Abstract
Circular RNAs (circRNAs), a novel type of endogenous RNAs with covalently closed-loop structures, have become a new research hotspot in the RNA world. Their diversity, stability, evolutionary conservation, and cell type- or tissue-specific expression patterns endow circRNAs with various important biological functions. As a consequence, circRNAs are emerging as important regulators of physiological development and disease pathogenesis. Growing evidence has shown that circRNAs can regulate parental gene expression through diverse mechanisms, such as transcription and splicing regulation, microRNA (miRNA) sponges, mRNA traps, translational modulation, and post-translational modification. The study of circRNAs and how circRNAs regulate the expression of parental genes will facilitate a deeper understanding of their biological functions and provide new perspectives on their clinical application. Herein, we review the biogenesis of circRNAs, with a particular focus on the molecular mechanisms of circRNAs regulating their parental gene expression and the biological significance of such regulation.
Keywords: circular RNA, gene expression, parental gene, regulatory mechanism
Graphical Abstract

circRNAs are covalently closed continuous loops that exhibit multiple biological functions. This review summarizes recent advances in circRNAs’ regulatory roles in parental gene expression, including transcription and splicing regulation, ceRNA mechanisms, mRNA traps, and translational/post-translational regulation. It may provide new perspectives for the understanding and further investigation of circRNAs.
Main text
Circular RNAs (circRNAs) are a novel group of endogenous transcripts generally characterized by their covalently closed-loop structures. The existence of circRNAs was first reported in 1976 when a group discovered circRNA molecules naturally existing in plant viroids.1 With advancements in RNA sequencing and bioinformatics analysis, large numbers of previously unannotated circRNAs have been identified in different organisms. Unlike linear RNAs, circRNAs lack free terminal structures since their 3′ and 5′ ends are joined together by covalent bonds.2 This unique structure enhances their stability and protects them from degradation by RNA exonuclease or RNase R.3,4 While most circRNAs are low in abundance, some are ubiquitously expressed and are present at higher copy numbers as compared to their linear transcripts.4 Moreover, most circRNAs are conserved across species and often exhibit cell type- or tissue-specific expression, suggesting potential regulatory roles.4, 5, 6
Based on the source of the genome and biogenesis patterns, circRNAs are mainly divided into three groups: exonic circRNAs (EcircRNAs), exonic-intronic circRNAs (EIciRNAs), and circular intronic RNAs (ciRNAs).7, 8, 9 circRNAs have been reported to directly bind proteins, sponge microRNAs (miRNAs), and translate into proteins.10, 11, 12, 13, 14 However, emerging studies demonstrate that circRNAs can modulate the expression of their parental genes at multiple levels and are involved in the regulation of various physiological or pathological processes, including embryogenesis, atherosclerosis, and tumorigenesis.15, 16, 17, 18 In this review, we briefly introduce the biogenesis of circRNAs, summarize their functional roles in regulating parental gene expression, and discuss the biological significance of their regulatory functions.
Biogenesis of circRNAs
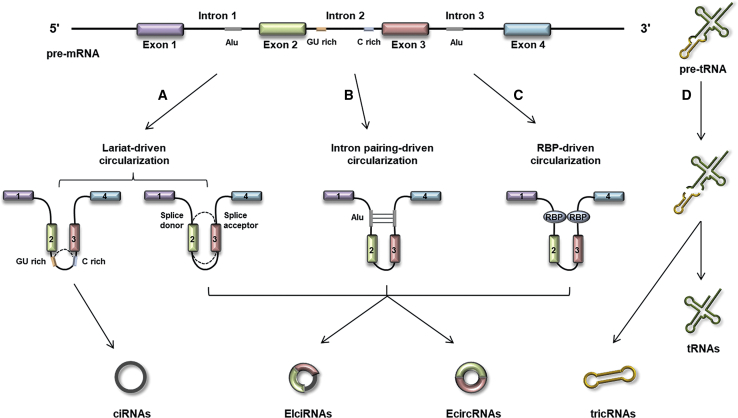
Precursor (pre-)mRNAs are synthesized by RNA polymerase II (RNA Pol II) and canonically spliced into linear mRNAs. However, circRNAs are derived from back-splicing of pre-mRNAs, a novel mode of RNA splicing involving joining of the 3′ splice site to the 5′ splice site.3,19 Several mechanisms have been recently proposed for the formation of circRNAs (Figure 1).4,9,20, 21, 22, 23 In 2013, Jeck et al.4 first put forward two models of circRNA circularization that were widely accepted, that is, lariat-driven circularization and intron pairing-driven circularization. The former requires covalent joining of 5′ donor splice sites and 3′ acceptor splice sites to form a lariat structure that contains at least one exon.4,20 In some cases, an intronic lariat, which is formed when an intron is removed during pre-mRNA splicing, can produce ciRNA.9 Intron pairing-driven circularization is mostly related to complementary inverted sequences within the flanking introns of the back-spliced exons. In this model, the pairing between two introns can bracket the back-spliced exons and induce their circularization.4,7 In addition to the complementary sequences, some RNA-binding proteins (RBPs), such as QKI and FUS, can directly bind to specific RNA motifs in the introns on both sides of the circularized exons.4,21,22 Through the dimerization of these RBPs, the splice sites are brought into close proximity, and the spliceosome engages in a back-splicing reaction. A special class of circRNAs called tRNA intronic circRNAs (tricRNAs) were found to be generated during pre-tRNA splicing. In this case, the tRNA splicing endonuclease complex cleaves an intron at the bulge-helix-bulge motif of the pre-tRNA, and the released ends are ligated to form a tRNA and a tricRNA.23
Figure 1.
The biogenesis of circRNAs
(A) In lariat-driven circularization, the intron lariats depend on the GU-rich sequences close to the 5′ splice site and the C-rich sequences near the branch point to form ciRNA, while the exon-containing lariats are processed by internal splicing to release EcircRNAs or EIciRNAs. (B) In intron pairing-driven circularization, the back-splicing event is guided by repeated complementary elements, such as Alu repeats. (C) In RBP-driven circularization, the back-splicing event can be promoted by RBPs. (D) TricRNAs are synthesized from introns spliced from pre-tRNA.
Regulation of parental gene expression by circRNAs
As circRNAs can interface with DNA or RNA at the sequence level and fold into a unique tertiary structure capable of specific interactions with proteins, they are particularly well suited to regulate gene expression at multiple levels. Indeed, recent studies have uncovered many specific examples illustrating the involvement of circRNAs in their parental gene expression at different regulatory levels (Figure 2), which are summarized in Table 1.
Figure 2.
Regulation of parental gene expression by circRNAs
(A) circRNAs interact with the transcriptional complex to control their parental gene transcription. (B) circRNA biogenesis competes with linear mRNA splicing. (C) circRNAs act as miRNA sponges to facilitate their parental gene expression. (D) circRNAs serve as mRNA traps to inhibit their parental gene expression. (E) circRNAs regulate the translation of parental genes through interaction with proteins. (F) circRNAs regulate post-translational modification of parental genes by encoding proteins.
Table 1.
circRNAs involved in parental gene expression
| Mechanistic classification | circRNA | Parental gene | Mechanism | References |
|---|---|---|---|---|
| Transcriptional regulation | circEIF3J circPAIP2 | EIF3J PAIP2 | interact with RNA Pol II and U1 snRNA | 8 |
| ci-ankrd52 | ANKRD52 | interacts with elongation RNA Pol II | 9 | |
| sisR-4 | dpn | activates an intronic enhancer | 15 | |
| circ-HuR | HuR | interacts with transcriptional factor CNBP | 24 | |
| FECR1 | FLI1 | induces DNA demethylation in the promoter | 25 | |
| circITGA7 | ITGA7 | suppresses the transcriptional factor RREB1 | 26 | |
| circ-DAB1 | DAB1 | upregulates the transcriptional factor RBPJ | 27 | |
| circ-STAT3 | STAT3 | upregulates the transcriptional factor Gli2 | 28 | |
| Splicing regulation | circMbl | MBL | competes with linear mRNA splicing | 29 |
| circSEP3 | SEP3 | favors alternative splicing of SEP3 mRNA | 30 | |
| ceRNA | circ-Sirt1 | SIRT1 | sponges miR-132/212 | 16 |
| circ-ENO1 | ENO1 | sponges miR-22-3p | 17 | |
| cTFRC | TFRC | sponges miR-107 | 31 | |
| circFBLIM1 | FBLIM1 | sponges miR-346 | 32 | |
| circGFRA1 | GFRA1 | sponges miR-34a | 33 | |
| circAmotl1 | Amotl1 | sponges miR-485-5p | 34 | |
| circ-VANGL1 | VANGL1 | sponges miR-605-3p | 35 | |
| cir-ITCH | ITCH | sponges miR-7 and miR-20a | 36 | |
| circSMO742 | SMO | sponges miR-338-3p | 37 | |
| circ-AKT1 | AKT1 | sponges miR-942-5p | 38 | |
| circ-TFF1 | TFF1 | sponges miR-326 | 39 | |
| mRNA trap | HIPK2/3 circRNAs | HIPK2/3 | sequester ATG translation start site | 4 |
| circular Fmn | Fmn | sequesters 5′ UTR exons | 40 | |
| dystrophin exonic circRNAs | dystrophin | lead to inactive dystrophin transcripts | 41 | |
| Translational regulation | circ-Dnmt1 | Dnmt1 | promotes nuclear translocation of AUF1 | 18 |
| circYap | Yap | competitively interacts with eIF4G and PABP | 42 | |
| circPABPN1 | PABPN1 | competitively interacts with HuR | 43 | |
| circ-MMP9 | MMP9 | competitively interacts with AUF1 | 44 | |
| Post-translational regulation | circFBXW7 | FBXW7 | encodes the FBXW7-185aa protein | 45 |
| circ-SPHRH | SPHRH | encodes the SHPRH-146aa protein | 46 | |
| circβ-catenin | β-catenin | encodes the β-catenin-370aa protein | 47 | |
| circ-AKT3 | AKT3 | encodes the AKT3-174aa protein | 48 |
circRNAs regulate the transcription of parental genes
In eukaryotes, transcription of protein-coding genes is performed by RNA Pol II.49, 50, 51 Genes transcribed by RNA Pol II typically require transcriptional regulatory elements (core promoters, proximal promoters, distal enhancers, and silencers) and the molecular machinery (general transcription factors, activators, and coactivators) that interacts with the regulatory elements to mediate precisely controlled patterns of gene expression. A growing body of evidence supports the role of circRNAs in the regulation of their parental gene transcription mediated by the transcriptional complex.8,9,25,26
Some nuclear circRNAs can interact with RNA Pol II or transcriptional factors. For example, ci-ankrd52 accumulates to its sites of transcription, interacts with an elongation RNA Pol II complex, and acts as a positive transcriptional regulator of its parental gene.9 EIciRNAs, such as circEIF3J and circPAIP2, have been found to hold factors such as U1 small nuclear ribonucleoprotein (snRNP) through RNA-RNA interaction between U1 small nuclear RNA (snRNA) and EIciRNA, and the EIciRNA-U1 snRNP complexes subsequently interact with the RNA Pol II transcription complex at the promoters of parental genes to enhance gene expression.8 Another circRNA, circ-HuR, can inhibit the transcription of HuR by repressing the binding of CCHC-type zinc finger nucleic acid-binding protein (CNBP) to the HuR promoter and suppress the growth and aggressiveness of gastric cancer.24
Certain nuclear circRNAs may activate parental gene transcription by inducing DNA hypomethylation in the promoter or by regulating intronic enhancer. For instance, a novel FLI1 exonic circRNA, FECR1, utilizes a positive feedback mechanism to activate FLI1 transcription by inducing DNA hypomethylation in the CpG islands of its promoter.25 Thus, FLI1 can regulate metastasis in breast cancer by using epigenetic mechanisms mediated by its exonic circRNA.25 In Drosophila, a maternally inherited circular stable intronic sequence RNA (sisR-4) promotes transcription of its parental gene by activating an intronic enhancer during embryogenesis.15
Recent studies have shown that several cytoplasmic circRNAs can regulate the expression of transcriptional factors.26, 27, 28 A study in 2018 showed that circITGA7 could promote the transcription of its host gene ITGA7 by suppressing the transcription factor RREB1 via the Ras pathway.26 Chia et al.27 have suggested that circ-DAB1 upregulates the expression of recombination signal-binding protein for the immunoglobulin kappa J region (RBPJ), an important transcriptional factor in the NOTCH pathway, to activate the parental gene DAB1 transcription. Gli2 was previously reported as the transcription factor to activate the expression of its downstream genes.52 More recently, circ-STAT3 was also found to elevate STAT3 expression by upregulating the transcription factor Gli2 via sponging miR-29a/b/c-3p.28 Studies cited above illustrate that circRNA can regulate the transcription of its parental gene and impact distinct steps in the transcription cycle. Given that both cytoplasmic and nuclear circRNAs are involved, transcriptional control could represent a more general role of circRNAs than is currently appreciated. It will be interesting to determine how circRNAs influence their host gene transcription and elucidate the mechanisms underlying these phenomena.
circRNAs affect the splicing of their linear counterparts
As linear splicing and back-splicing mostly use the same pool of canonical splice acceptors and donors, it is not surprising that the processing of circRNAs can affect linear splicing of pre-mRNAs, potentially leading to altered gene expression. Two different mechanisms have been proposed for how circRNAs regulate the splicing of their linear counterparts. (1) circRNA biogenesis competes with linear mRNA splicing. For instance, when spliceosome components are depleted or inhibited, the steady-state levels of circRNAs increase while the expression of their associated linear mRNAs concomitantly decreases.53 This is partly because nascent RNAs are directed to alternative pathways that lead to circRNA production. A study in 2018 revealed that cardiac circRNAs were mostly produced from constitutive exons, indicating that these circRNAs were generated at the expense of their linear counterparts.54 Another example is circMbl.29 This circRNA and its flanking introns contain conserved binding sites of the splicing factor muscleblind (MBL). When the MBL protein is in excess, it binds to the mbl pre-mRNA and causes it to back-splice into circMbl.29 The circMbl then binds to and sequesters MBL protein, lowering its free cellular concentration so that it can no longer produce circMbl transcripts, consequently lowering its own level. Therefore, the circularization and linear splicing of mbl compete against each other.29 (2) circRNA can skew splicing preference and favor alternative splicing of its host gene. For example, circSEP3 is capable of binding SEP3 genomic DNA to form an RNA:DNA hybrid or R-loop, which may physically slow transcriptional elongation, and thereby lead to the formation of alternatively spliced SEP3 mRNA with exon skipping.30
circRNAs act as miRNA sponges to facilitate parental gene expression
miRNAs are important gene expression regulators, binding to the 3′ UTR of target mRNA to prevent its translation or promote its degradation. In the cytoplasm, some circRNAs may function as miRNA sponges to enhance target gene expression by inhibiting miRNA activity. In 2013, two research groups provided the first evidence that the mouse Sry and human CDR1as circRNAs may function as miRNA sponges through their binding sites.3,12 With the extensive studies of circRNAs, an increasing number of circRNAs have been found to act as miRNA sponges and play important roles in disease initiation and progression.55, 56, 57, 58
Considering the homolog of circRNAs with parental genes, it is possible that circRNAs can serve as competing endogenous RNAs (ceRNAs) to regulate their linear counterparts. A recent study reported that circ-Sirt1 binds to miR-132/212 that interferes with SIRT1 mRNA, and it facilitates the expression of host gene SIRT1 in vascular smooth muscle cells (VSMCs).16 As a consequence, circ-Sirt1 exhibits beneficial protective effects against the inflammatory phenotype of VSMCs, suggesting that circ-Sirt1 is involved in the pathogenesis of vascular diseases and may act as a novel potential biomarker in the detection of atherosclerosis.16 Additionally, cTFRC promotes bladder carcinoma progression as a sponge of miR-107 to enhance the oncogenic effect of its parental gene TFRC through the cTFRC/miR-107/TFRC axis.31 In another example, circFBLIM1 may function as a ceRNA to regulate FBLIM1 expression through sponging miR-346 to exert regulatory functions in hepatocellular cancer.32 Besides the above circRNAs, circ-ENO1, circGFRA1, circAmotl1, circ-VANGL1, cir-ITCH, circSMO742, circ-AKT1, and circ-TFF1 have been found to function as miRNA sponges, regulating the expression of their cancer-related parental genes to alter the progression of human cancers.17,33, 34, 35, 36, 37, 38, 39,59 However, the release of target gene repression by competitive binding of circRNA to miRNA requires the levels of both circRNA and miRNA to be comparable. Given that most circRNAs are expressed at low levels, further investigation in diverse cell types or tissues is necessary to determine whether there is sufficient evidence for the regulation of parental gene expression by circRNAs acting as ceRNAs.
circRNAs serve as “mRNA traps” to inhibit parental gene expression
Translation is a complex process of gene expression that determines the abundance of the cellular proteome. Under most conditions, initiation is the rate-limiting step of translation, and it allows rapid, reversible, and spatial control over gene expression. During translation initiation, the cytosolic ribosome is recruited to the mRNA and scans its 5′ UTR for the presence of the translation start codon. However, when the circRNA contains the translation start site of the host gene, it may act as an mRNA trap by sequestering the translation start site, leaving a noncoding linear transcript and thereby reducing the expression level of the functional protein. For example, paralogous kinases HIPK2 and HIPK3 produce abundant circRNAs from their second exon that contains an ATG translation start site, yet the circularization of HIPK2/3 precludes the production of the usual protein-encoding transcript, as it lacks the ATG and significant N-terminal sequence.4 The formin (Fmn) gene, which is essential for limb development in mice, was reported to produce circular Fmn RNAs comprising 5′ UTR exons.40 Targeted deletion of the relevant exons in an animal model abrogates the production of circular transcripts and causes aberrant expression of Fmn proteins.40 Thus, the formation of circular Fmn RNAs may trap the transcripts arising from the Fmn gene in a nonfunctional form and prevent the existence of the linear Fmn transcripts that could be translated. The mRNA trap phenomenon has also been found in patients with Duchenne muscular dystrophy (DMD). A study from Gualandi et al.41 showed that the increased production of the dystrophin EcircRNAs may lead to inactive dystrophin transcripts in individuals with certain deletion mutations and reduce the levels of functional proteins.
circRNAs regulate the translation of parental genes
The synthesis and degradation of circRNAs are regulated by some RBPs.13,60,61 Conversely, circRNAs can interact with different proteins to form specific circRNA-protein complexes that influence the activities of the associated proteins. Certain circRNAs may function as protein sponges to block protein activity by working as competing elements. For instance, circANRIL disrupts pre-rRNA processing and ribosome biogenesis by competitively binding to pescadillo homolog 1, leading to nucleolar stress and activation of p53.10 Additionally, circRNAs may serve as molecular scaffolds for the assembly of protein complexes. A study by Du et al.62 found that circ-Foxo3 could interact with both CDK2 and p21 to form a ternary complex, which suppressed cell cycle progression and cell proliferation.
In some cases, circRNAs can interact with specific proteins and modulate the translation of their cognate mRNAs. Wu et al.42 demonstrated that circYap decreased Yap protein levels by inhibiting Yap translation in cancer cells via its interaction with Yap mRNA and the translation initiation associated proteins, eIF4G and PABP. The complex containing circYap abolished the interaction of PABP on the poly(A) tail with eIF4G on the 5′ cap of the Yap mRNA, which functionally led to the suppression of Yap translation initiation.42 Similarly, the extensive binding of circPABPN1 to HuR prevents HuR binding to PABPN1 mRNA and suppresses PABPN1 translation.43 A recent study showed that circ-MMP9 physically bound to AUF1 to block its inhibitory effect, resulting in enhanced MMP9 mRNA stability, thereby facilitating oral squamous cell carcinoma metastasis.44 Additionally, ectopic circ-Dnmt1 can interact with both p53 and AUF1 to promote their nuclear translocation.18 Nuclear translocation of p53 induces cellular autophagy while AUF1 nuclear translocation increases Dnmt1 mRNA stability and translation.18 Therefore, the ability of circRNAs to bind, sequester, or translocate these translation-associated proteins to particular subcellular fractions allows circRNAs to dynamically regulate the translation of their parental genes.
circRNAs regulate post-translational modifications of parental genes
Most circRNAs cannot be loaded into ribosomes and translated into proteins due to their lack of a 5′ cap structure.6,63 However, given that most circRNAs are cytosolic and originate from protein-coding exons, translation of circRNAs is a natural possibility. The presence of 5′ cap-independent mechanisms of ribosomal recruitment allows synthetic circRNAs to be translated in vivo and in vitro.64, 65, 66, 67, 68 For example, the UTR element of circ-ZNF609, spanning from the termination to the initiation codons, can drive internal ribosome entry site (IRES)-dependent translation.69 In other contexts, N6-methyladenosine (m6A), the most abundant base modification of RNA, was found to promote efficient initiation of protein translation from circRNAs.14
Because the translated product of a circRNA shares part of its amino acid sequence with that of the protein encoded by the parental gene, it may competitively interact with enzymes to regulate post-translational modification of the full-length protein. More recently, circFBXW7 was found to encode the FBXW7-185aa protein to suppress triple-negative breast cancer progression.45 Mechanistic studies demonstrated that FBXW7-185aa competitively interacted with the deubiquitinating enzyme USP28, preventing USP28 from binding to FBXW7, thereby upregulating FBXW7 expression.45 SHPRH-146aa encoded by circ-SPHRH can increase the levels of the full-length SHPRH protein by protecting it from ubiquitination.46 Stabilized SHPRH ubiquitinates proliferating cell nuclear antigen (PCNA), leading to suppression of cell proliferation and tumorigenicity.46 Another study showed that β-catenin-370aa, generated by circβ-catenin, competitively interacted with GSK3β and served as a decoy that prevented GSK3β from binding to full-length β-catenin, leading to the antagonization of GSK3β-induced β-catenin degradation.47 Moreover, Xia et al.48 showed that AKT3-174aa, encoded by circ-AKT3, competed with AKT isoforms to bind to pPDK1 and reduce AKT-Thr308 phosphorylation, suggesting that AKT3-174aa played a negative regulatory role in modulating the phosphatidylinositol 3-kinase (PI3K)/AKT signal intensity. With research on circRNA translation being at an early stage, more new circRNA-encoded proteins will be discovered in the near future, whose roles in regulating post-translational modification of parental genes will be worthy of further study.
Conclusions
To date, the molecular functions of circRNAs remain largely enigmatic. Nonetheless, research during the past decades has clearly demonstrated that circRNAs have roles in multiple aspects of parental gene expression through different mechanisms of action. This review comprehensively summarizes these regulatory mechanisms, including transcription and splicing regulation, ceRNA mechanisms, mRNA traps, and translational/post-translational regulation. In addition, circRNA can also inhibit degradation of proteins encoded by parental genes, as has been proposed for circFoxo3 and circ-GLI1.70,71 Accordingly, these regulatory mechanisms provide a further layer of complexity to the functions of circRNAs and make them potential regulators of specific cellular functions.
While the role of circRNAs in regulating parental gene expression has been elucidated to some extent, many questions remain unanswered. Future studies are required to determine whether this role may be a more general role for circRNAs and whether there may be additional regulatory mechanisms such as affecting chromatin structure and function. To address these questions, further improvements in techniques are necessary to study these circRNAs without affecting their host genes within cells. As their parental genes control a large set of biological processes, the upstream roles of corresponding circRNAs will also affect these pathways. Coupled with the stability and conserved nature of their structure, circRNAs might be a future target for therapies, either to decrease the circularization of functional transcripts or to inhibit the expression of dysfunctional transcripts through the regulatory function of circRNAs. A better understanding of the mechanisms underlying the functions of circRNAs will help us to understand multiple physiological and pathological processes, and to pave the way for circRNA-based therapeutic intervention and diagnostics for human diseases in the future.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81871120 and 82071576), the Natural Science Foundation of Guangdong Province (2019A1515010334, 2019KZDXM059, and 2017KZDXM039), the Yangfan Training Program of Guangdong Province (4YF16006G), and by the Program for Training High-Level Talents of Dongguan (201901019).
Author contributions
Both T.S. and X.X. conceived the idea for the article. T.S. performed the literature search and wrote the manuscript. X.X. and Y.P. revised the manuscript. X.X. supervised the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vicens Q., Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 4.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X.-O., Wang H.-B., Zhang Y., Lu X., Chen L.-L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Zhang X.-O., Chen T., Xiang J.-F., Yin Q.-F., Xing Y.-H., Zhu S., Yang L., Chen L.-L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du W.W., Yang W., Chen Y., Wu Z.-K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 12.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay M.L., Pek J.W. Maternally inherited stable intronic sequence RNA triggers a self-reinforcing feedback loop during development. Curr. Biol. 2017;27:1062–1067. doi: 10.1016/j.cub.2017.02.040. [DOI] [PubMed] [Google Scholar]
- 16.Kong P., Yu Y., Wang L., Dou Y.Q., Zhang X.H., Cui Y., Wang H.Y., Yong Y.T., Liu Y.B., Hu H.J. circ-Sirt1 controls NF-κB activation via sequence-specific interaction and enhancement of SIRT1 expression by binding to miR-132/212 in vascular smooth muscle cells. Nucleic Acids Res. 2019;47:3580–3593. doi: 10.1093/nar/gkz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J., Zhang S., Chen Z., He Z., Xu Y., Li Z. circRNA-ENO1 promoted glycolysis and tumor progression in lung adenocarcinoma through upregulating its host gene ENO1. Cell Death Dis. 2019;10:885. doi: 10.1038/s41419-019-2127-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du W.W., Yang W., Li X., Awan F.M., Yang Z., Fang L., Lyu J., Li F., Peng C., Krylov S.N. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 19.Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. doi: 10.1016/j.celrep.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Eger N., Schoppe L., Schuster S., Laufs U., Boeckel J.N. Circular RNA splicing. Adv. Exp. Med. Biol. 2018;1087:41–52. doi: 10.1007/978-981-13-1426-1_4. [DOI] [PubMed] [Google Scholar]
- 21.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Errichelli L., Dini Modigliani S., Laneve P., Colantoni A., Legnini I., Capauto D. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat. Commun. 2017;8:14741. doi: 10.1038/ncomms14741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Z., Filonov G.S., Noto J.J., Schmidt C.A., Hatkevich T.L., Wen Y., Jaffrey S.R., Matera A.G. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA. 2015;21:1554–1565. doi: 10.1261/rna.052944.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F., Hu A., Li D., Wang J., Guo Y., Liu Y., Li H., Chen Y., Wang X., Huang K. circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer. 2019;18:158. doi: 10.1186/s12943-019-1094-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., Song W., Li X., Li L., Du Z. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Wang J., Zhang C., Lin C., Zhang J., Zhang W., Zhang W., Lu Y., Zheng L., Li X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018;246:166–179. doi: 10.1002/path.5125. [DOI] [PubMed] [Google Scholar]
- 27.Chia W., Liu J., Huang Y.-G., Zhang C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11:372. doi: 10.1038/s41419-020-2572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Song J., Liu Y., Zhou Z., Wang X. Transcription activation of circ-STAT3 induced by Gli2 promotes the progression of hepatoblastoma via acting as a sponge for miR-29a/b/c-3p to upregulate STAT3/Gli2. J. Exp. Clin. Cancer Res. 2020;39:101. doi: 10.1186/s13046-020-01598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Conn V.M., Hugouvieux V., Nayak A., Conos S.A., Capovilla G., Cildir G., Jourdain A., Tergaonkar V., Schmid M., Zubieta C., Conn S.J. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 31.Su H., Tao T., Yang Z., Kang X., Zhang X., Kang D., Wu S., Li C. Circular RNA cTFRC acts as the sponge of microRNA-107 to promote bladder carcinoma progression. Mol. Cancer. 2019;18:27. doi: 10.1186/s12943-019-0951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai N., Peng E., Qiu X., Lyu N., Zhang Z., Tao Y., Li X., Wang Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J. Exp. Clin. Cancer Res. 2018;37:172. doi: 10.1186/s13046-018-0838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He R., Liu P., Xie X., Zhou Y., Liao Q., Xiong W., Li X., Li G., Zeng Z., Tang H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017;36:145. doi: 10.1186/s13046-017-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou R., Lv J., Zhang Q., Lin F., Zhu L., Huang F., Li X., Li T., Zhao L., Ren Y., Xu Y. circAMOTL1 motivates AMOTL1 expression to facilitate cervical cancer growth. Mol. Ther. Nucleic Acids. 2020;19:50–60. doi: 10.1016/j.omtn.2019.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng Z., Zhou W., Duan L., Zhang J., Lu X., Jin L., Yu Y. Circular RNA circ-VANGL1 as a competing endogenous RNA contributes to bladder cancer progression by regulating miR-605-3p/VANGL1 pathway. J. Cell. Physiol. 2019;234:3887–3896. doi: 10.1002/jcp.27162. [DOI] [PubMed] [Google Scholar]
- 36.Huang G., Zhu H., Shi Y., Wu W., Cai H., Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS ONE. 2015;10:e0131225. doi: 10.1371/journal.pone.0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong Z., Zhou C., Wang L., Zhu R., Zhong L., Wan D., Wang Q. Circular RNA SMO sponges miR-338-3p to promote the growth of glioma by enhancing the expression of SMO. Aging (Albany NY) 2019;11:12345–12360. doi: 10.18632/aging.102576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou R., Mo L., Tang H., Leng S., Zhu H., Zhao L., Ren Y., Xu Y. circRNA-AKT1 sequesters miR-942-5p to upregulate AKT1 and promote cervical cancer progression. Mol. Ther. Nucleic Acids. 2020;20:308–322. doi: 10.1016/j.omtn.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan G., Mao A., Liu J., Lu J., Ding J., Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53:e12720. doi: 10.1111/cpr.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chao C.W., Chan D.C., Kuo A., Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 1998;4:614–628. [PMC free article] [PubMed] [Google Scholar]
- 41.Gualandi F., Trabanelli C., Rimessi P., Calzolari E., Toffolatti L., Patarnello T., Kunz G., Muntoni F., Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J. Med. Genet. 2003;40:e100. doi: 10.1136/jmg.40.8.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu N., Yuan Z., Du K.Y., Fang L., Lyu J., Zhang C., He A., Eshaghi E., Zeng K., Ma J. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019;26:2758–2773. doi: 10.1038/s41418-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia B., Hong T., He X., Hu X., Gao Y. A circular RNA derived from MMP9 facilitates oral squamous cell carcinoma metastasis through regulation of MMP9 mRNA stability. Cell Transplant. 2019;28:1614–1623. doi: 10.1177/0963689719875409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye F., Gao G., Zou Y., Zheng S., Zhang L., Ou X., Xie X., Tang H. circFBXW7 inhibits malignant progression by sponging miR-197-3p and encoding a 185-aa protein in triple-negative breast cancer. Mol. Ther. Nucleic Acids. 2019;18:88–98. doi: 10.1016/j.omtn.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 47.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M., Zhang J.F. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia X., Li X., Li F., Wu X., Zhang M., Zhou H., Huang N., Yang X., Xiao F., Liu D. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent kinase-1. Mol. Cancer. 2019;18:131. doi: 10.1186/s12943-019-1056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das G., Hinkley C.S., Herr W. Basal promoter elements as a selective determinant of transcriptional activator function. Nature. 1995;374:657–660. doi: 10.1038/374657a0. [DOI] [PubMed] [Google Scholar]
- 50.Roeder R.G., Rutter W.J. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 51.Juven-Gershon T., Kadonaga J.T. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang D., Wang Y., Xu L., Chen L., Cheng M., Shi W., Xiong H., Zalli D., Luo S. GLI2 promotes cell proliferation and migration through transcriptional activation of ARHGEF16 in human glioma cells. J. Exp. Clin. Cancer Res. 2018;37:247. doi: 10.1186/s13046-018-0917-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.-L., Cherry S., Wilusz J.E. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aufiero S., van den Hoogenhof M.M.G., Reckman Y.J., Beqqali A., van der Made I., Kluin J., Khan M.A.F., Pinto Y.M., Creemers E.E. Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA. 2018;24:815–827. doi: 10.1261/rna.064394.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X., Li H.-D., Bu F.-T., Li X.-F., Chen Y., Zhu S., Wang J.-N., Chen S.-Y., Sun Y.-Y., Pan X.-Y. Circular RNA circFBXW4 suppresses hepatic fibrosis via targeting the miR-18b-3p/FBXW7 axis. Theranostics. 2020;10:4851–4870. doi: 10.7150/thno.42423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Han B., Zhang Y., Zhang Y., Bai Y., Chen X., Huang R., Wu F., Leng S., Chao J., Zhang J.H. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng Z., Yu C., Cui S., Wang H., Jin H., Wang C., Li B., Qin M., Yang C., He J. circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nat. Commun. 2019;10:3200. doi: 10.1038/s41467-019-11162-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang Q., Liu C., Li C.-P., Xu S.-S., Yao M.-D., Ge H.-M., Sun Y.-N., Li X.-M., Zhang S.-J., Shan K. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 2020;130:3833–3847. doi: 10.1172/JCI123353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jia C., Yao Z., Lin Z., Zhao L., Cai X., Chen S., Deng M., Zhang Q. circNFATC3 sponges miR-548I acts as a ceRNA to protect NFATC3 itself and suppressed hepatocellular carcinoma progression. J. Cell. Physiol. 2021;236:1252–1269. doi: 10.1002/jcp.29931. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S.K., Garg A., Bär C., Chatterjee S., Foinquinos A., Milting H., Streckfuß-Bömeke K., Fiedler J., Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park O.H., Ha H., Lee Y., Boo S.H., Kwon D.H., Song H.K., Kim Y.K. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 62.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kozak M. Inability of circular mRNA to attach to eukaryotic ribosomes. Nature. 1979;280:82–85. doi: 10.1038/280082a0. [DOI] [PubMed] [Google Scholar]
- 64.Chen C.Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 65.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., Abe H. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perriman R., Ares M., Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA. 1998;4:1047–1054. doi: 10.1017/s135583829898061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen J., Zhou X., Yang J., Sun Q., Liu Y., Li N., Zhang Z., Xu H. circ-GLI1 promotes metastasis in melanoma through interacting with p70S6K2 to activate Hedgehog/GLI1 and Wnt/β-catenin pathways and upregulate Cyr61. Cell Death Dis. 2020;11:596. doi: 10.1038/s41419-020-02799-x. [DOI] [PMC free article] [PubMed] [Google Scholar]