et al. (2014). Association between polyclonal and mixed mycobacterial Mycobacterium avium complex infection and environmental exposure. Ann. Am. Thorac. Soc. 11, 45–53. 10.1513/AnnalsATS.201309-297OC [DOI] [PubMed] [Google Scholar]
- Furphy C., Costello E., Murphy D., Corner L. A. L., Gormley E. (2012). DNA typing of Mycobacterium bovis isolates from badgers (Meles meles) culled from areas in Ireland with different levels of tuberculosis prevalence. Vet. Med. Int. 2012:742478. 10.1155/2012/742478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan M., Liu Q., Yang C., Gao Q., Luo T. (2016). Deep whole-genome sequencing to detect mixed infection of Mycobacterium tuberculosis. PLoS ONE 11:e0159029. 10.1371/journal.pone.0159029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García De Viedma D., Marín M., Ruiz Serrano M. J., Alcalá L., Bouza E. (2003). Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J. Infect. Dis. 187, 695–699. 10.1086/368368 [DOI] [PubMed] [Google Scholar]
- García De Viedma D., Rodriguez N. A., Andrés S., Ruiz Serrano M. J., Bouza E. (2005). Characterization of clonal complexity in tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat typing. J. Clin. Microbiol. 43, 5660–5664. 10.1128/JCM.43.11.5660-5664.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Pedrazuela M., Frutos J. M., Muñoz-Egea M. C., Alcaide F., Tórtola T., Vitoria A., et al. (2015). Polyclonality among clinical strains of non-pigmented rapidly growing mycobacteria: phenotypic and genotypic differences and their potential implications. Clin. Microbiol. Infect. 21, 348.e1–348.e4. 10.1016/j.cmi.2014.12.004 [DOI] [PubMed] [Google Scholar]
- Gardy J. L., Johnston J. C., Ho Sui S. J., Cook V. J., Shah L., Brodkin E., et al. (2011). Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364, 730–739. 10.1056/NEJMoa1003176 [DOI] [PubMed] [Google Scholar]
- Gengenbacher M., Kaufmann S. H. E. (2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532. 10.1111/j.1574-6976.2012.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerritsmann H., Stalder G. L., Spergser J., Hoelzl F., Deutz A., Kuebber-Heiss A., et al. (2014). Multiple strain infections and high genotypic diversity among Mycobacterium avium subsp. paratuberculosis field isolates from diseased wild and domestic ruminant species in the eastern Alpine region of Austria. Infect. Genet. Evol. 21, 244–251. 10.1016/j.meegid.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Ghielmetti G., Coscolla M., Ruetten M., Friedel U., Loiseau C., Feldmann J., et al. (2017). Tuberculosis in Swiss captive Asian elephants: microevolution of Mycobacterium tuberculosis characterized by multilocus variable-number tandem-repeat analysis and whole-genome sequencing. Sci. Rep. 7:14647. 10.1038/s41598-017-15278-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioffré A., Muñoz M. C., Alvarado Pinedo M. F., Vaca R., Morsella C., Fiorentino M. A., et al. (2015). Molecular typing of Argentinian Mycobacterium avium subsp. paratuberculosis isolates by multiple-locus variable number-tandem repeat analysis. Braz. J. Microbiol. 46, 557–564. 10.1590/S1517-838246220140283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo-Asensio J., Pérez I., Aguiló N., Uranga S., Picó A., Lampreave C., et al. (2018). New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis complex lineages. PLoS Genet. 14:e1007282. 10.1371/journal.pgen.1007282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal M., Lawn S., Afful B., Acheampong J. W., Griffin G., Shaw R. (1999). Spoligotyping in molecular epidemiology of tuberculosis in Ghana. J. Infect. 38, 171–175. 10.1016/S0163-4453(99)90246-3 [DOI] [PubMed] [Google Scholar]
- Grange J. M. (2001). Mycobacterium bovis infection in human beings. Tuberculosis 81, 71–77. 10.1054/tube.2000.0263 [DOI] [PubMed] [Google Scholar]
- Griffith D. E. (2010). Nontuberculous mycobacterial lung disease. Curr. Opin. Infect. Dis. 23, 185–190. 10.1097/QCO.0b013e328336ead6 [DOI] [PubMed] [Google Scholar]
- Griffith D. E., Aksamit T., Brown-Elliott B. A., Catanzaro A., Daley C., Gordin F., et al. (2007). An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175, 367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- Guernier-Cambert V., Diefenbach-Elstob T., Klotoe B. J., Burgess G., Pelowa D., Dowi R., et al. (2019). Diversity of Mycobacterium tuberculosis in the middle fly district of western province, Papua New Guinea: microbead-based spoligotyping using DNA from Ziehl-Neelsen-stained microscopy preparations. Sci. Rep. 9, 1–10. 10.1038/s41598-019-51892-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra-Assuncaõ J. A., Houben R. M. G. J., Crampin A. C., Mzembe T., Mallard K., Coll F., et al. (2015). Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow-up. J. Infect. Dis. 211, 1154–1163. 10.1093/infdis/jiu574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C., Bernasconi C., Burki D., Bodmer T., Telenti A. (1995). A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33, 304–307. 10.1128/JCM.33.2.304-307.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthertz L. S., Damsker B., Bottone E. J., Ford E. G., Midura T. E., Michael Janda J. (1989). Mycobacterium avium and Mycobacterium intracellulare infections in patients with and without AIDS. J. Infect. Dis. 160, 1037–1041. 10.1093/infdis/160.6.1037 [DOI] [PubMed] [Google Scholar]
- Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. (2005). A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1, 0474–0483. 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajimiri E. S., Masoomi M., Ebrahimzadeh N., Fateh A., Hadizadeh Tasbiti A., Rahimi Jamnani F., et al. (2016). High prevalence of Mycobacterium tuberculosis mixed infection in the capital of moderate tuberculosis incidence country. Microb. Pathog. 93, 213–218. 10.1016/j.micpath.2016.02.015 [DOI] [PubMed] [Google Scholar]
- Han X. Y., Tarrand J. J., Infante R., Jacobson K. L., Truong M. (2005). Clinical significance and epidemiologic analyses of Mycobacterium avium and Mycobacterium intracellulare among patients without AIDS. J. Clin. Microbiol. 43, 4407–4412. 10.1128/JCM.43.9.4407-4412.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanekom M., Streicher E. M., Van de Berg D., Cox H., McDermid C., Bosman M., et al. (2013). Population Structure of Mixed Mycobacterium tuberculosis Infection Is Strain Genotype and Culture Medium Dependent. PLoS ONE 8:e70178. 10.1371/journal.pone.0070178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherell H. A., Colijn C., Stagg H. R., Jackson C., Winter J. R., Abubakar I. (2016). Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med. 14:21. 10.1186/s12916-016-0566-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlik J. A., Robert Horsburgh C., Metchock B., Williams P. P., Alan Fann S., Thompson S. E. (1992). Disseminated Mycobacterium avium complex infection: clinical identification and epidemiologic trends. J. Infect. Dis. 165, 577–580. 10.1093/infdis/165.3.577 [DOI] [PubMed] [Google Scholar]
- Hermans P. W. M., Van Soolingen D., Bik E. M., De Haas P. E. W., Dale J. W., Van Embden J. D. A. (1991). Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun. 59, 2695–2705. 10.1128/IAI.59.8.2695-2705.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermans P. W. M., Van Soolingen D., Dale J. W., Schuitema A. R. J., McAdam R. A., Catty D., et al. (1990). Insertion element IS986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28, 2051–2058. 10.1128/JCM.28.9.2051-2058.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill V., Zozio T., Sadikalay S., Viegas S., Streit E., Kallenius G., et al. (2012). MLVA Based Classification of Mycobacterium tuberculosis complex lineages for a robust phylogeographic snapshot of its worldwide molecular diversity. PLoS ONE 7:e41991. 10.1371/journal.pone.0041991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingley-Wilson S. M., Casey R., Connell D., Bremang S., Evans J. T., Hawkey P. M., et al. (2013). Undetected multidrug-resistant tuberculosis amplified by first-line therapy in mixed infection. Emerg. Infect. Dis. 19, 1138–1141. 10.3201/eid1907.130313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocqueloux L., Lesprit P., Herrmann J. L., De La Blanchardiere A., Zagdanski A. M., Decazes J. M., et al. (1998). Pulmonary Mycobacterium avium complex disease without dissemination in HIV-lnfected patients. Chest 113, 542–548. 10.1378/chest.113.2.542 [DOI] [PubMed] [Google Scholar]
- Hoefsloot W., van Ingen J., Peters E. J. G., Magis-Escurra C., Dekhuijzen P. N. R., Boeree M. J., et al. (2013). Mycobacterium genavense in the Netherlands: an opportunistic pathogen in HIV and non-HIV immunocompromised patients. An observational study in 14 cases. Clin. Microbiol. Infect. 19, 432–437. 10.1111/j.1469-0691.2012.03817.x [DOI] [PubMed] [Google Scholar]
- Hofmann-Thiel S., Van Ingen J., Feldmann K., Turaev L., Uzakova G. T., Murmusaeva G., et al. (2009). Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur. Respir. J. 33, 368–374. 10.1183/09031936.00089808 [DOI] [PubMed] [Google Scholar]
- Homolka S., Projahn M., Feuerriegel S., Ubben T., Diel R., Nübel U., et al. (2012). High resolution discrimination of clinical Mycobacterium tuberculosis complex strains based on single nucleotide polymorphisms. PLoS ONE 7:e39855. 10.1371/journal.pone.0039855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoop R. K., Bottger E. C., Ossent P., Salfinger M. (1993). Mycobacteriosis due to Mycobacterium genavense in six pet birds. J. Clin. Microbiol. 31, 990–993. 10.1128/JCM.31.4.990-993.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Zhao Q., Werngren J., Hoffner S., Diwan V. K., Xu B. (2016). Drug resistance characteristics and cluster analysis of M. tuberculosis in Chinese patients with multiple episodes of anti-tuberculosis treatment. BMC Infect. Dis. 16:4. 10.1186/s12879-015-1331-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. Y., Tsai Y. S., Lee J. J., Chiang M. C., Chen Y. H., Chiang C. Y., et al. (2010). Mixed infection with Beijing and non-Beijing strains and drug resistance pattern of Mycobacterium tuberculosis. J. Clin. Microbiol. 48, 4474–4480. 10.1128/JCM.00930-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. S., Ball N. W., Love D. N., Canfield P. J., Wigney D. I., Dawson D., et al. (1999). Disseminated Mycobacterium genavense infection in a FIV-positive cat. J. Feline Med. Surg. 1, 23–29. 10.1016/S1098-612X(99)90006-2 [DOI] [PubMed] [Google Scholar]
- Huyen M. N. T., Kremer K., Lan N. T. N., Cobelens F. G. J., Buu T. N., Dung N. H., et al. (2012). Mixed tuberculosis infections in rural South Vietnam. J. Clin. Microbiol. 50, 1586–1592. 10.1128/JCM.00434-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama S., Shimokata K., Tsukamura M. (1988). The isolation of Mycobacterium avium Complex from soil, water, and dusts. Microbiol. Immunol. 32, 733–739. 10.1111/j.1348-0421.1988.tb01434.x [DOI] [PubMed] [Google Scholar]
- Jacobson M. A., Hopewell P. C., Yajko D. M., Hadley W. K., Lazarus E., Mohanty P. K., et al. (1991). Natural history of disseminated Mycobacterium avium complex infection in AIDS. J. Infect. Dis. 164, 994–998. 10.1093/infdis/164.5.994 [DOI] [PubMed] [Google Scholar]
- Jajou R., Kamst M., van Hunen R., de Zwaan C. C., Mulder A., Supply P., et al. (2018). Occurrence and nature of double alleles in variable-number tandem-repeat patterns of more than 8,000 Mycobacterium tuberculosis complex isolates in The Netherlands. J. Clin. Microbiol. 56:e00761–17. 10.1128/JCM.00761-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T. B., Agdestein A., Lium B., Jørgensen A., Djønne B. (2014). Mycobacterium avium subsp. hominissuis infection in swine associated with peat used for bedding. Biomed Res. Int. 2014:189649. 10.1155/2014/189649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T. B., Djønne B., Jensen M. R., Olsen I. (2005). Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J. Clin. Microbiol. 43, 2500–2502. 10.1128/JCM.43.5.2500-2502.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. D. (1988). Bacteriophage typing of Mycobacterium tuberculosis cultures from incidents of suspected laboratory cross-contamination. Tubercle 69, 43–46. 10.1016/0041-3879(88)90039-6 [DOI] [PubMed] [Google Scholar]
- Jones W. D., Greenberg J. (1978). Modification of methods used in bacteriophage typing of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 7, 467–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I. K., Rogozin I. B., Wolf Y. I., Koonin E. V. (2002). Microevolutionary genomics of bacteria. Theor. Popul. Biol. 61, 435–447. 10.1006/tpbi.2002.1588 [DOI] [PubMed] [Google Scholar]
- Källenius G., Koivula T., Ghebremichael S., Hoffner S. E., Norberg R., Svensson E., et al. (1999). Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 37, 3872–3878. 10.1128/JCM.37.12.3872-3878.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakoli M. K., Farmanfarmaei G., Masoumi M., Khanipour S., Gharibzadeh S., Sola C., et al. (2020a). Prediction of the hidden genotype of mixed infection strains in Iranian tuberculosis patients. Int. J. Infect. Dis. 95, 22–27. 10.1016/j.ijid.2020.03.056 [DOI] [PubMed] [Google Scholar]
- Kamakoli M. K., Hadifar S., Khanipour S., Farmanfarmaei G., Fateh A., Siadat S. D., et al. (2019). Comparison of MIRU-VNTR genotyping between old and fresh clinical samples in tuberculosis. Infect. Dis. 51, 659–667. 10.1080/23744235.2019.1638963 [DOI] [PubMed] [Google Scholar]
- Kamakoli M. K., Khanipour S., Hadifar S., Ghajavand H., Farmanfarmaei G., Fateh A., et al. (2018). Challenge in direct spoligotyping of Mycobacterium tuberculosis: a problematic issue in the region with high prevalence of polyclonal infections. BMC Res. Notes 11:486. 10.1186/s13104-018-3579-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakoli M. K., Khanipour S., Masoumi M., Ghajavand H., Farmanfarmaei G., Fateh A., et al. (2020b). Application of MIRU–VNTR on smear slides: a shortcut for detection of polyclonal infections in tuberculosis patients. Mol. Biol. Rep. 47, 1681–1689. 10.1007/s11033-020-05257-1 [DOI] [PubMed] [Google Scholar]
- Kamakoli M. K., Sadegh H. R., Farmanfarmaei G., Masoumi M., Fateh A., Javadi G., et al. (2017). Evaluation of the impact of polyclonal infection and heteroresistance on treatment of tuberculosis patients. Sci. Rep. 7, 1–4. 10.1038/srep41410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J., Schouls L., Kolk A., Van Agterveld M., Van Soolingen D., Kuijper S., et al. (1997). Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35, 907–914. 10.1128/JCM.35.4.907-914.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay G. L., Sergeant M. J., Zhou Z., Chan J. Z. M., Millard A., Quick J., et al. (2015). Eighteenth-century genomes show that mixed infections were common at time of peak tuberculosis in Europe. Nat. Commun. 6, 1–9. 10.1038/ncomms7717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A. D., Shahraki A. H., Dezfuli S. K., Hashemzadeh M., Goodarzi H., Mohajeri P. (2017). Genetic diversity of multidrug-resistant Mycobacterium tuberculosis strains isolated from tuberculosis patients in Iran using MIRU-VNTR technique. Kaohsiung J. Med. Sci. 33, 550–557. 10.1016/j.kjms.2017.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehn T. E., Hoefer H., Bottger E. C., Ross R., Wong M., Edwards F., et al. (1996). Mycobacterium genavense infections in pet animals. J. Clin. Microbiol. 34, 1840–1842. 10.1128/JCM.34.7.1840-1842.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimizuka Y., Hoshino Y., Nishimura T., Asami T., Sakakibara Y., Morimoto K., et al. (2019). Retrospective evaluation of natural course in mild cases of Mycobacterium avium complex pulmonary disease. PLoS ONE 14:e0216034 10.1371/journal.pone.0216034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh W. J., Jeong B. H., Jeon K., Lee N. Y., Lee K. S., Woo S. Y., et al. (2012). Clinical significance of the differentiation between Mycobacterium avium and Mycobacterium intracellulare in M. avium complex lung disease. Chest 142, 1482–1488. 10.1378/chest.12-0494 [DOI] [PubMed] [Google Scholar]
- Kontsevaya I., Nikolayevskyy V., Kovalyov A., Ignatyeva O., Sadykhova A., Simak T., et al. (2017). Tuberculosis cases caused by heterogeneous infection in Eastern Europe and their influence on outcomes. Infect. Genet. Evol. 48, 76–82. 10.1016/j.meegid.2016.12.016 [DOI] [PubMed] [Google Scholar]
- Krebs T., Zimmerli S., Bodmer T., Lämmle B. (2000). Mycobacterium genavense infection in a patient with long-standing chronic lymphocytic leukaemia. J. Intern. Med. 248, 343–348. 10.1046/j.1365-2796.2000.00730.x [DOI] [PubMed] [Google Scholar]
- Lamine-Khemiri H., Martínez R., García-Jiménez W. L., Benítez-Medina J. M., Cortés M., Hurtado I., et al. (2014). Genotypic characterization by spoligotyping and VNTR typing of Mycobacterium bovis and Mycobacterium caprae isolates from cattle of Tunisia. Trop. Anim. Health Prod. 46, 305–311. 10.1007/s11250-013-0488-y [DOI] [PubMed] [Google Scholar]
- Lazzarini L. C. O., Rosenfeld J., Huard R. C., Hill V., Lapa e Silva J. R., DeSalle R., et al. (2012). Mycobacterium tuberculosis spoligotypes that may derive from mixed strain infections are revealed by a novel computational approach. Infect. Genet. Evol. 12, 798–806. 10.1016/j.meegid.2011.08.028 [DOI] [PubMed] [Google Scholar]
- Le Flèche P., Fabre M., Denoeud F., Koeck J. L., Vergnaud G. (2002). High resolution, on-line identification of strains from the Mycobacterium tuberculosis complex based on tandem repeat typing. BMC Microbiol. 2, 1–12. 10.1186/1471-2180-2-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. R., Sheng W. H., Hung C. C., Yu C. J., Lee L. N., Hsueh P. R. (2015). Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 21, 1638–1646. 10.3201/2109.141634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand E., Devallois A., Horgen L., Rastogi N. (2000a). A molecular epidemiological study of Mycobacterium simiae isolated from AIDS patients in Guadeloupe. J. Clin. Microbiol. 38, 3080–3084. 10.1128/JCM.38.8.3080-3084.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand E., Sola C., Verdol B., Rastogi N. (2000b). Genetic diversity of Mycobacterium avium recovered from AIDS patients in the Caribbean as studied by a consensus IS1245-RFLP method and pulsed-field gel electrophoresis. Res. Microbiol. 151, 271–283. 10.1016/S0923-2508(00)00147-9 [DOI] [PubMed] [Google Scholar]
- Ley S. D., de Vos M., Van Rie A., Warren R. M. (2019). Deciphering Within-Host Microevolution of Mycobacterium tuberculosis through Whole-Genome Sequencing: the Phenotypic Impact and Way Forward. Microbiol. Mol. Biol. Rev. 83:e00062-18. 10.1128/MMBR.00062-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipworth S., Jajou R., De Neeling A., Bradley P., Van Der Hoek W., Maphalala G., et al. (2019). SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg. Infect. Dis. 25, 482–488. 10.3201/eid2503.180894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Via L. E., Luo T., Liang L., Liu X., Wu S., et al. (2015). Within patient microevolution of Mycobacterium tuberculosis correlates with heterogeneous responses to treatment. Sci. Rep. 5:17507. 10.1038/srep17507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok K. H., Benjamin W. H., Kimerling M. E., Pruitt V., Lathan M., Razeq J., et al. (2002). Molecular differentiation of Mycobacterium tuberculosis strains without IS6110 insertions. Emerg. Infect. Dis. 8, 1310–1313. 10.3201/eid0811.020291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenço M. C. S., Grinsztejn B., Fandinho-Montes F. C. O., Da Silva M. G., Saad M. H. F., Fonseca L. S. (2000). Genotypic patterns of multiple isolates of M. tuberculosis from tuberculous HIV patients. Trop. Med. Int. Heal. 5, 488–494. 10.1046/j.1365-3156.2000.00583.x [DOI] [PubMed] [Google Scholar]
- Lucas A., Furber H., James G., Hughes M. S., Martin P., Chen S. C. A., et al. (2000). Mycobacterium genavense infection in two aged ferrets with conjunctival lesions. Aust. Vet. J. 78, 685–689. 10.1111/j.1751-0813.2000.tb10406.x [DOI] [PubMed] [Google Scholar]
- Maghradze N., Jugheli L., Borrell S., Tukvadze N., Aspindzelashvili R., Avaliani Z., et al. (2019). Classifying recurrent Mycobacterium tuberculosis cases in Georgia using MIRU-VNTR typing. PLoS ONE 14:e0223610. 10.1371/journal.pone.0223610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard K., McNerney R., Crampin A. C., Houben R., Ndlovu R., Munthali L., et al. (2010). Molecular detection of mixed infections of Mycobacterium tuberculosis strains in sputum samples from patients in Karonga District, Malawi. J. Clin. Microbiol. 48, 4512–4518. 10.1128/JCM.01683-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankiewicz E., Liivak M. (1975). Phage types of Mycobacterium tuberculosis in cultures isolated from Eskimo patients. Amer.Rev.Resp.Dis. 111, 307–312. [DOI] [PubMed] [Google Scholar]
- Martín A., Herránz M., Serrano M. J. R., Bouza E., De Viedma D. G. (2007). Rapid clonal analysis of recurrent tuberculosis by direct MIRU-VNTR typing on stored isolates. BMC Microbiol. 7:73. 10.1186/1471-2180-7-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx F. M., Dunbar R., Enarson D. A., Williams B. G., Warren R. M., Van Der Spuy G. D., et al. (2014). The temporal dynamics of relapse and reinfection tuberculosis after successful treatment: a retrospective cohort study. Clin. Infect. Dis. 58, 1676–1683. 10.1093/cid/ciu186 [DOI] [PubMed] [Google Scholar]
- McGowan J., Sampson M., Salzwedel D. M., Cogo E., Foerster V., Lefebvre C. (2016). PRESS peer review of electronic search strategies: 2015 guideline statement. J. Clin. Epidemiol. 75, 40–46. 10.1016/j.jclinepi.2016.01.021 [DOI] [PubMed] [Google Scholar]
- McNaughton A. L., Sreenu V. B., Wilkie G., Gunson R., Templeton K., Leitch E. C. M. (2018). Prevalence of mixed genotype hepatitis C virus infections in the UK as determined by genotype-specific PCR and deep sequencing. J. Viral Hepat. 25, 524–534. 10.1111/jvh.12849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Z., Sun Z., Bai D., Xu Y., Li Z., Huang H., et al. (2015). Discrepancies in drug susceptibility test for tuberculosis patients resulted from the mixed infection and the testing system. Biomed Res. Int. 2015:651980. 10.1155/2015/651980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijs W., de Haas P., Rossau R., Van der Laan T., Rigouts L., Portaels F., et al. (2002). Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and “M. avium subsp. hominissuis” for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52, 1505–1518. 10.1099/00207713-52-5-1505 [DOI] [PubMed] [Google Scholar]
- Mokrousov I., Otten T., Vyazovaya A., Limeschenko E., Filipenko M. L., Sola C., et al. (2003). PCR-based methodology for detecting multidrug-resistant strains of Mycobacterium tuberculosis Beijing family circulating in Russia. Eur. J. Clin. Microbiol. Infect. Dis. 22, 342–348. 10.1007/s10096-003-0944-0 [DOI] [PubMed] [Google Scholar]
- Mokrousov I., Valcheva V., Sovhozova N., Aldashev A., Rastogi N., Isakova J. (2009). Penitentiary population of Mycobacterium tuberculosis in Kyrgyzstan: exceptionally high prevalence of the Beijing genotype and its Russia-specific subtype. Infect. Genet. Evol. 9, 1400–1405. 10.1016/j.meegid.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Moravkova M., Mrlik V., Parmova I., Kriz P., Pavlik I. (2013). High incidence of Mycobacterium avium subspecies hominissuis infection in a zoo population of bongo antelopes (Tragelaphus eurycerus). J. Vet. Diagnostic Investig. 25, 531–534. 10.1177/1040638713490689 [DOI] [PubMed] [Google Scholar]
- Morris R. S., Pfeiffer D. U., Jackson R. (1994). The epidemiology of Mycobacterium bovis infections. Vet. Microbiol. 40, 153–177. 10.1016/0378-1135(94)90053-1 [DOI] [PubMed] [Google Scholar]
- Mulenga C., Shamputa I. C., Mwakazanga D., Kapata N., Portaels F., Rigouts L. (2010). Diversity of Mycobacterium tuberculosis genotypes circulating in Ndola, Zambia. BMC Infect. Dis. 10:177. 10.1186/1471-2334-10-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Roberts C. A., Brown T. A. (2014). Genotyping of ancient Mycobacterium tuberculosis strains reveals historic genetic diversity. Proc. R. Soc. B Biol. Sci. 281:20133236. 10.1098/rspb.2013.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa S., Javed H., Hashmi J., Jamil N., Tahir Z., Akhtar A. M. (2016). Emergence of mixed infection of Beijing/non-Beijing strains among multi-drug resistant Mycobacterium tuberculosis in Pakistan. 3 Biotech 6, 1–9. 10.1007/s13205-016-0423-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muwonge A., Kankya C., Olea-Popelka F., Biffa D., Ssengooba W., Berit D., et al. (2013). Molecular investigation of multiple strain infections in patients with tuberculosis in Mubende district, Uganda. Infect. Genet. Evol. 17, 16–22. 10.1016/j.meegid.2013.03.039 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., et al. (1987). Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235, 1616–1622. 10.1126/science.3029872 [DOI] [PubMed] [Google Scholar]
- Namasivayam S., Diarra B., Diabate S., Sarro Y., dit S., Kone A., et al. (2020). Patients infected with Mycobacterium africanum versus Mycobacterium tuberculosis possess distinct intestinal microbiota. PLoS Negl. Trop. Dis. 14, 1–20. 10.1371/journal.pntd.0008230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namasivayam S., Kauffman K. D., McCulloch J. A., Yuan W., Thovarai V., Mittereder L. R., et al. (2019). Correlation between disease severity and the intestinal microbiome in Mycobacterium tuberculosis-infected rhesus macaques. mBio 10:e01018-19. 10.1128/mBio.01018-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathavitharana R. R., Shi C. X., Chindelevitch L., Calderon R., Zhang Z., Galea J. T., et al. (2017). Polyclonal pulmonary tuberculosis infections and risk for multidrug resistance, Lima, Peru. Emerg. Infect. Dis. 23, 1887–1890. 10.3201/eid2311.170077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Y., Herranz M., Pérez-Lago L., Lirola M. M., Ruiz-Serrano M. J., Bouza E., et al. (2011). Systematic survey of clonal complexity in tuberculosis at a populational level and detailed characterization of the isolates involved. J. Clin. Microbiol. 49, 4131–4137. 10.1128/JCM.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro Y., Romero B., Copano M. F., Bouza E., Domínguez L., de Juan L., et al. (2015). Multiple sampling and discriminatory fingerprinting reveals clonally complex and compartmentalized infections by M. bovis in cattle. Vet. Microbiol. 175, 99–104. 10.1016/j.vetmic.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Nicol M. P., Cox H. (2019). Recent developments in the diagnosis of drug-resistant tuberculosis. Microbiol. Aust. 40, 82–86. 10.1071/MA19023 [DOI] [Google Scholar]
- Nimmo C., Brien K., Millard J., Grant A. D., Padayatchi N., Pym A. S., et al. (2020). Dynamics of within-host Mycobacterium tuberculosis diversity and heteroresistance during treatment. EBioMed. 55:102747. 10.1016/j.ebiom.2020.102747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivin B., Driscoll J., Glaser T., Bifani P., Munsiff S. (2000). Use of spoligotype analysis to detect laboratory cross-contamination. Infect. Control Hosp. Epidemiol. 21, 525–527. 10.1086/501799 [DOI] [PubMed] [Google Scholar]
- Oakey J., Gavey L., Singh S. V., Platell J., Waltisbuhl D. (2014). Variable-number tandem repeats genotyping used to aid and inform management strategies for a bovine Johne's disease incursion in tropical and subtropical Australia. J. Vet. Diagn. Invest. 26, 651–657. 10.1177/1040638714547257 [DOI] [PubMed] [Google Scholar]
- O'Donnell M. R., Larsen M. H., Brown T. S., Jain P., Munsamy V., Wolf A., et al. (2019). Early detection of emergent extensively drug-resistant tuberculosis by flow cytometry-based phenotyping and whole-genome sequencing. Antimicrob. Agents Chemother. 63:e01834-18. 10.1128/AAC.01834-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkusu K., Bermudez L. E., Nash K. A., MacGregor R. R., Inderlied C. B. (2004). Differential virulence of Mycobacterium avium strains isolated from HIV-infected patients with disseminated M. avium complex disease. J. Infect. Dis. 190, 1347–1354. 10.1086/424488 [DOI] [PubMed] [Google Scholar]
- Oliveira R. S., Sircili M. P., Ueki S. Y. M., Telles M. A. S., Schnabel B., Briones M. R. S., et al. (2000). PCR-restriction enzyme analysis of a bone marrow isolate from a human immunodeficiency virus-positive patient discloses polyclonal infection with two Mycobacterium avium strains. J. Clin. Microbiol. 38, 4643–4645. 10.1128/JCM.38.12.4643-4645.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overduin P., Schouls L., Roholl P., Van Der Zanden A., Mahmmod N., Herrewegh A., et al. (2004). Use of multilocus variable-number tandem-repeat analysis for typing Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42, 5022–5028. 10.1128/JCM.42.11.5022-5028.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Zhou Y., Wang S., Song Y., Ou X., Zhao B., et al. (2015). Prevalence and risk factors of mixed Mycobacterium tuberculosis complex infections in China. J. Infect. 71, 231–237. 10.1016/j.jinf.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Panunto A. C., Villares M. C. B., Ramos M. C. (2003). IS1245 RFLP fingerprinting of Mycobacterium avium from patients admitted to a reference hospital in Campinas, Brazil. Brazilian J. Med. Biol. Res. 36, 1397–1401. 10.1590/S0100-879X2003001000017 [DOI] [PubMed] [Google Scholar]
- Parte A. C. (2018). LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 68, 1825–1829. 10.1099/ijsem.0.002786 [DOI] [PubMed] [Google Scholar]
- Pate M., Žolnir-Dovč M., Krt B., Ocepek M. (2008). IS1245 RFLP-based genotyping study of Mycobacterium avium subsp. hominissuis isolates from pigs and humans. Comp. Immunol. Microbiol. Infect. Dis. 31, 537–550. 10.1016/j.cimid.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Pavlic M., Allerberger F., Dierich M. P., Prodinger W. M. (1999). Simultaneous infection with two drug-susceptible Mycobacterium tuberculosis strains in an immunocompetent host. J. Clin. Microbiol. 37, 4156–4157. 10.1128/JCM.37.12.4156-4157.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Yang C., Li X., Luo T., Li F., Gao Q. (2013). Multiple samples improve the sensitivity for detection of mixed Mycobacterium infections. Tuberculosis 93, 548–550. 10.1016/j.tube.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Pérez-Lago L., Comas I., Navarro Y., González-Candelas F., Herranz M., Bouza E., et al. (2014). Whole Genome Sequencing Analysis of Intrapatient Microevolution in Mycobacterium tuberculosis: potential impact on the inference of tuberculosis transmission. J. Infect. Dis. 209, 98–108. 10.1093/infdis/jit439 [DOI] [PubMed] [Google Scholar]
- Pérez-Lago L., Navarro Y., Herranz M., Bouza E., García-de-Viedma D. (2013). Differences in gene expression between clonal variants of Mycobacterium tuberculosis emerging as a result of microevolution. Int. J. Med. Microbiol. 303, 674–677. 10.1016/j.ijmm.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Pfeiffer W., Braun J., Burchell J., Witte C. L., Rideout B. A. (2017). Whole-genome analysis of mycobacteria from birds at the San Diego Zoo. PLoS ONE 12:e0173464. 10.1371/journal.pone.0173464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardeau M., Varnerot A., Lecompte T., Brel F., May T., Vincent V. (1997). Use of different molecular typing techniques for bacteriological follow- up in a clinical trial with AIDS patients with Mycobacterium avium bacteremia. J. Clin. Microbiol. 35, 2503–2510. 10.1128/JCM.35.10.2503-2510.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picardeau M., Véronique V. (1996). Typing of Mycobacterium avium Isolates by PCR. J. Clin. Microbiol. 34, 389–392. 10.1128/JCM.34.2.389-392.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersimoni C., Scarparo C. (2008). Pulmonary infections associated with non-tuberculous mycobacteria in immunocompetent patients. Lancet Infect. Dis. 8, 323–334. 10.1016/S1473-3099(08)70100-2 [DOI] [PubMed] [Google Scholar]
- Podder M. P., Banfield S. E., Keefe G. P., Whitney H. G., Tahlan K. (2015). Typing of Mycobacterium avium subspecies paratuberculosis isolates from newfoundland using fragment analysis. PLoS ONE 10:e0126071. 10.1371/journal.pone.0126071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primm T. P., Lucero C. A., Falkinham J. O. (2004). Health impacts of environmental mycobacteria. Clin. Microbiol. Rev. 17, 98–106. 10.1128/CMR.17.1.98-106.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C., Von Reyn C. F., Chamblee S., Ellerbrock T. V., Johnson J. W., Marsh B. J., et al. (2006). Environmental risk factors for infection with Mycobacterium avium complex. Am. J. Epidemiol. 164, 32–40. 10.1093/aje/kwj159 [DOI] [PubMed] [Google Scholar]
- Reis A. C., Albuquerque T., Botelho A., Cunha M. V. (2020). Polyclonal infection as a new scenario in Mycobacterium caprae epidemiology. Vet. Microbiol. 240:108533. 10.1016/j.vetmic.2019.108533 [DOI] [PubMed] [Google Scholar]
- Richardson M., Carroll N. M., Engelke E., Van der Spuy G. D., Salker F., Munch Z., et al. (2002). Multiple Mycobacterium tuberculosis strains in early cultures from patients in a high-incidence community setting. J. Clin. Microbiol. 40, 2750–2754. 10.1128/JCM.40.8.2750-2754.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe W. D., Lenting B., Kokosinska A., Hunter S., Duignan P. J., Gartrell B., et al. (2019). Pathology and molecular epidemiology of Mycobacterium pinnipedii tuberculosis in native New Zealand marine mammals. PLoS ONE 14:e0212363. 10.1371/journal.pone.0212363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetzer A., Diel R., Kohl T. A., Rückert C., Nübel U., Blom J., et al. (2013). Whole Genome Sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis Outbreak: a longitudinal molecular epidemiological study. PLoS Med. 10:e1001387. 10.1371/journal.pmed.1001387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roring S., Scott A., Brittain D., Walker I., Hewinson G., Neill S., et al. (2002). Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40, 2126–2133. 10.1128/JCM.40.6.2126-2133.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad M. H. F., Telles M. A., Porfirio F., Ferrazoli L., De Souza Fonseca L., Johnson W., et al. (2000). Multiple isolates from aids patients: aspects of an analysis by a genotypic marker and antimicrobial susceptibilities variations. Mem. Inst. Oswaldo Cruz 95, 729–732. 10.1590/S0074-02762000000500021 [DOI] [PubMed] [Google Scholar]
- Sadegh H., Kargarpour Kamakoli M., Farmanfarmaei G., Masoumi M., Abdolrahimi F., Fateh A., et al. (2017). Pros and cons of direct genotyping on tuberculosis clinical samples. Microb. Pathog. 103, 135–138. 10.1016/j.micpath.2016.12.025 [DOI] [PubMed] [Google Scholar]
- Sassi M., Drancourt M. (2014). Genome analysis reveals three genomospecies in Mycobacterium abscessus. BMC Genomics 15:359. 10.1186/1471-2164-15-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Röbbecke R. (1993). Mycobacteria in the environment. Immun. Infekt. 21, 126–131. [PubMed] [Google Scholar]
- Scollard D. M., Adams L. B., Gillis T. P., Krahenbuhl J. L., Truman R. W., Williams D. L. (2006). The continuing challenges of leprosy. Clin. Microbiol. Rev. 19, 338–381. 10.1128/CMR.19.2.338-381.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamputa I. C., Jugheli L., Sadradze N., Willery E., Portaels F., Supply P., et al. (2006). Mixed infection and clonal representativeness of a single sputum sample in tuberculosis patients from a penitentiary hospital in Georgia. Respir. Res. 7:99. 10.1186/1465-9921-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamputa I. C., Lee J., Allix-Béguec C., Cho E. J., Lee J. I., Rajan V., et al. (2010). Genetic diversity of Mycobacterium tuberculosis isolates from a tertiary care tuberculosis hospital in South Korea. J. Clin. Microbiol. 48, 387–394. 10.1128/JCM.02167-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamputa I. C., Rigouts L., Eyongeta L. A., El Aila N. A., Van Deun A., Salim A. H., et al. (2004). Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J. Clin. Microbiol. 42, 5528–5536. 10.1128/JCM.42.12.5528-5536.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. S., Modongo C., Baik Y., Allender C., Lemmer D., Colman R. E., et al. (2018). Mixed Mycobacterium tuberculosis-strain infections are associated with poor treatment outcomes among patients with newly diagnosed tuberculosis, independent of pretreatment heteroresistance. J. Infect. Dis. 218, 1974–1982. 10.1093/infdis/jiy480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. S., Modongo C., Ncube R., Sepako E., Klausner J. D., Zetola N. M. (2015). Advanced immune suppression is associated with increased prevalence of mixed-strain Mycobacterium tuberculosis infections among persons at high risk for drug-resistant tuberculosis in Botswana. J. Infect. Dis. 211, 347–351. 10.1093/infdis/jiu421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitaye J. E., Matlova L., Horvathova A., Moravkova M., Dvorska-Bartosova L., Treml F., et al. (2008). Mycobacterium avium subsp. avium distribution studied in a naturally infected hen flock and in the environment by culture, serotyping and IS901 RFLP methods. Vet. Microbiol. 127, 155–164. 10.1016/j.vetmic.2007.07.026 [DOI] [PubMed] [Google Scholar]
- Sia I. G., Wieland M. L. (2011). Current concepts in the management of tuberculosis. Mayo Clin. Proc. 86, 348–361. 10.4065/mcp.2010.0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichewo P. R., Hlokwe T. M., Etter E. M. C., Michel A. L. (2020). Tracing cross species transmission of Mycobacterium bovis at the wildlife/livestock interface in South Africa. BMC Microbiol. 20:49. 10.1186/s12866-020-01736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi S. H. (1978). Atypical mycobacteria. J. Pakistan Med. Assoc. 28, 102–110. [PubMed] [Google Scholar]
- Silva-Pereira T. T., Ikuta C. Y., Zimpel C. K., Camargo N. C. S. S., De Souza Filho A. F., Ferreira Neto J. S., et al. (2019). Genome sequencing of Mycobacterium pinnipedii strains: genetic characterization and evidence of superinfection in a South American sea lion (Otaria flavescens). BMC Genomics 20:1030. 10.1186/s12864-019-6407-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Verma R., Shah D. H. (2004). Molecular fingerprinting of clinical isolates of Mycobacterium bovis and Mycobacterium tuberculosis from India by restriction fragment length polymorphism (RFLP). J. Vet. Sci. 5, 331–335. 10.4142/jvs.2004.5.4.331 [DOI] [PubMed] [Google Scholar]
- Skuce R. A., McCorry T. P., McCarroll J. F., Roring S. M. M., Scott A. N., Brittain D., et al. (2002). Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148, 519–528. 10.1099/00221287-148-2-519 [DOI] [PubMed] [Google Scholar]
- Slutsky A. M., Arbeit R. D., Barber T. W., Rich J., Von Reyn C. F., Pieciak W., et al. (1994). Polyclonal infections due to Mycobacterium avium complex in patients with AIDS detected by pulsed-field gel electrophoresis of sequential clinical isolates. J. Clin. Microbiol. 32, 1773–1778. 10.1128/JCM.32.7.1773-1778.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small P. M., Hopewell P. C., Singh S. P., Paz A., Parsonnet J., Ruston D. C., et al. (1994). The epidemiology of tuberculosis in san francisco: a Population-based study using conventional and molecular methods. N. Engl. J. Med. 330, 1703–1709. 10.1056/NEJM199406163302402 [DOI] [PubMed] [Google Scholar]
- Smittipat N., Palittapongarnpim P. (2000). Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tuber. Lung Dis. 80, 69–74. 10.1054/tuld.2000.0236 [DOI] [PubMed] [Google Scholar]
- Snider D. E., Jones W. D., Good R. C. (1984). The usefulness of phage typing Mycobacterium tuberculosis isolates. Am. Rev. Respir. Dis. 130, 1095–1099. [DOI] [PubMed] [Google Scholar]
- Sobkowiak B., Glynn J. R., Houben R. M. G. J., Mallard K., Phelan J. E., Guerra-Assunção J. A., et al. (2018). Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics 19:613. 10.1186/s12864-018-4988-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreevatsan S., Pan X., Stockbauer K. E., Connell N. D., Kreiswirth B. N., Whittam T. S., et al. (1997). Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. U.S.A. 94, 9869–9874. 10.1073/pnas.94.18.9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ssengooba W., Cobelens F. G., Nakiyingi L., Mboowa G., Armstrong D. T., Manabe Y. C., et al. (2015). High genotypic discordance of concurrent Mycobacterium tuberculosis isolates from sputum and blood of HIV-infected individuals. PLoS ONE 10:e0132581. 10.1371/journal.pone.0132581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ssengooba W., de Jong B. C., Joloba M. L., Cobelens F. G., Meehan C. J. (2016). Whole genome sequencing reveals mycobacterial microevolution among concurrent isolates from sputum and blood in HIV infected TB patients. BMC Infect. Dis. 16:371 10.1186/s12879-016-1737-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrum R., Mphahlele M., Øvreås K., Muthivhi T., Fourie P. B., Weyer K., et al. (2009). High diversity of Mycobacterium tuberculosis genotypes in South Africa and preponderance of mixed infections among ST53 isolates. J. Clin. Microbiol. 47, 1848–1856. 10.1128/JCM.02167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels D., Fauville-Dufaux M., Boie J., De Beenhouwera H. (2013). Failure of PCR-based IS6110 analysis to detect vertebral spondylodiscitis caused by Mycobacterium bovis. J. Clin. Microbiol. 51, 366–368. 10.1128/JCM.02524-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit E., Millet J., Rastogi N. (2015). Mycobacterium tuberculosis polyclonal infections and microevolution identified by MIRU-VNTRs in an epidemiological study. Int. J. Mycobacteriology 4, 222–227. 10.1016/j.ijmyco.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Supply P., Allix C., Lesjean S., Cardoso-Oelemann M., Rüsch-Gerdes S., Willery E., et al. (2006). Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44, 4498–4510. 10.1128/JCM.01392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P., Lesjean S., Savine E., Kremer K., Van Soolingen D., Locht C. (2001). Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39, 3563–3571. 10.1128/JCM.39.10.3563-3571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P., Magdalena J., Himpens S., Locht C. (1997). Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26, 991–1003. 10.1046/j.1365-2958.1997.6361999.x [DOI] [PubMed] [Google Scholar]
- Sweeney R. W., Collins M. T., Koets A. P., Mcguirk S. M., Roussel A. J. (2012). Paratuberculosis (Johne's disease) in cattle and other susceptible species. J. Vet. Intern. Med. 26, 1239–1250. 10.1111/j.1939-1676.2012.01019.x [DOI] [PubMed] [Google Scholar]
- Taylor L. H., Walliker D., Read A. F. (1997). Mixed-genotype infections of malaria parasites: within-host dynamics and transmission success of competing clones. Proc. R. Soc. B Biol. Sci. 264, 927–935. 10.1098/rspb.1997.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239. 10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen A., Reichel C., Rüsch-Gerdes S., Haas W. H., Rockstroh J. K., Spengler U., et al. (1995). Mixed-strain infection with a drug-sensitive and multidrug-resistant strain of Mycobacterium tuberculosis. Lancet 345, 1512–1513. 10.1016/S0140-6736(95)91073-5 [DOI] [PubMed] [Google Scholar]
- Thierry D., Brisson-Noel A., Vincent-Levy-Frebault V., Nguyen S., Guesdon J. L., Gicquel B. (1990a). Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J. Clin. Microbiol. 28, 2668–2673. 10.1128/JCM.28.12.2668-2673.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry D., Cave M. D., Eisenach K. D., Crawford J. T., Bates J. H., Gicquel B., et al. (1990b). IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 18:188. 10.1093/nar/18.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel M. F., Krichevsky M., Levy-Frebault V. V. (1990). Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. Silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40, 254–260. 10.1099/00207713-40-3-254 [DOI] [PubMed] [Google Scholar]
- Tortoli E. (2014). Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev. 27, 727–752. 10.1128/CMR.00035-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umubyeyi A. N., Shamputa I. C., Rigouts L., Dediste A., Karita E., Struelens M. J., et al. (2007). Molecular investigation of recurrent tuberculosis in patients from Rwanda. Int. J. Tuberc. Lung Dis. 11, 860–867. [PubMed] [Google Scholar]
- Van Den Berg R. J., Ameen H. A. A., Furusawa T., Claas E. C. J., Van Der Vorm E. R., Kuijper E. J. (2005). Coexistence of multiple PCR-ribotype strains of Clostridium difficile in faecal samples limits epidemiological studies. J. Med. Microbiol. 54, 173–179. 10.1099/jmm.0.45825-0 [DOI] [PubMed] [Google Scholar]
- Van Der Zanden A. G. M., Rahim Z., Fedder G., Vos-van Adrichem J., Sebens F. W., Heilmann F. G. C., et al. (2007). Multiple Mycobacterium tuberculosis infections in an HIV-infected patient. Southeast Asian J. Trop. Med. Public Health 38, 704–705. [PubMed] [Google Scholar]
- Van Embden J. D. A., Cave M. D., Crawford J. T., Dale J. W., Eisenach K. D., Gicquel B., et al. (1993). Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31, 406–409. 10.1128/JCM.31.2.406-409.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rie A., Victor T. C., Richardson M., Johnson R., Van Der Spuy G. D., Murray E. J., et al. (2005). Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am. J. Respir. Crit. Care Med. 172, 636–642. 10.1164/rccm.200503-449OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soolingen D., De Haas P. E. W., Hermans P. W. M., Groenen P. M. A., Van Embden J. D. A. (1993). Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J. Clin. Microbiol. 31, 1987–1995. 10.1128/JCM.31.8.1987-1995.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soolingen D., Hermans P. W. M., De Haas P. E. W., Soll D. R., Van Embden J. D. A. (1991). Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29, 2578–2586. 10.1128/JCM.29.11.2578-2586.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soolingen D., Qian L., De Haas P. E. W., Douglas J. T., Traore H., Portaels F., et al. (1995). Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33, 3234–3238. 10.1128/JCM.33.12.3234-3238.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Reyn C. F., Jacobs N. J., Arbeit R. D., Maslow J. N., Niemczyk S. (1995). Polyclonal Mycobacterium avium infections in patients with AIDS: variations in antimicrobial susceptibilities of different strains of M. avium isolated from the same patient. J. Clin. Microbiol. 33, 1008–1010. 10.1128/JCM.33.4.1008-1010.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Reyn C. F., Kimambo S., Mtei L., Arbeit R. D., Maro I., Bakari M., et al. (2011). Disseminated tuberculosis in human immunodeficiency virus infection: ineffective immunity, polyclonal disease and high mortality. Int. J. Tuberc. Lung Dis. 15, 1087–1092. 10.5588/ijtld.10.0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Reyn C. F., Waddell R. D., Eaton T., Arbeit R. D., Maslow J. N., Barber T. W., et al. (1993). Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J. Clin. Microbiol. 31, 3227–3230. 10.1128/JCM.31.12.3227-3230.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. M., Ip C. L. C., Harrell R. H., Evans J. T., Kapatai G., Dedicoat M. J., et al. (2013). Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect. Dis. 13, 137–146. 10.1016/S1473-3099(12)70277-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. J., Zhang Y., Brown B. A., Dawson D., Murphy D. T., Wilson R., et al. (1998). Polyclonal Mycobacterium avium complex infections in patients with nodular bronchiectasis. Am. J. Respir. Crit. Care Med. 158, 1235–1244. 10.1164/ajrccm.158.4.9712098 [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Hsu H. L., Yu M. C., Chiang C. Y., Yu F. L., Yu C. J., et al. (2011). Mixed infection with Beijing and non-Beijing strains in pulmonary tuberculosis in Taiwan: prevalence, risk factors, and dominant strain. Clin. Microbiol. Infect. 17, 1239–1245. 10.1111/j.1469-0691.2010.03401.x [DOI] [PubMed] [Google Scholar]
- Wang X., Liu H., Wei J., Wu X., Yu Q., Zhao X., et al. (2015). An investigation on the population structure of mixed infections of Mycobacterium tuberculosis in Inner Mongolia, China. Tuberculosis 95, 695–700. 10.1016/j.tube.2015.08.006 [DOI] [PubMed] [Google Scholar]
- Warren R. M., Streicher E. M., Charalambous S., Churchyard G., Van der Spuy G. D., Grant A. D., et al. (2002). Use of spoligotyping for accurate classification of recurrent tuberculosis. J. Clin. Microbiol. 40, 3851–3853. 10.1128/JCM.40.10.3851-3853.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. M., Victor T. C., Streicher E. M., Richardson M., Beyers N., Gey Van Pittius N. C., et al. (2004). Patients with active tuberculosis often have different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169, 610–614. 10.1164/rccm.200305-714OC [DOI] [PubMed] [Google Scholar]
- Wayne L. G., Kubica G. P. (1986). Genus Mycobacterium, in Bergey's Manual of Systematic Bacteriology, eds Sneath H. A., Mair N. S., Sharpe H. E., Holt J. G. (Baltimore: Williams and Wilkins; ), 1436–1457. [Google Scholar]
- Whitlock R. H., Buergelt C. (1996). Preclinical and clinical manifestations of paratuberculosis (including pathology). Vet. Clin. North Am. Food Anim. Pract. 12, 345–356. 10.1016/S0749-0720(15)30410-2 [DOI] [PubMed] [Google Scholar]
- WHO (2019). Global Tuberculosis Report 2019. World Health Organization. [Google Scholar]
- Witney A. A., Bateson A. L. E., Jindani A., Phillips P. P. J., Coleman D., Stoker N. G., et al. (2017). Use of whole-genome sequencing to distinguish relapse from reinfection in a completed tuberculosis clinical trial. BMC Med. 15:71. 10.1186/s12916-017-0834-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg K., Harris M., Gabrielian A., Ciobanu N., Chesov D., Long A., et al. (2020). A retrospective genomic analysis of drug-resistant strains of M. tuberculosis in a high-burden setting, with an emphasis on comparative diagnostics and reactivation and reinfection status. BMC Infect. Dis. 20:17. 10.1186/s12879-019-4739-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajko D. M., Chin D. P., Gonzalez P. C., Nassos P. S., Hopewell P. C., Reingold A. L., et al. (1995). Mycobacterium avium complex in water, food, and soil samples collected from the environment of HIV-infected individuals. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9, 176–182. [PubMed] [Google Scholar]
- Yeh R. W., Hopewell P. C., Daley C. L. (1999). Simultaneous infection with two strains of Mycobacterium tuberculosis identified by restriction fragment length polymorphism analysis. Int. J. Tuberc. Lung Dis. 3, 537–539. [PubMed] [Google Scholar]
- Yoshida S., Tsuyuguchi K., Kobayashi T., Tomita M., Inoue Y., Hayashi S., et al. (2018). Association between sequevar and antibiotic treatment outcome in patients with Mycobacterium abscessus complex infections in Japan. J. Med. Microbiol. 67, 74–82. 10.1099/jmm.0.000661 [DOI] [PubMed] [Google Scholar]
- Yuen L. K. W., Ross B. C., Jackson K. M., Dwyer B. (1993). Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J. Clin. Microbiol. 31, 1615–1618. 10.1128/JCM.31.6.1615-1618.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetola N. M., Shin S. S., Tumedi K. A., Moeti K., Ncube R., Nicol M., et al. (2014). Mixed Mycobacterium tuberculosis complex infections and false-negative results for rifampin resistance by genexpert MTB/RIF are associated with poor clinical outcomes. J. Clin. Microbiol. 52, 2422–2429. 10.1128/JCM.02489-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Rajagopalan M., Brown B. A., Wallace R. J. (1997). Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 35, 3132–3139. 10.1128/JCM.35.12.3132-3139.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Li S., Luo Z., Pi R., Sun H., He Q., et al. (2015). Mixed infections and rifampin heteroresistance among Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 53, 2138–2147. 10.1128/JCM.03507-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.