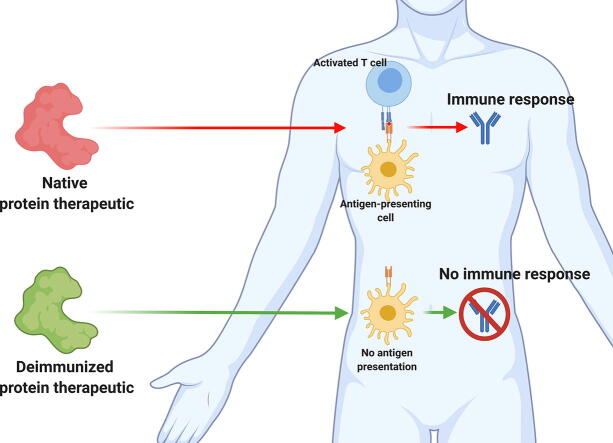
Graphical abstract
Keywords: Protein therapeutic, Immunogenicity, Anti-drug-antibody, T cell epitope, B cell epitope
Abbreviations: ABR, Antigen-binding region; ADA, Anti-drug antibody; ANN, Artificial neural network; APC, Antigen-presenting cell; Bab, Binding antibody; BCR, B cell receptor; CDR, Complementarity determining region; CRISPR, Clustered regularly interspaced short palindromic repeats; DC, Dendritic cell; ELP, Elastin-like polypeptide; EPO, Erythropoietin; ER, Endoplasmatic reticulum; GLK, Gelatin-like protein; HAP, Homo-amino-acid polymer; HLA, Human leukocyte antigen; HMM, Hidden Markov model; Ig, Immunoglobulin; IL, Interleukin; LPS, Lipopolysaccharide; MHC, Major histocompatibility complex; Nab, Neutralizing antibody; NMR, Nuclear magnetic resonance; PAMP, Pathogen-associated molecular pattern; PAS, Polypeptide composed of proline, alanine, and/or serine; PBMC, Peripheral blood mononuclear cell; PD, Pharmacodynamics; PEG, Polyethylene glycol; PK, Pharmacokinetics; PRR, Pattern recognition receptor; PSA, Sialic acid polymers; RNN, Recurrent artificial neural network; SVM, Support vector machine; TAP, Transporter associated with antigen processing; TCR, T cell receptor; TLR, Toll-like receptor; XTEN, “Xtended” recombinant polypeptide
Abstract
Biotherapeutics, and antimicrobial proteins in particular, are of increasing interest for human medicine. An important challenge in the development of such therapeutics is their potential immunogenicity, which can induce production of anti-drug-antibodies, resulting in altered pharmacokinetics, reduced efficacy, and potentially severe anaphylactic or hypersensitivity reactions. For this reason, the development and application of effective deimmunization methods for protein drugs is of utmost importance. Deimmunization may be achieved by unspecific shielding approaches, which include PEGylation, fusion to polypeptides (e.g., XTEN or PAS), reductive methylation, glycosylation, and polysialylation. Alternatively, the identification of epitopes for T cells or B cells and their subsequent deletion through site-directed mutagenesis represent promising deimmunization strategies and can be accomplished through either experimental or computational approaches. This review highlights the most recent advances and current challenges in the deimmunization of protein therapeutics, with a special focus on computational epitope prediction and deletion tools.
1. Introduction
The immune systems of humans and other mammals evolved to defend them from pathogenic microbes, invading viruses or other substances that are not beneficial or may cause harm. Additionally, the immune system has the ability to discriminate self from non-self [1]. Also, tolerance of commensal organisms, indispensable for our body, is an essential competence of the immune system [2].
In order to use proteins, including enzymes, as therapeutic agents to tackle complex and persistent diseases, it is necessary to avoid a strong immune response to these agents as a result of the treatment.
Systemic application of therapeutic proteins could not only lead to unpredictable pharmacokinetics (PK) and pharmacodynamics (PD) due to an immune response, the induction of anti-drug antibodies (ADAs) [3] and loss of efficacy, but also to anaphylactic responses, hypersensitivity, and other complications [4], [5].
The immune system can be divided into the innate and adaptive immune system. Components of both systems interact and play important roles in an immune response [6]. The innate immune response describes an unspecific defense mechanism, which is germ-line encoded and includes physical barriers, soluble proteins as well as cellular components such as phagocytes and natural killer cells [7]. In contrast, the adaptive immune response is characterized by its high specificity for target antigens [8].
1.1. Innate immune system
The vanguard of every specific immune reaction is a non-specific one. Antigen-presenting cells (APCs) play a crucial role and can be found in tissues where they endocytose extracellular material to scan the environment for harmful substances [9]. Pattern recognition receptors (PRR) such as toll-like receptors (TLR) expressed by dendritic cells (DCs) or macrophages enable the innate immune system to differentiate harmful from harmless substances. PRRs recognize conserved structures of microbial or viral origin called pathogen-associated molecular patterns (PAMPs) [10]. PAMPs detected by TLRs include, for example, bacterial lipopolysaccharides (LPSs), bacterial flagellin, peptidoglycan, and viral DNA, which are recognized by TLR4, TLR5, TLR2, and TLR9, respectively [11]. Since potential pathogens are able to reside either extra- or intracellularly, PRRs can be found on the cell surface as well as in the cytoplasm of host cells. While important for the immune response, the innate immune system is not a target of deimmunization strategies based on T or B cell epitope prediction and deletion, which represent the major focus of this review.
1.2. Adaptive immune system
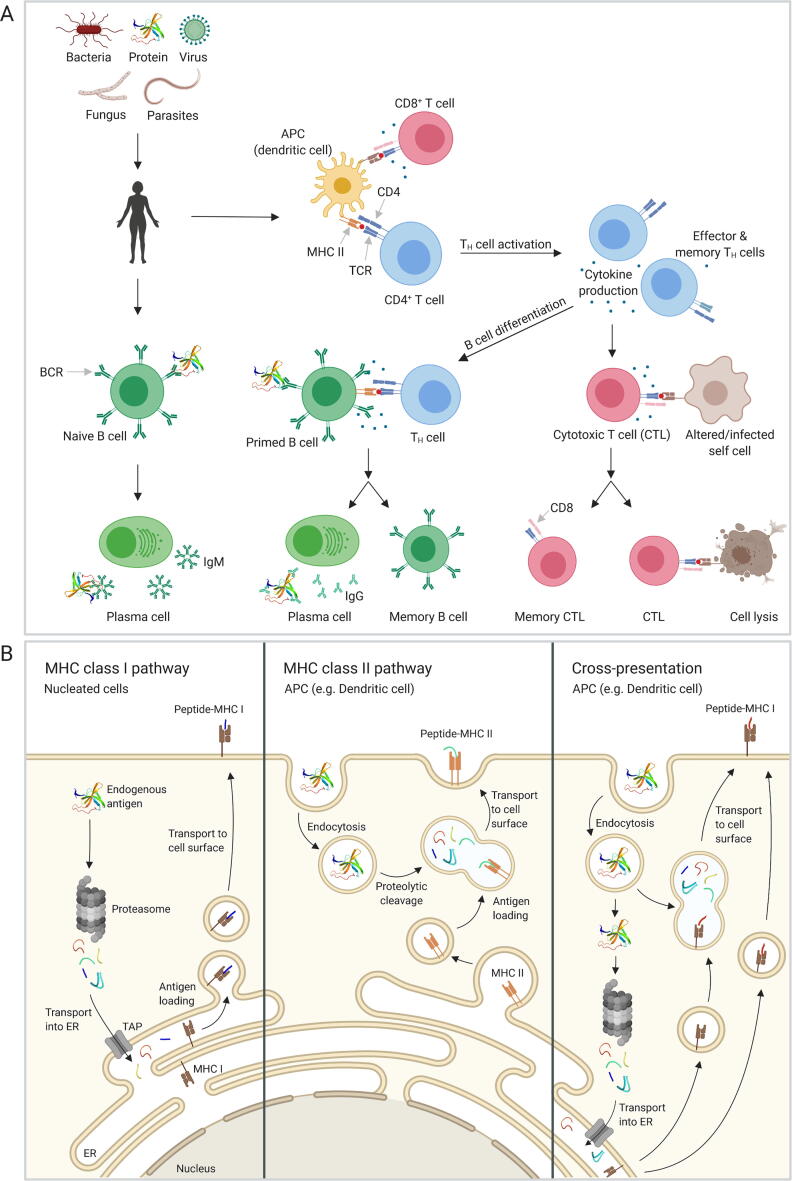
The adaptive immunity is controlled and activated through the three-signal paradigm which includes the antigen, co-stimulation, and inflammatory cytokines [12] though recent studies suggest an expansion of this model, including factors such as innate cytokines (IL-1 family), commensal-derived metabolites and tissue-specific parameters [13]. The initial activation of the adaptive immune system is mediated by APCs, which present antigen bound to the major histocompatibility complex (MHC) to cells of the adaptive immune system, which includes T and B cells (Fig. 1A). The main concern when designing non-immunogenic biotherapeutics is the activation of those cell types.
Fig. 1.
Overview of the adaptive immune system and antigen processing pathways. A) Antigen presenting cells (APCs) are constantly sampling their extracellular environment. Antigens are endocytosed, proteolytically cleaved, and presented on the cell surface bound to MHC class II. CD4 T helper cells with the matching T cell receptor (TCR) are activated through the interaction with the antigen-MHC class II complex. This leads to cytokine production and differentiation and activation of cytotoxic T cells. Activated CD4 T cells and cytokine secretion lead to differentiation and class switch of B cells into plasma cells secreting highly specific antibodies and long-lived memory B cells. B) In the MHC class I pathway, endogenous proteins are proteolytically processed by the proteasome and transported into the endoplasmic reticulum (ER) via the TAP channel. Antigens are loaded onto MHC class I molecules and transported via the Golgi to the cell surface. In the MHC class II pathway, endosomes containing an endocytosed protein fuse with lysosomes leading to degradation of the protein by proteases activated with decreasing pH. The endolysosme fuses with a vesicle containing the MHC class II molecule, and the antigen is loaded. The complex is transported to the cell surface and exposed for interaction with CD4 T cells. The cross-presentation pathway is only found in APCs and allows to present exogenous antigens bound to MHC class I molecules. The peptides are generated either by degradation through the proteasome or by fusion of the vesicle with a lysosome and cleaved by proteases. (Created with BioRender.com).
1.2.1. T cell response
T cells originate from bone marrow-derived progenitor cells and undergo further positive and negative selection in the thymus eventually resulting in naïve CD4+ and CD8+ T cells [14]. CD8+ T cells are activated by interaction of the antigen presented by the MHC class I on the cell surface of all nucleated cells with the T cell receptor (TCR). In contrast, CD4+ T cells are activated by the interaction of the TCR with MHC class II present on APCs such as DCs, macrophages, or B cells. A crucial part in T cell reactivity is the way proteins are processed into peptides that are eventually presented as antigens (Fig. 1B) and the binding specificity of MHC towards these antigens.
1.2.1.1. Antigen processing and MHC presentation
The MHC class I complex presents processed cytosolic antigens, typically the result of viral infection and replication, on the surface of nucleated cells (Fig. 1B) [7]. Endogenous proteins in the cytosol are degraded by the proteasome. When stimulated by interferons that are released during the onset of an immune response, the proteasome is converted to an immunoproteasome by incorporation of three alternative β subunits. The immunoproteasome favors the production of peptides with hydrophobic or basic C termini, which constitute important anchor residues for binding to MHC class I [15]. The generated protein fragments are transported into the endoplasmic reticulum (ER) by the TAP (transporter associated with antigen processing) channel [16]. In the ER, the peptide associates with MHC class I and is transported via the Golgi and transport vesicles to the cell surface to allow interaction with T lymphocytes [17]. The average length of peptides produced by the proteasome is 7–9 amino acids, but peptides can range from 4 to 25 residues [16]. The binding of the antigen to the MHC complex is another important step in antigen presentation. The average length of antigens that are loaded onto the MHC class I complex range from 8 to 10 amino acids [16]. In general, MHC class I prefers hydrophobic C-terminal residues which, as stated above, are more likely to be generated by the immunoproteasome [18]. The identities of residues lining the peptide-binding groove and thereby conferring specificity are dependent on the allelic variant of the MHC class I [19].
In contrast, MHC class II presents extracellular proteins on the surface of APCs (Fig. 1B). APCs take up proteins by endocytosis into endosomes. These endosomes fuse with lysosomes, which results in cleavage of the protein into peptides. Lysosomes contain more than 60 different enzymes, which are synthesized by the ER [20], [21]. Therefore, a precise analysis and the attempt to predict antigen cleavage sites and preferences is an almost impossible task. The strength of the immune response against some peptides depends on the allelic variations of the MHC class II molecules that confer peptide specificity. Peptides eliciting a strong immune response are called immunodominant peptides [22]. In contrast, epitopes that are targeted to a lower degree are called subdominant epitopes. The size of peptides bound to MHC class II varies between 11 and 30 amino acids [23]. The positions and identities of peptide amino acid side chains interacting with the peptide-binding groove are dependent on the allelic variant of the MHC class II. An important difference between MHC class I and MHC class II are the closed and open binding grooves, respectively. This also influences length of the peptide and binding specificity [24].
Besides the classical MHC class I and MHC class II pathways, extracellular antigens (typically class II antigens), which have made it into an APC, can also undergo a special pathway termed cross-presentation, which results in their surface presentation by the MHC class I complex (Fig. 1B) [25].
In summary, T cells possess the ability to react to substances acquired intra- or extracellularly. Moreover, T cells are able to kill these infected cells, bacteria or even parasites. However, of higher relevance for the scope of this review is the negative effect of T cells on biotherapeutics due to their key upstream role in the generation of ADAs [26].
1.2.2. B cell response
B cells are mainly involved in the production and secretion of either short-lived antibodies with lower affinity (e.g., IgM) or longer-lived antibodies with much higher affinity (e.g., IgG). This part of the immunity belongs to the humoral immunity comprising antibodies in blood plasma and lymph, in addition to the complement system. The immune response to proteins is mediated through specific antibodies, which can be T cell-dependent or -independent [27]. The T cell-independent response is induced by B cells recognizing repeated patterns, such as polymeric repeats or carbohydrate molecules, in a foreign protein or other molecule and react with the production of low affinity IgM antibodies [28]. A T cell-dependent response refers to specific antibody production, foremost IgG, by B cells, which are stimulated by cytokines secreted by T cells. A major difference between B and T cells is the antigen type recognized by the cells. While T cells recognize processed antigens presented by the MHC complexes, B cells recognize proteins in their native state with their B cell receptor (BCR) [29]. When the BCR encounters its antigen, the antigen is taken up by the B cell through receptor-mediated endocytosis and digested [30]. The antigen is then displayed on the surface bound to MHC class II as described above for other APCs. This enables the B cell to get T cell help and induce class switching (i.e., production of other antibody classes such as IgG, IgA, or IgE instead of IgM) and differentiation into a plasma cell. While IgG is important for antibody-based immunity against invading pathogens, IgA prevents colonization of mucosal areas by pathogens, and IgE is involved in triggering allergic reactions [31]. These cells produce high amounts of antibodies against the presented antigen. Little is known about the antigen-processing pathways and peptide-MHC class II presentation in B cells. Therefore, it is not possible to make predictions regarding preferred cleavage sites and processing details on which deimmunization of biotherapeutics could be based.
1.3. Immunogenicity of biologics
Immune response-mediated reactions represent an increasing concern for the pharmaceutical industry, predominantly for biotherapeutics that possess enzymatic or regulatory activity (e.g. Insulin, protein C, adenosine deaminase, human albumin) and are used for long-term treatments. Adverse reactions towards protein drugs, such as immunogenicity and the production of ADAs against biotherapeutics can have various negative effects. ADAs can either be binding (BAb) or neutralizing Antibodies (NAb) [32] both of which can result in decreased efficacy and induction of allergic reactions [33]. Differences are that NAbs bind to epitopes which are functionally relevant, whereas BAbs bind to epitopes which are generally not participating in the receptor/target interaction of the therapeutic [34]. Patient factors such as age, disease and immune status, as well as therapeutic factors including molecular structure, amino acid sequence, nature of the target protein, degree of humanization and impurities play a role in how fast and efficiently ADAs are produced against a certain biotherapeutic [32].
In general, the detection of ADAs defines the immunogenicity of a certain drug. Reduction of efficacy and bioavailability or even neutralization of the drug are effects that foremost interfere with the treatment. More severe and dangerous for the patient are life-threatening immune reactions, hypersensitivity or cross-reactions with endogenous proteins, which could lead to deficiency syndromes [35]. Numerous factors influence the immunogenicity of a drug that can be classified into three main groups: treatment-, patient-, and drug property-associated factors [36]. The route and frequency of administration are treatment-associated factors, whereas compromised immune system function and general health condition are part of the patient-associated factors. Also, the polymorphism of the MHC genes that are affecting the intensity of the T cell-dependent immune response is a patient-associated factor [37]. Glycosylation patterns, concealment or removal of MHC epitopes, and impurities and contaminants in the production steps are drug property-associated factors influencing immunogenicity [36].
2. Protein deimmunization methods
Several strategies for deimmunization of biotherapeutics, mainly aiming at mitigating immunogenicity, are available. These include shielding approaches such as PEGylation, XTENylation or PASylation as well as deletion of T or B cell epitopes.
2.1. Shielding methods
2.1.1. PEGylation
The attachment of polyethylene glycol (PEG) polymers to drugs and larger biotherapeutics can be used to mask surface epitopes of the agent from the host immune system. PEGylation is achieved by incubation of chemically activated derivatives of PEG with the target molecule. Strands of the polymer are bound to the structure of the target resulting in circulatory half-life extension [38] increased water solubility due to a stable hydration layer through hydrogen bonding to water molecules [39] and reduced immunogenicity of the shielded therapeutic [40]. The extension of circulatory half-life is mostly attributed to the increase of the hydrodynamic volume leading to a decrease in glomerular filtration [38]. Despite several pharmacological advantages, some drawbacks of PEGylation have also been reported. A possible decrease in biological activity due to steric hindrance imposed by the PEG molecules [38] is one of them. More severe are altered biological properties of the conjugated species [40]. In addition, the prolonged use of PEGylated drugs can lead to accumulation of non-biodegradable PEG in the liver and other organs [41] and treatment with PEGylated drugs can give rise to anti-PEG antibodies, which have been correlated with loss of therapeutic efficacy and a general increase in adverse effects [42]. The limited understanding of anti-PEG immunity demands both efforts in research and approaches beyond PEGylation such as nanogel encapsulation [43] or polypeptide fusion [44].
2.1.2. XTENylation
In contrast to PEGylation, XTENylation is achieved by genetic fusion of XTEN polypeptides to the target protein [44]. These polypeptides are negatively charged, consist of proline, alanine, serine, threonine, glycine, and glutamine in a non-repetitive manner, and increase the hydrodynamic radius of the target protein. They were developed to mimic the beneficial properties of PEG on the one hand and avoid disadvantages such as production of anti-XTEN antibodies and the lack of biodegradability on the other hand [45]. XTEN was reported to be biodegradable in several mammalian species and therefore, the risk of accumulation could be eliminated in contrast to PEG-derived conjugates [44]. Among the advantages of this genetic fusion approach are expression, purification and characterization of the modified proteins as single molecules [46]. More recently, chemical XTENylation has been achieved with high selectivity [46]. One potential challenge of XTENylation is the possible introduction of new epitopes at the linker region between the protein and the polypeptide.
2.1.3. PASylation
Another polypeptide increasing the hydrodynamic volume and used for protein shielding is PAS, which consists of the amino acids proline, alanine and serine [47]. Despite this conceptual similarity, PAS and XTEN differ in amino acid composition and charge, which results in different pharmacological behaviors [47]. A drawback of PASylated therapeutics is the observed tendency for aggregation at higher concentrations [46]. The effect of PAS aggregates on PD and PK have to be assessed in more detailed studies. An advantage compared to PEGylation is that no immunogenicity has been been reported to date [48]. Depending on the length of the linker between PAS and a therapeutic enzyme, the catalytic activity of the enzymes was reduced only slightly compared to the non-PASylated version [49].
Similar polypeptide fusion techniques inlcude ELPylation [50] HAPylation [51] or treatment with gelatin-like protein (GLK) [52].
2.1.4. Reductive methylation
With reductive methylation, a conversion of primary to tertiary dimethyl amine is achieved. The aim is primarily to stabilize the therapeutic protein in the cytosol and decelerate the degradation by evading host-mediated ubiquitination [53]. The stability of many proteins in the cytosol follows the N-end rule, stating that the susceptibility of proteins and peptides to ubiquitination is dependent on the N-terminal residue identity [54]. Furthermore, it was shown that proteins with lower levels of lysine evade host-mediated ubiquitination more efficiently [55]. Therefore, reductive methylation of lysines can increase the cytosolic stability of protein drugs. Simultaneously, it can also reduce their neutralization by antibodies, as has been shown for an anti-cancer fusion protein consisting of the anthrax lethal factor and the catalytic domain of Pseudomonas exotoxin A [53]. However, the applicability of reductive methylation to reduce immunogenicity of therapeutic enzymes has yet to be demonstrated.
2.1.5. Glycosylation
The covalent attachment of carbohydrates to proteins is a natural way for protein stabilization in biological systems [56]. The carbohydrates can be covalently attached to either asparagine (N-glycosylation) or threonine/serine residues (O-glycosylation). Both N- and O-linked glycosylation have been used to extend the half-lives of approved drugs, and N-glycosylation was shown to reduce immunogenicity [51]. The N-glycosylated erythropoietin (EPO) analog Darbepoietin alfa has a 3-fold increased half-live compared to the native EPO [57]. A reduction of immunogenicity due to the interaction of N-glycans with multiple glycan-binding proteins, such as the asialoglycoprotein receptor or the mannose 6-phosphate receptor, has been demonstrated [58]. However, aberrant glycosylation of new potential therapeutic proteins could lead to pathological conditions [59] such as choriocarcinoma [60]. Furthermore, production of recombinant proteins in nonhuman cells can lead to the recognition of nonhuman glycans by pre-existing antibodies in the serum such as antibodies against α-1,3-linked galactose or α-1,3-linked fructose [61], [62].
2.1.6. Polysialylation
Sialic acid polymers (PSA) are naturally occurring and highly biodegradable. The recognition of sialic acids by sialic acid-binding immunoglobulin-type lectins (SIGLECs) can lead to an altered (and possibly reduced) immune response [63]. Several drugs and enzymes, such as asparaginase [64] or tetrameric butyrylcholinesterase [65] have been successfully polysialylated and show reduced immunogenicity as well as improved PK and PD. Also the polysialylation of EPO and DNAse I resulted in improved stability and/or an increased circulation half life, whereas effects on immunogenicity of these therapeutics remain to be shown [66]. In this context, it should also be noted that extension of the half-life of a therapeutic protein can impact its immunogenicity.
Despite their benefits, protein therapeutics modified by shielding methods can provoke immune reactions against the shielding moieties, which have to be assessed in extensive PK and PD studies. Also, shielding methods as listed above, albeit viable for therapeutic proteins, peptides, and hormones such as insulin [51], [67] could cause a reduction in activity of therapeutic enzymes, e.g. due to steric hindrance. Furthermore, the primary objective of the methods mentioned above is the extension of circulation half-life and not the reduction of immunogenicity of a biotherapeutic.
2.2. Humanization of therapeutic proteins
While not a major focus of this review, humanization is another strategy to deimmunize therapeutic proteins and in particular, therapeutic antibodies [68]. When antibodies produced by a non-human immune system (e.g., by rodents) are used as therapeutics in humans, they can elicit an immune response. To circumvent this problem, such exogenous antibodies can be “humanized”, e.g., through various grafting approaches aiming at the transfer of the antigen binding function of an exogenous antibody to a human antibody scaffold [69]. This can be achieved by inserting portions of the complementarity determining region (CDR) of the exogenous donor antibody to a human acceptor antibody by recombinant DNA techniques. This is not to be confused with the construction of chimeric antibodies, in which the entire variable region of a human antibody is exchanged with that of an exogenous antibody [68]. While classical CDR-based grafting approaches for therapeutic antibody humanization have been employed successfully in the past, they also face certain challenges such as possible reductions in stability or antigen affinity. These problems could possibly be mitigated by computational approaches such as “Computationally-Driven Antibody Humanization” (CoDAH), which aim at maintaining structural stability and achieving humanization simultaneously by selecting and incorporating sets of amino acids from human germline sequences [69]. Using DNA shuffling or other protein engineering techniques, humanization has not only been accomplished for antibodies but also for other proteins such as therapeutic enzymes [70], [71].
2.3. T cell epitope prediction and deletion
Immune responses against biotherapeutics often result in ADAs. T cell-dependent antibody responses can evoke class switch, affinity maturation and induction of long-lived memory B cells and plasma cells [72]. Therefore, identification, prediction and deletion of T cell epitopes of a biotherapeutic protein can not only reduce its immunogenicity but moreover increase half-live, bioavailability and efficacy, resulting in overall better pharmacodynamics and -kinetics. The recognition of specific T cell epitopes by an individual depends on both genetic factors (due to the high allelic polymorphism in the human leukocyte antigen (HLA) locus, which encodes the MHCs), and the environmental exposure history of the individual [73], [74]. Distinct binding specificities lead to the presentation of diverse epitopes by different individuals of the human population, which makes it hard for pathogens but also therapeutic proteins to evade recognition by our immune system [73].
2.3.1. T cell epitope prediction
T cell epitope prediction in general aims to identify the shortest peptides within an antigen that are able to stimulate T cells [75]. Analysis of epitope binding showed that some positions in the peptide only tolerate very similar amino acids in order to retain binding affinity [76], [77]. These positions are called anchor positions and have similar spacing in various epitopes recognized by the same MHC. Therefore, MHC ligand motifs are defined as the sum of anchor position spacing and specificity [77]. The anchor residues are usually at positions P2 and P9 for MHC class I molecules [78] and at positions P1, P4, P6, and P9 for MHC class II molecules [79]. The MHC ligand motif is defined by the binding groove forming the epitope-binding site in MHC molecules, which is highly variable in the population [80], [81], [82]. Elution data of MHC class I ligands revealed conserved anchor positions and homogenous peptide length [76]. This allowed fast advances to define various MHC class I ligand motifs based on eluted epitopes. In contrast, the open peptide-binding groove of MHC class II leads to ligands with variable length. The anchor positions of eluted ligands are therefore out of frame and much harder to interpret [74]. MHC ligands need to have a certain affinity for their corresponding MHC in order to be recognized as a T cell epitope. The affinity measurement values defined as thresholds for MHC class I and MHC class II amount to an IC50 < 500 nM [83] and an IC50 < 1000 nM [84] respectively.
The vast amount of possible peptide-MHC combinations quickly led to the need of computational prediction tools (Table 1). The various prediction algorithms developed over the last thirty years have been reviewed in detail by Peters et al., 2020 [74]. The development of computational tools to predict T cell epitopes started in 1989 with a program that was able to identify putative ligands in a protein of interest using allele-specific ligand motifs [85]. Another way to approach T cell epitope prediction was the use of matrices containing quantitative scores. For every amino acid, a score was assigned for each position in the binding groove. The values for each position in a peptide were summed up giving a score for the specific combination of peptide and MHC [74]. Well known is the SYFPEITHI score based on ligand elution data [86]. More complex methods to predict MHC class I epitopes including artificial neural networks (ANN) and hidden Markov models (HMM) followed shortly after [87], [88], [89]. Due to only limited data available to train those algorithms, prediction was only successful for a small range of alleles [74]. The need of larger datasets to efficiently train the algorithms led to the initiation of several databases [86], [90]. The Immune Epitope Database (IEBD) contains information on T cell and B cell epitopes compiled from experiments and literature and is updated regularly [91], [92]. The IEDB-Analysis Resource (IEDB-AR) contains several T and B cell prediction and epitope analysis tools, which were trained on the IEDB [93]. The most recent database SysteMHC collects raw and analyzed mass spectrometry data of MHC ligand elution [94]. Those databases allowed training and evaluation of machine learning-based prediction tools. Advances in MHC class I epitope prediction were faster than MHC class II as the open binding groove of MHC class II adds complexity to the prediction [74]. The ANN-based NetMHC algorithm outperformed other MHC class I prediction tools, especially when training data sets were small. The difference was explained by the additional information on amino acid similarity included in the NetMHC tool allowing for extrapolation of residues not found in the training data [74], [95]. NetMHCII is the equivalent tool for MHC class II epitope prediction based on the NN-align algorithm [96], [97].
Table 1.
T cell epitope prediction tools.
| Name of the tool (latest version) | Method | Year of development | Latest version |
|---|---|---|---|
| Unnamed | MHC-specific motifs | 1989 [85] | |
| SYFPEITHI | Ligand elution data, average relative binding matrices | 1999 [86] | |
| TEPITOPE | Virtual matrices | 1999 [99] | |
| ProPred | Virtual matrices | 2001 [128] | |
| NetChop 3.1 | ANN | 2002 [191] | 2005 [107] |
| Unnamed | HMM | 2002 [192] | |
| NetMHC 4.0 | BLOSUM matrices, ANN | 2003 [193] | 2016 [78] |
| Unnamed | Gibbs sampling approach | 2004 [194] | |
| SMM | Stabilized matrix method (SMM) | 2005 [195] | |
| DynaPred | SVM-trained, quantitative matrix-based method | 2006 [196] | |
| SVMHC | Support vector machine | 2006 [197] | |
| SVRMHC | Support vector machine regression (SVR) models | 2006 [198] | |
| NetMHCII 2.3 | ANN | 2007 [199] | 2017 [97] |
| NetMHCpan 4.1 | ANN | 2007 [100] | 2020 [200] |
| NetMCHIIpan 4.0 | ANN | 2008 [102] | 2020 [201] |
| SMM-PMBEC | Amino acid similarity matrix | 2009 [202] | |
| PickPocket 1.1 | Position-specific weight matrices, binding pocket matrix extrapolation | 2009 [203] | |
| EpiSweep | Combines epitope prediction and deletion | 2010 [129] | 2017 [137] |
| NetMHCcons 1.1 | ANN, matrix based | 2012 [204] | |
| Unnamed | Immunogenicity data | 2013 [114] | |
| NetTepi 1.0 | ANN, matrix based, immunogenicity data | 2014 [205] | |
| NetMHCstabpan 1.0 | ANN | 2016 [206] |
ANN, artificial neural network; HMM, hidden Markov model;
To date, over 27′000 HLA alleles are described in the IMGT-HLA database [98]. This enormous variation makes it impossible to generate enough experimental data for every allele to train prediction algorithms. Therefore, pan-specific prediction tools able to extrapolate to experimentally uncharacterized MHC molecules were developed. The first tool, which successfully predicted binding for HLA-DR alleles computationally was TEPITOPE [99]. It uses virtual matrices constructed by comparing the sequence of the binding pocket to the pockets of MHC molecules with previously defined binding specificity [99]. The best performing pan-specific algorithm for MHC class I epitopes is NetMHCpan including information about the amino acids in the binding groove in addition to the peptide binding data [100]. Similar tools have been developed for MHC class II, including binding data and information on binding environment in order to predict epitopes for all class II alleles with known sequence [101], [102], [103]. The vast amount of HLA alleles is also challenging when it comes to the decision on which alleles to include in a specific study. In most cases, studies attempt to cover a broad range of the human population. It was found that MHC molecules can be grouped into supertypes based on similar binding specificities leading to ten MHC class I and ten MHC class II supertypes [104], [105]. Including representatives of different supertypes allowed to cover the most common allelic variants of most populations [106].
Efforts were made to analyze the benefit of including information regarding antigen processing (e.g., by the proteasome) in epitope prediction tools. It seems that MHC molecules have co-evolved to efficiently bind peptides generated by the proteasome and transported by TAP [107]. Therefore, improvements for MHC class I were not significant when antigen processing information was included in the prediction algorithms [107]. The same was found for MHC class II antigens, as the terminal regions of the epitopes are not well defined [108]. Furthermore, mass spectrometry data on ligand elution has led to the identification of unusual peptides. Those peptides seem to be spliced together [109], [110] or are longer than typical MHC class I ligands [111], [112]. Those unusual ligands are most likely missed by prediction tools, but conventional T cell epitopes seem to represent the majority of the ligands and can therefore be prioritized for prediction [74].
Limitations of in silico T cell epitope prediction include factors difficult to implement in algorithms such as aspects of APC function, including antigen processing and presentation of peptides to T cells, and T cell activation. One recent report described a method named ITCell, which integrates information about three consecutive stages of the antigen presentation and T cell activation pathway, i.e., (i) antigen cleavage, (ii) MHC class II binding, and (iii) recognition of peptide-MHC-II complexes by a given TCR. This integrated approach proved to be more accurate than current single-stage epitope prediction tools [113]. Still, in vitro and ex vivo assays, testing for APC function and T cell response are very important to account for incomputable biological factors. First attempts were made to divide peptides into immunogenic and nonimmunogenic peptides [114], [115]. A combination of in silico prediction methods with experimental approaches including assays with peripheral blood mononuclear cells (PBMCs) from healthy donors is probably the most promising path for epitope detection.
2.3.2. T cell epitope deletion
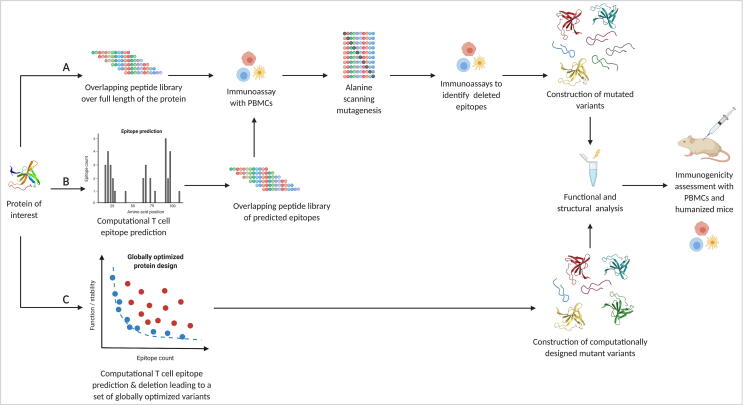
Suppressing antidrug antibodies by disruption of the immune response pathway by T cell epitope deletion has profiled itself as a solid strategy. A selective substitution of amino acids in a biotherapeutic without changing its properties as a drug or an enzyme can disrupt the delicate binding interfaces between the antigen and the T cell and therefore delete these epitopes. Such substitutions can either be introduced and tested experimentally or designed computationally [116] (Fig. 2).
Fig. 2.
Overview of T cell epitope deletion workflows. T cell epitope deletion transitioned from being purely experimental to more and more computational. A) This pathway illustrates the steps for experimental T cell epitope deletion. The most common way was to synthesize an overlapping peptide library over the full length of the protein of interest. Epitopes among those peptides are identified through ex vivo immunoassays with peripheral blood mononuclear cells (PBMCs) from a broad range of healthy donors. The immunogenic peptides are subjected to alanine scanning mutagenesis and then tested again for immunogenicity with the PBMCs. Confirmed deimmunizing mutations can then be combined and introduced in the full length protein. Introducing mutations comes with a high risk of misfolding and inactivation of the protein. Therefore, functional and structural characterisation is needed. Deimmunization success of a functional protein can be tested and validated in humanized mice and with PBMCs. B) The use of epitope prediction reduces the number of peptides to be tested for immunogenicity by only testing overlapping peptides in predicted epitope regions. The subsequent steps are identical to the experimental strategy. C) The computational approach combines epitope prediction with epitope deletion in one program by introducing mutations in silico while taking into account the global effect of those changes. Independent of the strategy chosen, deimmunized variants have to be produced and tested experimentally. (Created with BioRender.com).
2.3.2.1. Experimental
Strategies to substitute amino acids in order to disrupt antigen binding are largely based on scanning alanine or trial and error mutagenesis. The experimental approach includes several steps (Fig. 2): First, a complete panel of overlapping peptide fragments has to be synthesized in order to map epitopes by immunoassays. PBMCs from several different donors are needed in order to cover a certain genetic diversity and obtain a representative picture of the different HLA alleles present in the population. Second, alanine scanning or similar mutagenic deletions of identified epitopes are performed followed by further PBMC immunoassays. The confirmed epitope-deleting mutations are then combined and introduced into the protein. Tests for functional and structural consequences of these mutations are performed as final step [117]. Mutagenic substitution of lysine, arginine, glutamine and glutamic acid residues, which have been shown to contribute to antibody epitopes, helped to successfully deimmunize the diphtheria toxin [118]. Therapeutic enzymes and cofactors with engineered reduced immunogenicity such as E. coli type II asparaginase proved that the deimmunization of enzymes by mutation is possible without losing its biological activity. Introducing two mutations (K228S and Y176F) was enough to get rid of undesirable glutaminase activity, increasing cytotoxicity towards leukemic cells and in addition reduce reactivity with immune serum [119]. Experimental approaches for T cell epitope deletion have produced several partially deimmunized biotherapeutics such as Factor VIII domain C1 [120] interferon beta [121] or β-lactamase [122] to mention some early examples. One study combined in silico prediction and experimental approaches to successfully remove an immunodominant T cell epitope in the Factor VIII C2 domain [123]. A more recent achievement in deimmunizing a biotherapeutic was the reduction of immunogenicity of Hirudin III [124]. This approach was based on in silico sequence analysis to decrease HLA-DR-binding affinity. This was followed by confirmation of these epitopes by in vitro biochemical evaluations and confirmation of bioactivity. Hirudin III showed a reduction in immunogenicity for several alleles of the HLA-DR group [124].
The discovery of CRISPR-Cas9 and the possibilities that it opened for genome engineering and editing [125] was hoped to bring research closer towards personalized gene therapies for complex diseases. However, the administration of Cas9 proteins in mice often resulted in immune responses [126] and therefore, raised concerns regarding its safety as a therapeutic protein. A recent study [127] demonstrated that the Cas9 protein could be modified to eliminate immunodominant MHC class I epitopes through targeted mutation of the anchor residues while preserving its function and specificity. However, the deimmunization and prediction algorithm was optimized only for the HLA-A*02:01 allele. A reduction in immunogenicity covering a broader range of HLA alleles remains to be shown. This study combined in silico T cell epitope prediction with ex vivo epitope mapping [127].
Although improvements have been made over the past years by increasing throughput and efficiency, experimental T cell epitope deletion is still time-intensive, work-intensive, and expensive. Therefore, combining T cell epitope prediction tools with computational protein design methods could reduce the costs significantly and increase the hit rate. For instance, instead of relying on merely experimental alanine scanning, in silico methods can be used to guide amino acid substitution decisions, and then only the best in silico candidates need to be tested experimentally. Moreover, it is almost impossible to fully account experimentally for the allelic diversity of HLA class II present in a population of potential patients by relying on donor blood samples from random individuals. However, incorporating the supertype allele concept described in the previous section into the epitope prediction and deletion process can make this challenge more manageable. Overall, the trend towards the replacement of experimental work by computer-based models is inevitable.
2.3.2.2. Computational
Introducing deimmunizing mutations into a protein is coupled with the risk of a potential loss of function and altered structure [116]. Computational tools for deimmunization aim to find optimal trade-offs between epitope deletion and protein activity and stability. DP2 (dynamic programming for deimmunizing proteins) was the first implementation combining the two epitope prediction tools ProPred [128] and SMM-align [100] with three different stability evaluation methods [129]. With this approach, variants of staphylokinase (SakSTAR), EPO, and three therapeutic antibodies with high similarity to previously developed deimmunized variants were successfully designed [129]. The computational deimmunization approach was further developed to combine the prediction tool ProPred with the Rosetta flexible backbone design package [130] and again tested with SakSTAR and EPO [131]. In 2014, King et al. described a method also combining the Rosetta protein design software [130] with a customized T cell epitope predictor successfully deimmunizing GFP and domain III of the Pseudomonas exotoxin A (PE38) [132]. Mice were challenged with the deimmunized GFP variant and Freund’s adjuvant and after six days, spleens were analyzed by flow cytometry. In four out of the five mice, no T cell signal above background level was detected [132]. For PE38, reduced immunogenicity was shown ex vivo measuring IL-2 response from PBMCs stimulated with the mutant variants [132]. A combination of several algorithmic papers [133], [134], [135], [136] laid the basis for EpiSweep [134], [137]. In this computational deimmunization tool, target sequence, multiple sequence alignments and, if available, protein structure are used as input data. The user can specify the design strategy (sequence-based or structure-based) as well as mutational load, number of Pareto optimal and near-optimal frontiers, and a threshold for epitope prediction [137]. EpiSweep was first applied to SakSTAR and EPO, and the new designs were predicted to outperform previous experimentally designed variants [134]. More recent deimmunization studies with EpiSweep were performed on lysostaphin, a highly active bacteriocin specifically cleaving the peptidoglycan of Staphylococcus aureus [138], [139]. Lysostaphin is a promising therapeutic protein to treat drug-resistant S. aureus infections, but due to its bacterial origin, it comes with immunogenicity [140]. Two different approaches were performed to deimmunize lysostaphin: individual structure-based design [141] and combinatorial library design [142]. Both approaches generated deimmunized variants that showed reduced immunogenicity in vivo. The lysostaphin variant Lib5 designed with the combinatorial library approach successfully suppressed ADA formation in humanized mice leading to better therapeutic efficacy in a methicillin-resistant S. aureus infection model [142]. Combinatorial library design has also been applied to an agent for anticancer therapy, P99 β-lactamase (P99βL) [143]. The study resulted in a highly active and stable 14-mutation variant, which showed reduced ex vivo immunogenicity [143]. Computational tools such as EpiSweep increase the hit rate of deimmunization attempts and decrease the amount of time and resources necessary to develop therapeutic proteins with reduced immunogenicity. The great potential of protein therapeutics will likely lead to increased interest and use of computational approaches in order to accelerate the development of novel therapeutics.
2.4. B cell epitope prediction and deletion
The induction of long-lived memory B cells and plasma cells by T cells can produce high titers and high affinity of often neutralizing ADAs [27]. Epitopes recognized by B cells can be divided into linear and conformational epitopes. Linear epitopes consist of a continuous peptide segment whereas conformational epitopes consist of discontinuous stretches of amino acid residues brought into proximity through protein folding [144], [145], [146]. Discontinuous epitopes rely on proper folding and can therefore not be isolated and assessed for binding experimentally on their own [147]. They make up the majority of B cell epitopes, whereas linear epitopes amount to only around 10% [145]. It was found that discontinuous epitopes range from 6 to 29 residues in size, whereas linear epitopes can vary between 2 and several hundred residues, with the majority of epitopes being 6 to 30 amino acids long [144], [145], [148].
To identify the amino acids of a protein that interact with antibodies, the structure of antigen–antibody complexes can be solved with X-ray crystallography or nuclear magnetic resonance (NMR) analysis [149]. An alternative way to identify interacting residues is the use of hydrogen–deuterium exchange coupled to mass spectrometry [150]. Solvent-exposed residues will exchange their hydrogen of the backbone amide against deuterium present in the solvent. The exchange rate is reduced for interacting residues and therefore, comparing deuterium levels in antigens with and without antibody allows identification of residues involved in the interaction [150], [151], [152]. As an alternative to these precise mapping approaches, synthetic peptide libraries (similar to what has been described for T cell epitopes) or random phage libraries [153], [154] can be used to detect B cell epitopes. However, while these strategies can identify continuous epitopes, they will most likely miss more complex conformational epitopes [150]. Another experimental approach allowing to screen a vast range of residues in the antigen is (random) mutagenesis followed by high throughput screening [150], [155]. Besides these experimental approaches, there has been an increasing number of computational tools for prediction of B cell epitopes, which will be described in the following subsections.
Once putative B cell epitopes on a target protein have been identified, the deletion of these epitopes follows similar procedures as what has been described above for T cell epitopes. Mutations are introduced into the protein in an effort to destroy the epitopes and, at the same time, retain protein structure and function. Typically, alanine scanning mutagenesis is used, as alanine usually has only minor impact on protein conformation due to the absence of possibly interacting side chains [156]. However, it is still hard to know whether interaction is disrupted because of changes in protein structure or because a key interacting residue is targeted [150]. Once a collection of mutated variants of the target protein has been generated, these variants can be analyzed for function and immunogenicity using either in vitro immunogenicity assays or humanized mice. A summary of the various B cell prediction and deletion strategies is presented in Fig. 3.
Fig. 3.
Overview of B cell epitope prediction and deletion approaches. B cell epitopes can be identified experimentally or through various computational approaches (in silico epitope prediction). Experimental strategies include the use of synthetic peptide libraries, epitope mapping, and random mutagenesis combined with high throughput screening. While synthetic peptide libraries only identify continuous epitopes, the other approaches can also predict conformational epitopes. Once putative B cell epitopes have been identified, multiple mutated variants of the protein of interest presumably devoid of these epitopes are generated, analyzed for structural and functional integrity, and assessed for their immunogenicity. (Created with BioRender.com).
2.4.1. Antibody-independent B cell epitope prediction
As experimental approaches are time-consuming and expensive, computational tools are desirable. Early tools for B cell epitope prediction were primarily based on physiochemical properties such as hydrophilicity and surface accessibility [145]. In the early 1980ies, Hopp and Woods developed the first computational prediction tool to predict linear epitopes based on the assumption that regions containing charged and polar residues but lacking hydrophobic residues tend to be more antigenic [157], [158]. More methods were developed based on hydrophilicity, turns, solvent accessibility, and flexibility [159], [160], [161], [162]. Later, methods combining different physicochemical properties, such as PREDITOPE [163] PEOPLE [164] BEPITOPE [165] and BcePred [166] followed. In 2005, a study showed that prediction tools based on propensity scales were only somewhat better than random selection [167]. These findings led to the development of prediction tools based on more complex algorithms and machine learning. BepiPred was the first prediction tool combining a HMM with Parker’s hydrophilicity scale [168]. It was later updated to BepiPred-2.0 in 2017, which is trained only on crystal structures and achieves better prediction accuracy [169]. The first time a recurrent artificial neural network (RNN) was implemented to predict linear B cell epitopes from the amino acid sequence of a protein was the tool ABCPred by Saha and Raghava [170]. One of the main challenges was the variable length of B cell epitopes, which had to be fixed to train the RNN. Nonetheless, ABCPred achieved maximum accuracy of almost 66% when the network was trained on peptides of 16 amino acids in length [170]. FBCPred, which is based on support vector machine (SVM) classifiers with a string kernel, is able to predict linear B cell epitopes of varying length [171]. Another SVM-based method developed by Gupta et al. aims at predicting the specific antibody class induced by an antigen [172].
As most B cell epitopes are conformational, it is desirable to not only predict linear epitopes but also discontinuous epitopes. The conformational epitope predictor (CEP) was the first tool predicting linear and conformational epitopes based on spatial distance and accessibility of residues in an antigen with known structure [173]. For some of the prediction tools, newer versions are available by now. For example, DiscoTope-2.0 is also based on surface measure and spatial distance, but with the addition of amino acid statistics derived from the comparison between epitopes and non-epitopes, and achieves better prediction results than the first version [174]. ElliPro [175] which implements three algorithms, was shown to outperform CEP [173] and DiscoTope [176]. ElliPro uses an approximation of the protein shape as an ellipsoid [177] combined with a residue protrusion index (PI) [178] and a MODELLER-based clustering [179] of neighboring residues on the basis of the PI values [175]. Additional B cell prediction tools are summarized in Table 2. Overall, B cell epitope prediction remains challenging and has not been very successful so far [174], [180].
Table 2.
B cell epitope prediction tools.
| Name of the tool (latest version) | Epitope type | Year of first publication | Latest version |
|---|---|---|---|
| ABCpred | Continuous | 2006 [170] | |
| BCPred | Continuous | 2008 [207] | |
| FBCPred | Continuous | 2008 [171] | |
| COBEpro | Continuous | 2009 [208] | |
| LBtope | Continuous | 2013 [209] | |
| EPMLR | Continuous | 2014 [210] | |
| DRREP | Continuous | 2017 [211] | |
| BepiPred-2.0 | Continuous | 2006 [168] | 2017 [169] |
| iBCE-EL | Continuous | 2018 [212] | |
| CEP | Discontinuous | 2005 [173] | |
| DiscoTope-2.0 | Discontinuous | 2006 [176] | 2012 [174] |
| MIMOX | Discontinuous | 2006 [213] | |
| PEPITOPE | Discontinuous | 2007 [214] | |
| ElliPro | Discontinuous | 2008 [175] | |
| PEPITO | Discontinuous | 2008 [215] | |
| SEPPA | Discontinuous | 2009 [216] | |
| EPITOPIA | Discontinuous | 2009 [217] | |
| EPISEARCH | Discontinuous | 2009 [218] | |
| EPSVR | Discontinuous | 2010 [219] | |
| CBTOPE | Discontinuous | 2010 [220] | |
| MIMOPRO | Discontinuous | 2011 [221] | |
| PEPMAPPER | Discontinuous | 2012 [222] | |
| SnugDock | Antibody-specific epitopes | 2010 [185] | |
| ASEP | Antibody-specific epitopes | 2010 [223] | |
| ClusPro | Antibody-specific epitopes | 2012 [224] | |
| EpiPred | Antibody-specific epitopes | 2014 [186] | |
| PEASE | Antibody-specific epitopes | 2014 [187], [188] |
2.4.2. Antibody-specific B cell epitope prediction
The incredible number and diversity of antibodies circulating in the body could explain why B cell epitope prediction has been far from satisfactory, since any antigen surface region can possibly serve as an epitope [146]. Therefore, the research question was recently redefined and focused more on the prediction of epitopes for specific antibodies [181]. The first approaches in this direction used general protein–protein interaction algorithms to predict antibody-antigen interaction while only considering interactions with the CDRs of the antibody [181]. The CDRs, which correspond more or less to the antigen-binding regions (ABRs), define the specificity of the unique interface between antibody and antigen and can be identified computationally [182], [183]. It was shown that antibodies usually do not recognize typical protein–protein interfaces but rather regions identical to the rest of the surface [184]. Therefore, antigen–antibody specific docking approaches were developed, such as SnugDock [185]. SnugDock simultaneously optimizes the CDRs, the relative orientation of the antibody light and heavy chains and the antigen–antibody orientation. This allows for antibody flexibility and might capture intramolecular changes happening in the antibody upon antigen binding [185]. The flexibility introduced during docking allows to overcome inaccuracies originating from homology modeling of the antibody structure [185]. The EpiPred tool identifies possible epitopes on a given antigen structure for a specified antibody. Interestingly, no significant differences were observed between prediction based on an experimentally determined antibody structure and prediction based on a homology model of the antibody [186]. Antibody-specific epitope prediction based on antibody sequence and antigen structure or sequence was implemented in the tool PEASE [187], [188]. This machine learning algorithm was trained on known antibody-antigen complex data obtained from different experimental epitope mapping methods [188]. The increase in performance achieved through this is likely due to the combination of epitope prediction with cross-blocking experimental data [188]. Similarly, EpiScope also combines computational prediction with experimental validation [189]. This tool minimizes experimental effort by computationally predicting promising mutant variants of the antigen based on docking models with minimal prior knowledge. This approach narrows down the number of variants to be tested experimentally for validation [189]. Most recent advances in computational antibody-specific B cell epitope prediction were proposed by Jespersen et al. [146] who developed a Feed Forward Neural Network capable of differentiating between the epitope and other similar surface patches. The algorithm that is able to generate those surface patches was based on a statistical approach that identified correlation of structural, geometrical and physicochemical features of the interacting residues of antigen–antibody complexes [146]. Even though the tool performed better than other prediction methods, and correct epitopes were predicted and paired with correct antibodies, it is still far from perfect [146]. With an increasing number of known structures of antibody-antigen complexes, machine learning approaches trained on more extensive datasets, and new algorithms developed, further advances in tackling the complexity of B cell epitope prediction can be expected in the future.
3. Summary and outlook
The efficient deimmunization of biotherapeutics is of increasing interest in human medicine. The design and production of protein-based vaccines or enzyme-based antimicrobials are just a few examples of the diversity of applications that can profit from targeted deimmunization. The process of protein deimmunization can be divided into several steps, i.e., epitope prediction, epitope identification and epitope deletion, each of them holding their own complexities and challenges. Not only can epitope identification and prediction be useful to solve biomedical questions, but they could also shed light on immunological processes in general or be used for the prediction of epitopes in the diagnosis of disease. One trend common to all three steps of protein deimmunization is the desired and often successful transition from experimental to computational methods. Even though shielding methods have been proven to be an efficient tool to deimmunize peptides or proteins, they are possibly not the best choice for biotherapeutics with enzymatic activity due to the possible masking of the active sites. Instead, the alteration and deletion of predicted and identified epitopes on biotherapeutic drugs through exchange of amino acids appears to be a more promising approach, particularly when efficient computational tools are available. The implementation of machine learning techniques in the field of immunoinformatics for epitope prediction underpins the complexity of these processes. The problem is further complicated by the enormous diversity of the HLA alleles in humans and the resulting variety of epitopes that can possibly be identified by the immune systems of different patients. In order to produce deimmunized biotherapeutic drugs effective in all patients, all these different alleles would need to be considered. In this context, an interesting option may be the development of personalized deimmunized biotherapeutics, taking into consideration only individual HLA alleles. These personalized deimmunized drugs, however, would generate high costs for patients and/or healthcare systems (even though the use of computationally-driven methods can reduce costs for drug development significantly) and likely face regulatory obstacles. In general, computationally-driven methods for epitope prediction depend on the quality of the input data that are provided, the degrees of freedom allowed, and the algorithms used. Nevertheless, computational methods have already been reported to be more accurate than experimental approaches in epitope prediction in some cases, e.g., for the development of peptide-based vaccines [190]. In this context, it is worth mentioning that overprediction of T cell epitopes (i.e., the identification of false positive epitopes) by computational approaches is more of a problem in protein deimmunization than in peptide vaccine development, since a few non-immunogenic peptides within a collection that is enriched overall for immunogenic peptides will likely have no or only minor impacts on the efficacy of the vaccine. The importance of growing and available databases for epitopes as well as 3D protein and antigen structures is evident. In recent years, the improvement of B and T cell epitope prediction algorithms has been enormous. The current limiting factor is possibly not the capability of the present algorithms but rather the limited data available to train these algorithms on. If sufficiently extensive data sets were available, other problems and questions, such as which combination of classifiers would lead to better results, could be addressed more precisely. A problem that cannot be solved by the enlargement of data sets is the difficulty to determine whether a protein reacting with an antibody is changing its conformation, thereby distorting the tertiary structure that was used as a basis for computation. This is particularly problematic for the prediction of discontinuous epitopes, which represents the largest portion of all B cell epitopes. The incorporation of structural data from antigen-antibody complexes is an essential factor to further increase the prediction accuracy of these tools. To conclude, in order to advance efficient deimmunization of biotherapeutics, both T and B cell epitope prediction and deletion should be considered. The advance that T cell epitope prediction has gained to this date is mostly due to the higher complexity of B cell epitopes and the dependence on structural information. The deimmunization of enzymes compared to peptides and other proteins is more challenging, since both steric hindrance through unspecific shielding approaches and alterations of the amino acid sequence through mutations can reduce enzymatic activity significantly. The trend is going towards the development of increasingly sophisticated algorithms and programs that can (i) reliably predict epitopes and (ii) suggest possible mutations which are most effective for deimmunization and at the same time not detrimental for the activity of the enzyme.
CRediT authorship contribution statement
Léa V. Zinsli: Conceptualization, Writing - original draft, Writing - review & editing, Visualization. Noël Stierlin: Writing - original draft. Martin J. Loessner: Writing - review & editing. Mathias Schmelcher: Conceptualization, Writing - review & editing, Visualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to acknowledge Anja Keller for her critical input on the figures of this article.
The figures and the graphical abstract of this article were created with BioRender.com.
References
- 1.Guillet J., Lai M., Briner T., Buus S., Sette A., Grey H. Immunological self, nonself discrimination. Science. 1987;235(4791):865–870. doi: 10.1126/science.2433769. [DOI] [PubMed] [Google Scholar]
- 2.González S., González-Rodríguez A.P., López-Soto A., Huergo-Zapico L., López-Larrea C., Suárez-Álvarez B. Conceptual aspects of self and nonself discrimination. Self Nonself. 2011;2(1):19–25. doi: 10.4161/self.2.1.15094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Groot A.S., Scott D.W. Immunogenicity of protein therapeutics. Trends Immunol. 2007;28(11):482–490. doi: 10.1016/j.it.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Brinks V., Jiskoot W., Schellekens H. Immunogenicity of therapeutic proteins: the use of animal models. Pharm Res. 2011;28(10):2379–2385. doi: 10.1007/s11095-011-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster J., Koulov A., Mahler H.-C., Detampel P., Huwyler J., Singh S. In Vivo Stability of Therapeutic Proteins. Pharm Res. 2020;37(2) doi: 10.1007/s11095-019-2689-1. [DOI] [PubMed] [Google Scholar]
- 6.Dempsey P.W., Vaidya S.A., Cheng G. The art of war: Innate and adaptive immune responses. Cell Mol Life Sci. 2003;60(12):2604–2621. doi: 10.1007/s00018-003-3180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaplin D.D. Overview of the immune response. J Allergy Clin Immunol. 2010;125(2):S3–S23. doi: 10.1016/j.jaci.2009.12.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonilla F.A., Oettgen H.C. Adaptive immunity. J Allergy Clin Immunol. 2010;125:S33–S40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 9.Jiskoot W., Kersten G.F.A., Mastrobattista E., Slütter B. Pharmaceutical biotechnology: fundamentals and applications. Springer International Publishing; Cham: 2019. pp. 281–304. [Google Scholar]
- 10.Kawai T., Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar H., Kawai T., Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 12.Curtsinger J.M., Mescher M.F. Inflammatory cytokines as a third signal for T cell activation. Curr Opin Immunol. 2010;22(3):333–340. doi: 10.1016/j.coi.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain A., Pasare C. Innate control of adaptive immunity: beyond the three-signal paradigm. J Immunol. 2017;198(10):3791–3800. doi: 10.4049/jimmunol.1602000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberts B., Johnson A., Lewis J., Raff M., Roberts K. 5th ed. Garland Science; New York, NY: 2007. Molecular biology of the cell. [Google Scholar]
- 15.Chapiro J., Claverol S., Piette F., Ma W., Stroobant V., Guillaume B. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol. 2006;176(2):1053–1061. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 16.Pamer E., Cresswell P. Mechanisms of MHC class I–restricted antigen processing. Annu Rev Immunol. 1998;16(1):323–358. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 17.Rudd P.M., Elliott T., Cresswell P., Wilson I.A., Dwek R.A. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 18.Cascio P., Hilton C., Kisselev A.F., Rock K.L., Goldberg A.L. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20:2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy K., Weaver C. W.W. Norton & Company; New York, NY: 2016. Janeway's immunobiology. [Google Scholar]
- 20.Xu H., Ren D. Lysosomal physiology. Annu Rev Physiol. 2015;77(1):57–80. doi: 10.1146/annurev-physiol-021014-071649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saftig P., Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol. 2009;10(9):623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 22.Weaver J.M., Lazarski C.A., Richards K.A., Chaves F.A., Jenks S.A., Menges P.R. Immunodominance of CD4 T cells to foreign antigens is peptide intrinsic and independent of molecular context: implications for vaccine design. J Immunol. 2008;181(5):3039–3048. doi: 10.4049/jimmunol.181.5.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meydan C., Otu H.H., Sezerman O. Prediction of peptides binding to MHC class I and II alleles by temporal motif mining. BMC Bioinf. 2013;14(Suppl 2):S13. doi: 10.1186/1471-2105-14-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieczorek M, Abualrous ET, Sticht J, Alvaro-Benito M, Stolzenberg S, et al. Major histocompatibility complex (MHC) Class I and MHC Class II proteins: conformational plasticity in antigen presentation. Front Immunol 2017;8:292. [DOI] [PMC free article] [PubMed]
- 25.Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012;12(8):557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 26.Jawa V., Hokom M., Hu Z., El-Abaadi N., Zhuang Y., Berger D. Assessment of immunogenicity of romiplostim in clinical studies with ITP subjects. Ann Hematol. 2010;89(S1):75–85. doi: 10.1007/s00277-010-0908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jawa V., Cousens L.P., Awwad M., Wakshull E., Kropshofer H., De Groot A.S. T-cell dependent immunogenicity of protein therapeutics: Preclinical assessment and mitigation. Clin Immunol. 2013;149(3):534–555. doi: 10.1016/j.clim.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Sauerborn M., van de Vosse E., Delawi D., van Dissel J.T., Brinks V., Schellekens H. Natural antibodies against bone morphogenic proteins and interferons in healthy donors and in patients with infections linked to type-1 cytokine responses. J Interferon Cytokine Res. 2011;31(9):661–669. doi: 10.1089/jir.2010.0075. [DOI] [PubMed] [Google Scholar]
- 29.Janeway C.A., Travers P., Walport M., Shlomchik M.J. Garland Science; New York, NY: 2001. Immunobiology, 5th edition: the immune system in health and disease. [Google Scholar]
- 30.Mattila P.K., Vainio M., Perez S.H., Sustar V., Rajala J. Small GTPase Rab8 plays a critical role in B cell antigen presentation. J Immunol. 2019;202(1 Supplement):117.12. [Google Scholar]
- 31.Pier G.B., Lyczak J.B., Wetzler L.M. ASM Press; Washington, D.C.: 2004. Immunology, infection, and immunity. [Google Scholar]
- 32.Sethu S., Govindappa K., Alhaidari M., Pirmohamed M., Park K. Immunogenicity to biologics: mechanisms, prediction and reduction. Arch Immunol Ther Exp (Warsz.) 2012;60:331–344. doi: 10.1007/s00005-012-0189-7. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg A.S. Effects of protein aggregates: an immunologic perspective. AAPS J. 2006;8(3):E501–E507. doi: 10.1208/aapsj080359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertolotto A., Sala A., Malucchi S., Marnetto F., Caldano M. Biological activity of interferon betas in patients with multiple sclerosis is affected by treatment regimen and neutralising antibodies. J Neurol Neurosurg Psychiatry. 2004;75:1294–1299. doi: 10.1136/jnnp.2004.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Yang C, Xia Y, Bertino A, Glaspy J, et al. Thrombocytopenia caused by the development of antibodies to thrombopoietin. Blood 98;2001:3241–48. [DOI] [PubMed]
- 36.Pineda C., Castañeda Hernández G., Jacobs I.A., Alvarez D.F., Carini C. Assessing the immunogenicity of biopharmaceuticals. BioDrugs. 2016;30(3):195–206. doi: 10.1007/s40259-016-0174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.BARBOSA M., CELIS E. Immunogenicity of protein therapeutics and the interplay between tolerance and antibody responses. Drug Discov Today. 2007;12(15-16):674–681. doi: 10.1016/j.drudis.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Clark K.A., Tan Z. Methods for engineering therapeutic peptides. Chinese Chem Lett. 2018;29(7):1074–1078. [Google Scholar]
- 39.Branca C., Magazù S., Maisano G., Migliardo F., Migliardo P., Romeo G. Hydration study of PEG/water mixtures by quasi elastic light scattering, acoustic and rheological measurements. J Phys Chem B. 2002;106(39):10272–10276. [Google Scholar]
- 40.Veronese F.M., Pasut G. PEGylation, successful approach to drug delivery. Drug Discov Today. 2005;10(21):1451–1458. doi: 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- 41.Di L.i. Strategic approaches to optimizing peptide ADME properties. AAPS J. 2015;17(1):134–143. doi: 10.1208/s12248-014-9687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P., Sun F., Liu S., Jiang S. Anti-PEG antibodies in the clinic: current issues and beyond PEGylation. J Control Release. 2016;244:184–193. doi: 10.1016/j.jconrel.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang P., Sun F., Tsao C., Liu S., Jain P., Sinclair A. Zwitterionic gel encapsulation promotes protein stability, enhances pharmacokinetics, and reduces immunogenicity. Proc Natl Acad Sci U S A. 2015;112(39):12046–12051. doi: 10.1073/pnas.1512465112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schellenberger V., Wang C.-W., Geething N.C., Spink B.J., Campbell A., To W. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat Biotechnol. 2009;27(12):1186–1190. doi: 10.1038/nbt.1588. [DOI] [PubMed] [Google Scholar]
- 45.Podust V.N., Sim B.-C., Kothari D., Henthorn L., Gu C., Wang C.-w. Extension of in vivo half-life of biologically active peptides via chemical conjugation to XTEN protein polymer. Protein Eng Des Sel. 2013;26(11):743–753. doi: 10.1093/protein/gzt048. [DOI] [PubMed] [Google Scholar]
- 46.Anand R., Vallooran J. 11 - Polypeptides: PASylation and XTEN. In: Parambath A., editor. Engineering of biomaterials for drug delivery systems. Woodhead Publishing; 2018. pp. 299–315. [Google Scholar]
- 47.Brandl F., Merten H., Zimmermann M., Béhé M., Zangemeister-Wittke U., Plückthun A. Influence of size and charge of unstructured polypeptides on pharmacokinetics and biodistribution of targeted fusion proteins. J Control Release. 2019;307:379–392. doi: 10.1016/j.jconrel.2019.06.030. [DOI] [PubMed] [Google Scholar]
- 48.Binder U., Skerra A. PASylation®: a versatile technology to extend drug delivery. Cur Opin Colloid Interface Sci. 2017;31:10–17. [Google Scholar]
- 49.Lee H., DeLoache W.C., Dueber J.E. Spatial organization of enzymes for metabolic engineering. Metab Eng. 2012;14(3):242–251. doi: 10.1016/j.ymben.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Floss D.M., Schallau K., Rose-John S., Conrad U., Scheller J. Elastin-like polypeptides revolutionize recombinant protein expression and their biomedical application. Trends Biotechnol. 2010;28(1):37–45. doi: 10.1016/j.tibtech.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 51.Zaman R., Othman I., Chowdhury E. Carrier mediated systemic delivery of protein and peptide therapeutics. Curr Pharm Des. 2016;22(40):6167–6191. doi: 10.2174/1381612822666160720145328. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y.-S., Wen X.-F., Wu Y.-L., Wang Y.-F., Fan M., Yang Z.-Y. Engineering a pharmacologically superior form of granulocyte-colony-stimulating factor by fusion with gelatin-like-protein polymer. Eur J Pharm Biopharm. 2010;74(3):435–441. doi: 10.1016/j.ejpb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Bachran C., Gupta P.K., Bachran S., Leysath C.E., Hoover B., Fattah R.J. Reductive methylation and mutation of an anthrax toxin fusion protein modulates its stability and cytotoxicity. Sci Rep. 2015;4(1) doi: 10.1038/srep04754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20(8):1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.London E., Luongo C.L. Domain-specific bias in arginine/lysine usage by protein toxins. Biochem Biophys Res Commun. 1989;160(1):333–339. doi: 10.1016/0006-291x(89)91660-4. [DOI] [PubMed] [Google Scholar]
- 56.Qi Y., Chilkoti A. Protein-polymer conjugation-moving beyond PEGylation. Curr Opin Chem Biol. 2015;28:181–193. doi: 10.1016/j.cbpa.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Powell J., Gurk-Turner C. Darbepoetin alfa (Aranesp) Proc (Bayl Univ Med Cent) 2002;15(3):332–335. doi: 10.1080/08998280.2002.11927861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Q., Qiu H. The mechanistic impact of N-glycosylation on stability, pharmacokinetics, and immunogenicity of therapeutic proteins. J Pharm Sci. 2019;108(4):1366–1377. doi: 10.1016/j.xphs.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 59.Cole L.A. Hyperglycosylated hCG, a review. Placenta. 2010;31(8):653–664. doi: 10.1016/j.placenta.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 60.Cole L.A., Dai D., Butler S.A., Leslie K.K., Kohorn E.I. Gestational trophoblastic diseases: 1. Pathophysiology of hyperglycosylated hCG. Gynecol Oncol. 2006;102(2):145–150. doi: 10.1016/j.ygyno.2005.12.047. [DOI] [PubMed] [Google Scholar]
- 61.van Ree R., Cabanes-Macheteau M., Akkerdaas J., Milazzo J.P., Loutelier-Bourhis C. Beta(1,2)-xylose and alpha(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. J Biol Chem. 2000;275:11451–11458. doi: 10.1074/jbc.275.15.11451. [DOI] [PubMed] [Google Scholar]
- 62.Macher B.A., Galili U. The Galalpha 1,3Galbeta1,4GlcNAc-R (alpha-Gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim Biophys Acta. 2008;1780:75–88. doi: 10.1016/j.bbagen.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crocker P.R., Paulson J.C., Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 64.Fernandes A.I., Gregoriadis G. Polysialylated asparaginase: preparation, activity and pharmacokinetics. Biochim Biophys Acta. 1997;1341(1):26–34. doi: 10.1016/s0167-4838(97)00056-3. [DOI] [PubMed] [Google Scholar]
- 65.Terekhov SS, Smirnov IV, Shamborant OG, Bobik TV, Ilyushin DG, et al. Chemical polysialylation and in vivo tetramerization improve pharmacokinetic characteristics of recombinant human butyrylcholinesterase-based bioscavengers. Acta Naturae 7;2015:136–141. [PMC free article] [PubMed]
- 66.Meng H.e., Jain S., Lockshin C., Shaligram U., Martinez J., Genkin D. Clinical application of polysialylated deoxyribonuclease and erythropoietin. Recent Pat Drug Deliv Formul. 2019;12(3):212–222. doi: 10.2174/1872211312666180717164758. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y.J., Choi S., Koh J.J., Lee M., Ko K.S. Controlled release of insulin from injectable biodegradable triblock copolymer. Pharm Res. 2001;18:548–550. doi: 10.1023/a:1011074915438. [DOI] [PubMed] [Google Scholar]
- 68.Riechmann L., Clark M., Waldmann H., Winter G. Reshaping human antibodies for therapy. Nature. 1988;332(6162):323–327. doi: 10.1038/332323a0. [DOI] [PubMed] [Google Scholar]
- 69.Choi Y., Hua C., Sentman C.L., Ackerman M.E., Bailey-Kellogg C. Antibody humanization by structure-based computational protein design. MAbs. 2015;7(6):1045–1057. doi: 10.1080/19420862.2015.1076600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J., Jiang N., Wang T., Xie G., Zhang Z., Li H. DNA shuffling of uricase gene leads to a more “human like” chimeric uricase with increased uricolytic activity. Int J Biol Macromol. 2016;82:522–529. doi: 10.1016/j.ijbiomac.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 71.Cantor J.R., Panayiotou V., Agnello G., Georgiou G., Stone E.M. Engineering reduced-immunogenicity enzymes for amino acid depletion therapy in cancer. Methods Enzymol. 2012;502:291–319. doi: 10.1016/B978-0-12-416039-2.00015-X. [DOI] [PubMed] [Google Scholar]
- 72.Goodnow C.C., Vinuesa C.G., Randall K.L., Mackay F., Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11(8):681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- 73.Markov PV, Pybus OG. Evolution and diversity of the human leukocyte antigen (HLA). Evol Med Public Health 2015;2015:1. [DOI] [PMC free article] [PubMed]
- 74.Peters B., Nielsen M., Sette A. T cell epitope predictions. Annu Rev Immunol. 2020;38(1):123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ahmed R.K., Maeurer M.J. T-cell epitope mapping. Methods Mol Biol. 2009;524:427–438. doi: 10.1007/978-1-59745-450-6_31. [DOI] [PubMed] [Google Scholar]
- 76.Falk K., Rötzschke O., Stevanovié S., Jung G., Rammensee H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 77.Rammensee H.-G., Friede T., Stevanović S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41(4):178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 78.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–517. [DOI] [PMC free article] [PubMed]
- 79.Jones E.Y., Fugger L., Strominger J.L., Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6(4):271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 80.Benoist C.O., Mathis D.J., Kanter M.R., Williams V.E., 2nd, McDevitt H.O. Regions of allelic hypervariability in the murine A alpha immune response gene. Cell. 1983;34:169–177. doi: 10.1016/0092-8674(83)90147-2. [DOI] [PubMed] [Google Scholar]
- 81.Bjorkman P.J., Saper M.A., Samraoui B., Bennett W.S., Strominger J.L., Wiley D.C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 82.Brown J.H., Jardetzky T.S., Gorga J.C., Stern L.J., Urban R.G., Strominger J.L. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364(6432):33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 83.Sette A., Vitiello A., Reherman B., Fowler P., Nayersina R. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 84.Southwood S., Sidney J., Kondo A., del Guercio M.F., Appella E. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 85.Sette A., Buus S., Appella E., Smith J.A., Chesnut R., Miles C. Prediction of major histocompatibility complex binding regions of protein antigens by sequence pattern analysis. Proc Natl Acad Sci U S A. 1989;86(9):3296–3300. doi: 10.1073/pnas.86.9.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rammensee H.-G., Bachmann J., Emmerich N.P.N., Bachor O.A., Stevanović S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50(3-4):213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 87.Adams H.-P., Koziol J.A. Prediction of binding to MHC class I molecules. J Immunol Methods. 1995;185(2):181–190. doi: 10.1016/0022-1759(95)00111-m. [DOI] [PubMed] [Google Scholar]
- 88.Brusic V., Rudy G., Honeyman G., Hammer J., Harrison L. Prediction of MHC class II-binding peptides using an evolutionary algorithm and artificial neural network. Bioinformatics. 1998;14(2):121–130. doi: 10.1093/bioinformatics/14.2.121. [DOI] [PubMed] [Google Scholar]
- 89.Mamitsuka H. Predicting peptides that bind to MHC molecules using supervised learning of hidden Markov models. Proteins. 1998;33(4):460–474. doi: 10.1002/(sici)1097-0134(19981201)33:4<460::aid-prot2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 90.Korber B., Brander C., Haynes B., Moore J., Koup R. Los Alamos National Laboratory; Los Alamos, NM: 1999. HIV molecular immunology database 1999. [Google Scholar]
- 91.Peters B, Sidney J, Bourne P, Bui HH, Buus S, et al. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol 2005;3:e91. [DOI] [PMC free article] [PubMed]
- 92.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, et al. The immune epitope database (IEDB): 2018 update. Nucleic Acids Res 2019;47:D339–D343. [DOI] [PMC free article] [PubMed]
- 93.Dhanda SK, Mahajan S, Paul S, Yan Z, Kim H, et al. IEDB-AR: immune epitope database-analysis resource in 2019. Nucleic Acids Res 2019;47:W502–W506. [DOI] [PMC free article] [PubMed]
- 94.Shao W, Pedrioli PGA, Wolski W, Scurtescu C, Schmid E, et al. The SysteMHC Atlas project. Nucleic Acids Res 2018;46:D1237–D1247. [DOI] [PMC free article] [PubMed]
- 95.Lundegaard C., Lamberth K., Harndahl M., Buus S., Lund O. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nielsen M., Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinf. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jensen K.K., Andreatta M., Marcatili P., Buus S., Greenbaum J.A., Yan Z. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154(3):394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson J., Barker D.J., Georgiou X., Cooper M.A., Flicek P. IPD-IMGT/HLA database. Nucleic Acids Res. 2020;48:D948–D955. doi: 10.1093/nar/gkz950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sturniolo T., Bono E., Ding J., Raddrizzani L., Tuereci O., Sahin U. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17(6):555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 100.Nielsen M, Lundegaard C, Blicher T, Lamberth K, Harndahl M, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One 2007;2:e796. [DOI] [PMC free article] [PubMed]
- 101.Bordner A.J., Mittelmann H.D. MultiRTA: a simple yet reliable method for predicting peptide binding affinities for multiple class II MHC allotypes. BMC Bioinf. 2010;11:482. doi: 10.1186/1471-2105-11-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, et al. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol 2008;4:e1000107. [DOI] [PMC free article] [PubMed]
- 103.Pfeifer N, Kohlbacher O. Multiple instance learning allows MHC class II epitope predictions across alleles. In: Crandall KA, Lagergren J, editors. Algorithms in bioinformatics. WABI 2008. Lecture Notes in Computer Science, vol 5251. Berlin, Heidelberg: Springer; 2008. pp. 210–221.
- 104.Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63(6):325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9(1):1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKinney D.M., Southwood S., Hinz D., Oseroff C., Arlehamn C.S.L., Schulten V. A strategy to determine HLA class II restriction broadly covering the DR, DP, and DQ allelic variants most commonly expressed in the general population. Immunogenetics. 2013;65(5):357–370. doi: 10.1007/s00251-013-0684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nielsen M., Lundegaard C., Lund O., Keşmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57(1-2):33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 108.Hunt D., Michel H., Dickinson T., Shabanowitz J., Cox A., Sakaguchi K. Peptides presented to the immune system by the murine class II major histocompatibility complex molecule I-Ad. Science. 1992;256(5065):1817–1820. doi: 10.1126/science.1319610. [DOI] [PubMed] [Google Scholar]
- 109.Faridi P., Li C., Ramarathinam S.H., Vivian J.P., Illing P.T., Mifsud N.A. A subset of HLA-I peptides are not genomically templated: evidence for cis- and trans-spliced peptide ligands. Sci Immunol. 2018;3(28):eaar3947. doi: 10.1126/sciimmunol.aar3947. [DOI] [PubMed] [Google Scholar]
- 110.Liepe J., Marino F., Sidney J., Jeko A., Bunting D.E., Sette A. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science. 2016;354(6310):354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 111.Li X., Lamothe P.A., Walker B.D., Wang J.-H. Crystal structure of HLA-B*5801 with a TW10 HIV Gag epitope reveals a novel mode of peptide presentation. Cell Mol Immunol. 2017;14(7):631–634. doi: 10.1038/cmi.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pymm P., Illing P.T., Ramarathinam S.H., O'Connor G.M., Hughes V.A., Hitchen C. MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat Struct Mol Biol. 2017;24(4):387–394. doi: 10.1038/nsmb.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schneidman-Duhovny D, Khuri N, Dong GQ, Winter MB, Shifrut E, et al. Predicting CD4 T-cell epitopes based on antigen cleavage, MHCII presentation, and TCR recognition. PloS One 2018;13:e0206654. [DOI] [PMC free article] [PubMed]
- 114.Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 2013;9:e1003266. [DOI] [PMC free article] [PubMed]
- 115.Dhanda SK, Karosiene E, Edwards L, Grifoni A, Paul S, et al. Predicting HLA CD4 Immunogenicity in Human Populations. Front Immunol 2018;9:1369. [DOI] [PMC free article] [PubMed]
- 116.Griswold K.E., Bailey-Kellogg C. Design and engineering of deimmunized biotherapeutics. Curr Opin Struct Biol. 2016;39:79–88. doi: 10.1016/j.sbi.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lemmermann N.A.W., Kropp K.A., Seckert C.K., Grzimek N.K.A., Reddehase M.J. Reverse genetics modification of cytomegalovirus antigenicity and immunogenicity by CD8 T-cell epitope deletion and insertion. J Biomed Biotechnol. 2011;2011:1–15. doi: 10.1155/2011/812742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Onda M., Nagata S., FitzGerald D.J., Beers R., Fisher R.J., Vincent J.J. Characterization of the B cell epitopes associated with a truncated form of Pseudomonas exotoxin (PE38) used to make immunotoxins for the treatment of cancer patients. J Immunol. 2006;177(12):8822–8834. doi: 10.4049/jimmunol.177.12.8822. [DOI] [PubMed] [Google Scholar]
- 119.Mehta R.K., Verma S., Pati R., Sengupta M., Khatua B., Jena R.K. Mutations in subunit interface and B-cell epitopes improve antileukemic activities of Escherichia coli asparaginase-II: evaluation of immunogenicity in mice. J Biol Chem. 2014;289(6):3555–3570. doi: 10.1074/jbc.M113.486530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.JONES T.D., PHILLIPS W.J., SMITH B.J., BAMFORD C.A., NAYEE P.D., BAGLIN T.P. Identification and removal of a promiscuous CD4+ T cell epitope from the C1 domain of factor VIII. J Thromb Haemost. 2005;3(5):991–1000. doi: 10.1111/j.1538-7836.2005.01309.x. [DOI] [PubMed] [Google Scholar]
- 121.Yeung V.P., Chang J., Miller J., Barnett C., Stickler M. Elimination of an immunodominant CD4+ T cell epitope in human IFN-beta does not result in an in vivo response directed at the subdominant epitope. J Immunol. 2004;172:6658–6665. doi: 10.4049/jimmunol.172.11.6658. [DOI] [PubMed] [Google Scholar]
- 122.Harding F.A., Liu A.D., Stickler M., Razo O.J., Chin R. A beta-lactamase with reduced immunogenicity for the targeted delivery of chemotherapeutics using antibody-directed enzyme prodrug therapy. Mol Cancer Ther. 2005;4:1791–1800. doi: 10.1158/1535-7163.MCT-05-0189. [DOI] [PubMed] [Google Scholar]
- 123.Ettinger RA, Liberman JA, Gunasekera D, Puranik K, James EA, et al. FVIII proteins with a modified immunodominant T-cell epitope exhibit reduced immunogenicity and normal FVIII activity. Blood Adv 2018;2:309–322. [DOI] [PMC free article] [PubMed]
- 124.Asgari S., Ebrahim-Habibi A., Mahdavi M., Choopani M., Mirzahoseini H. Therapeutic protein deimmunization by T-cell epitope removal: antigen-specific immune responses in vitro and in vivo. APMIS. 2017;125(6):544–552. doi: 10.1111/apm.12682. [DOI] [PubMed] [Google Scholar]
- 125.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 126.Chew W.L., Tabebordbar M., Cheng J.K.W., Mali P., Wu E.Y., Ng A.H.M. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13(10):868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ferdosi S.R., Ewaisha R., Moghadam F., Krishna S., Park J.G., Ebrahimkhani M.R. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Singh H., Raghava G.P.S. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17(12):1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 129.Parker A.S., Zheng W., Griswold K.E., Bailey-Kellogg C. Optimization algorithms for functional deimmunization of therapeutic proteins. BMC Bioinf. 2010;11(1):180. doi: 10.1186/1471-2105-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang PS, Ban YE, Richter F, Andre I, Vernon R, et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One 2011;6:se24109. [DOI] [PMC free article] [PubMed]
- 131.Choi Y., Griswold K.E., Bailey-Kellogg C. Structure-based redesign of proteins for minimal T-cell epitope content. J Comput Chem. 2013;34(10):879–891. doi: 10.1002/jcc.23213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.King C., Garza E.N., Mazor R., Linehan J.L., Pastan I., Pepper M. Removing T-cell epitopes with computational protein design. Proc Natl Acad Sci U S A. 2014;111(23):8577–8582. doi: 10.1073/pnas.1321126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.He L.u., Friedman A.M., Bailey-Kellogg C. A divide-and-conquer approach to determine the Pareto frontier for optimization of protein engineering experiments. Proteins. 2012;80(3):790–806. doi: 10.1002/prot.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Parker A.S., Choi Y., Griswold K.E., Bailey-Kellogg C. Structure-guided deimmunization of therapeutic proteins. J Comput Biol. 2013;20(2):152–165. doi: 10.1089/cmb.2012.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parker A.S., Griswold K.E., Bailey-Kellogg C. Optimization of combinatorial mutagenesis. J Comput Biol. 2011;18(11):1743–1756. doi: 10.1089/cmb.2011.0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Verma D., Grigoryan G., Bailey-Kellogg C. Structure-based design of combinatorial mutagenesis libraries. Protein Sci. 2015;24(5):895–908. doi: 10.1002/pro.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Choi Y., Verma D., Griswold K.E., Bailey-Kellogg C. EpiSweep: computationally driven reengineering of therapeutic proteins to reduce immunogenicity while maintaining function. Methods Mol Biol. 2017;1529:375–398. doi: 10.1007/978-1-4939-6637-0_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schindler C.A., Schuhardt V.T. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A. 1964;51(3):414–421. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sugai M, Fujiwara T, Akiyama T, Ohara M, Komatsuzawa H, et al. Purification and molecular characterization of glycylglycine endopeptidase produced by Staphylococcus capitis EPK1. J Bacteriol 1997;179:1193–1202. [DOI] [PMC free article] [PubMed]
- 140.Kokai-Kun J. Lysostaphin: a Silver Bullet for Staph. In: Tegos A, Mylonakis E, editors. Antimicrobial Drug Discovery. Oxfordshire, Wallingford, UK: CABI Publishing; 2012. pp. 147–165.
- 141.Blazanovic K., Zhao H., Choi Y., Li W., Salvat R.S., Osipovitch D.C. Structure-based redesign of lysostaphin yields potent antistaphylococcal enzymes that evade immune cell surveillance. Mol Ther Methods Clin Dev. 2015;2:15021. doi: 10.1038/mtm.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhao H., Verma D., Li W., Choi Y., Ndong C. Depletion of T cell epitopes in lysostaphin mitigates anti-drug antibody response and enhances antibacterial efficacy in vivo. Chem Biol. 2015;22:629–639. doi: 10.1016/j.chembiol.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Salvat R.S., Verma D., Parker A.S., Kirsch J.R., Brooks S.A. Computationally optimized deimmunization libraries yield highly mutated enzymes with low immunogenicity and enhanced activity. Proc Natl Acad Sci U S A. 2017;114:E5085–E5093. doi: 10.1073/pnas.1621233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Barlow D.J., Edwards M.S., Thornton J.M. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322(6081):747–748. doi: 10.1038/322747a0. [DOI] [PubMed] [Google Scholar]
- 145.Wang H.W., Pai T.W. Machine learning-based methods for prediction of linear B-cell epitopes. Methods Mol Biol. 2014;1184:217–236. doi: 10.1007/978-1-4939-1115-8_12. [DOI] [PubMed] [Google Scholar]
- 146.Jespersen M.C., Mahajan S., Peters B., Nielsen M., Marcatili P. Antibody specific B-cell epitope predictions: leveraging information from antibody-antigen protein complexes. Front Immunol. 2019;10:298. doi: 10.3389/fimmu.2019.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Van Regenmortel M.H.V. What is a B-cell epitope? Methods Mol Biol. 2009;524:3–20. doi: 10.1007/978-1-59745-450-6_1. [DOI] [PubMed] [Google Scholar]
- 148.Kringelum J.V., Nielsen M., Padkjær S.B., Lund O. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol. 2013;53(1-2):24–34. doi: 10.1016/j.molimm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Votapka L, Amaro RE. Multistructural hot spot characterization with FTProd. Bioinformatics 2013;29:393–394. [DOI] [PMC free article] [PubMed]
- 150.Abbott W.M., Damschroder M.M., Lowe D.C. Current approaches to fine mapping of antigen-antibody interactions. Immunology. 2014;142(4):526–535. doi: 10.1111/imm.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Coales S.J., Tuske S.J., Tomasso J.C., Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23(5):639–647. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- 152.Pandit D., Tuske S.J., Coales S.J., SY E., Liu A. Mapping of discontinuous conformational epitopes by amide hydrogen/deuterium exchange mass spectrometry and computational docking. J Mol Recognit. 2012;25:114–124. doi: 10.1002/jmr.1169. [DOI] [PubMed] [Google Scholar]
- 153.Kern Karolin, Havenith Heide, Delaroque Nicolas, Rautenberger Paul, Lehmann Jörg, Fischer Markus. The immunome of soy bean allergy: Comprehensive identification and characterization of epitopes. Clin Exp Allergy. 2019;49(2):239–251. doi: 10.1111/cea.13285. [DOI] [PubMed] [Google Scholar]
- 154.Qi H, Ma M, Hu C, Xu ZW, Wu FL, et al. Antibody binding epitope Mapping (AbMap) of hundred antibodies in a single run. Mol Cell Proteom, 2020. doi:10.1074/mcp.RA120.002314. [DOI] [PMC free article] [PubMed]
- 155.Gustafsson E, Rosen A, Barchan K, van Kessel KP, Haraldsson K, et al. Directed evolution of chemotaxis inhibitory protein of Staphylococcus aureus generates biologically functional variants with reduced interaction with human antibodies. Protein Eng Des Sel 2010;23:91–101. [DOI] [PubMed]
- 156.Nagata Satoshi, Pastan Ira. Removal of B cell epitopes as a practical approach for reducing the immunogenicity of foreign protein-based therapeutics. Adv Drug Deliv Rev. 2009;61(11):977–985. doi: 10.1016/j.addr.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.EL-Manzalawy Yasser, Honavar Vasant. Recent advances in B-cell epitope prediction methods. Immun Res. 2010;6(Suppl 2):S2. doi: 10.1186/1745-7580-6-S2-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hopp T.P., Woods K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Karplus P.A., Schulz G.E. Prediction of chain flexibility in proteins. Naturwissenschaften. 1985;72(4):212–213. [Google Scholar]
- 160.Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276:172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 161.Parker J.M.R., Guo D., Hodges R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 162.Pellequer Jean-Luc, Westhof Eric, Van Regenmortel Marc H.V. Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol Lett. 1993;36(1):83–99. doi: 10.1016/0165-2478(93)90072-a. [DOI] [PubMed] [Google Scholar]
- 163.Pellequer J.L., Westhof E. PREDITOP: a program for antigenicity prediction. J Mol Graph. 1993;11(204–210):191–202. doi: 10.1016/0263-7855(93)80074-2. [DOI] [PubMed] [Google Scholar]
- 164.Alix Alain J.P. Predictive estimation of protein linear epitopes by using the program PEOPLE. Vaccine. 1999;18(3-4):311–314. doi: 10.1016/s0264-410x(99)00329-1. [DOI] [PubMed] [Google Scholar]
- 165.Odorico Michael, Pellequer Jean-Luc. BEPITOPE: predicting the location of continuous epitopes and patterns in proteins. J Mol Recognit. 2003;16(1):20–22. doi: 10.1002/jmr.602. [DOI] [PubMed] [Google Scholar]
- 166.Saha S, Raghava GPS. BcePred: prediction of continuous B-cell epitopes in antigenic sequences using physico-chemical properties, In: Nicosia G, Cutello V, Bentley PJ, Timmis J, editors. Artificial immune systems. ICARIS 2004. Lecture Notes in Computer Science, vol. 3239. Berlin, Heidelberg: Springer; 2004. https://doi.org/10.1007/978-3-540-30220-9_16.
- 167.Blythe M.J., Flower D.R. Benchmarking B cell epitope prediction: underperformance of existing methods. Protein Sci. 2005;14:246–248. doi: 10.1110/ps.041059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Larsen J.E., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immun Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45:W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Saha Sudipto, Raghava G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 171.El-Manzalawy Y., Dobbs D., Honavar V. Predicting flexible length linear B-cell epitopes. Comput Syst Bioinform Conf. 2008;7:121–132. [PMC free article] [PubMed] [Google Scholar]
- 172.Gupta Sudheer, Ansari Hifzur Rahman, Gautam Ankur, Raghava Gajendra PS. Identification of B-cell epitopes in an antigen for inducing specific class of antibodies. Biol Direct. 2013;8(1) doi: 10.1186/1745-6150-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kulkarni-Kale U., Bhosle S., Kolaskar A.S. CEP: a conformational epitope prediction server. Nucleic Acids Res. 2005;33(Web Server):W168–W171. doi: 10.1093/nar/gki460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kringelum JV, Lundegaard C, Lund O, Nielsen M. Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Comput Biol 2012;8:e1002829. [DOI] [PMC free article] [PubMed]
- 175.Ponomarenko Julia, Bui Huynh-Hoa, Li Wei, Fusseder Nicholas, Bourne Philip E, Sette Alessandro. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008;9(1) doi: 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Haste Andersen Pernille, Nielsen Morten, Lund Ole. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15(11):2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.R Taylor W, M Thornton J, G Turnell W. An ellipsoidal approximation of protein shape. J Mol Graph. 1983;1(2):30–38. [Google Scholar]
- 178.Thornton J.M., Edwards M.S., Taylor W.R., Barlow D.J. Location of 'continuous' antigenic determinants in the protruding regions of proteins. EMBO J. 1986;5(2):409–413. doi: 10.1002/j.1460-2075.1986.tb04226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinf 2006;54:5.6.1–5.6.37. [DOI] [PMC free article] [PubMed]
- 180.Yao B, Zheng D, Liang S, Zhang C. Conformational B-cell epitope prediction on antigen protein structures: a review of current algorithms and comparison with common binding site prediction methods. PLoS One 2013;8:e62249. [DOI] [PMC free article] [PubMed]
- 181.Sela-Culang Inbal, Ofran Yanay, Peters Bjoern. Antibody specific epitope prediction-emergence of a new paradigm. Curr Opin Virol. 2015;11:98–102. doi: 10.1016/j.coviro.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kunik V, Ashkenazi S, Ofran Y. Paratome: an online tool for systematic identification of antigen-binding regions in antibodies based on sequence or structure. Nucleic Acids Res 2012;40:W521–524. [DOI] [PMC free article] [PubMed]
- 183.Kunik V, Peters B, Ofran Y. Structural consensus among antibodies defines the antigen binding site. PLoS Comput Biol 2012;8:e1002388. [DOI] [PMC free article] [PubMed]
- 184.Kunik V, Ofran Y. The indistinguishability of epitopes from protein surface is explained by the distinct binding preferences of each of the six antigen-binding loops. Protein Eng Des Sel 2013;26:599–609. [DOI] [PubMed]
- 185.Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput Biol 2010;6:e1000644. [DOI] [PMC free article] [PubMed]
- 186.Krawczyk K., Liu X., Baker T., Shi J., Deane C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics. 2014;30:2288–2294. doi: 10.1093/bioinformatics/btu190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sela-Culang I., Ashkenazi S., Peters B., Ofran Y. PEASE: predicting B-cell epitopes utilizing antibody sequence. Bioinformatics. 2015;31(8):1313–1315. doi: 10.1093/bioinformatics/btu790. [DOI] [PubMed] [Google Scholar]
- 188.Sela-Culang Inbal, Benhnia Mohammed Rafii-El-Idrissi, Matho Michael H., Kaever Thomas, Maybeno Matt, Schlossman Andrew. Using a combined computational-experimental approach to predict antibody-specific B cell epitopes. Structure. 2014;22(4):646–657. doi: 10.1016/j.str.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 189.Hua CK, Gacerez AT, Sentman CL, Ackerman ME, Choi Y, et al. Computationally-driven identification of antibody epitopes. Elife 2017;6:e29023. [DOI] [PMC free article] [PubMed]
- 190.Raoufi E, EinAbadi H, Hemmati M, Fallahi H. Predicting candidate epitopes on Ebolaviruse for possible vaccine development. In: 2015 IEEE/ACM International Conference on Advances in Social Networks Analysis and Mining (ASONAM). Paris, France; 2015.
- 191.Kesmir C, Nussbaum AK, Schild H, Detours V, Brunak S. Prediction of proteasome cleavage motifs by neural networks. Protein Eng 2002;15:287–296. [DOI] [PubMed]
- 192.Noguchi Hideki, Kato Ryuji, Hanai Taizo, Matsubara Yukari, Honda Hiroyuki, Brusic Vladimir. Hidden Markov model-based prediction of antigenic peptides that interact with MHC class II molecules. J Biosci Bioeng. 2002;94(3):264–270. doi: 10.1263/jbb.94.264. [DOI] [PubMed] [Google Scholar]
- 193.Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, et al. Sensitive quantitative predictions of peptide-MHC binding by a 'Query by Committee' artificial neural network approach. Tissue Antigens 2003;62:378–384. [DOI] [PubMed]
- 194.Nielsen M., Lundegaard C., Worning P., Hvid C.S., Lamberth K., Buus S. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004;20(9):1388–1397. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- 195.Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinf. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Antes I., Siu S.W.I., Lengauer T. DynaPred: a structure and sequence based method for the prediction of MHC class I binding peptide sequences and conformations. Bioinformatics. 2006;22(14):e16–e24. doi: 10.1093/bioinformatics/btl216. [DOI] [PubMed] [Google Scholar]
- 197.Donnes P., Kohlbacher O. SVMHC: a server for prediction of MHC-binding peptides. Nucleic Acids Res. 2006;34(Web Server):W194–W197. doi: 10.1093/nar/gkl284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liu W., Meng X., Xu Q., Flower D.R., Li T. Quantitative prediction of mouse class I MHC peptide binding affinity using support vector machine regression (SVR) models. BMC Bioinf. 2006;7:182. doi: 10.1186/1471-2105-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinf. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Reynisson Birkir, Barra Carolina, Kaabinejadian Saghar, Hildebrand William H., Peters Bjoern, Nielsen Morten. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J Proteome Res. 2020;19(6):2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 202.Kim Yohan, Sidney John, Pinilla Clemencia, Sette Alessandro, Peters Bjoern. Derivation of an amino acid similarity matrix for peptide: MHC binding and its application as a Bayesian prior. BMC Bioinf. 2009;10(1):394. doi: 10.1186/1471-2105-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang H, Lund O, Nielsen M. The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics 2009;25:1293–1299. [DOI] [PMC free article] [PubMed]
- 204.Karosiene Edita, Lundegaard Claus, Lund Ole, Nielsen Morten. NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics. 2012;64(3):177–186. doi: 10.1007/s00251-011-0579-8. [DOI] [PubMed] [Google Scholar]
- 205.Trolle Thomas, Nielsen Morten. NetTepi: an integrated method for the prediction of T cell epitopes. Immunogenetics. 2014;66(7-8):449–456. doi: 10.1007/s00251-014-0779-0. [DOI] [PubMed] [Google Scholar]
- 206.Rasmussen Michael, Fenoy Emilio, Harndahl Mikkel, Kristensen Anne Bregnballe, Nielsen Ida Kallehauge, Nielsen Morten. Pan-Specific prediction of peptide-MHC Class I complex stability, a correlate of T cell immunogenicity. J Immunol. 2016;197(4):1517–1524. doi: 10.4049/jimmunol.1600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.EL‐Manzalawy Yasser, Dobbs Drena, Honavar Vasant. Predicting linear B-cell epitopes using string kernels. J Mol Recognit. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sweredoski MJ, Baldi P. COBEpro: a novel system for predicting continuous B-cell epitopes. Protein Eng Des Sel 2009;22:113–20. [DOI] [PMC free article] [PubMed]
- 209.Singh H, Ansari HR, Raghava GP. Improved method for linear B-cell epitope prediction using antigen's primary sequence. PLoS One 2013;8:e62216. [DOI] [PMC free article] [PubMed]
- 210.Lian Y., Ge M., Pan X.M. EPMLR: sequence-based linear B-cell epitope prediction method using multiple linear regression. BMC Bioinf. 2014;15:414. doi: 10.1186/s12859-014-0414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sher G., Zhi D., Zhang S. DRREP: deep ridge regressed epitope predictor. BMC Genom. 2017;18:676. doi: 10.1186/s12864-017-4024-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Manavalan B., Govindaraj R.G., Shin T.H., Kim M.O., Lee G. iBCE-EL: a new ensemble learning framework for improved linear B-cell epitope prediction. Front Immunol. 2018;9:1695. doi: 10.3389/fimmu.2018.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Huang J., Gutteridge A., Honda W., Kanehisa M. MIMOX: a web tool for phage display based epitope mapping. BMC Bioinf. 2006;7:451. doi: 10.1186/1471-2105-7-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mayrose I., Penn O., Erez E., Rubinstein N.D., Shlomi T., Freund N.T. Pepitope: epitope mapping from affinity-selected peptides. Bioinformatics. 2007;23(23):3244–3246. doi: 10.1093/bioinformatics/btm493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sweredoski MJ, Baldi P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 2008;24:1459–60. [DOI] [PubMed]
- 216.Sun J, Wu D, Xu T, Wang X, Xu X, et al. SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res 2009;37:W612–6. [DOI] [PMC free article] [PubMed]
- 217.Rubinstein Nimrod D, Mayrose Itay, Martz Eric, Pupko Tal. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinf. 2009;10(1):287. doi: 10.1186/1471-2105-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Negi S.S., Braun W. Automated detection of conformational epitopes using phage display peptide sequences. Bioinform Biol Insights. 2009;3:71–81. doi: 10.4137/bbi.s2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liang Shide, Zheng Dandan, Standley Daron M, Yao Bo, Zacharias Martin, Zhang Chi. EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinf. 2010;11(1):381. doi: 10.1186/1471-2105-11-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ansari Hifzur, Raghava Gajendra PS. Identification of conformational B-cell Epitopes in an antigen from its primary sequence. Immun Res. 2010;6(1):6. doi: 10.1186/1745-7580-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chen Wen, Sun Ping, Lu Yang, Guo William W, Huang Yan, Ma Zhi. MimoPro: a more efficient Web-based tool for epitope prediction using phage display libraries. BMC Bioinf. 2011;12(1):199. doi: 10.1186/1471-2105-12-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Chen W, Guo WW, Huang Y, Ma Z. PepMapper: a collaborative web tool for mapping epitopes from affinity-selected peptides. PLoS One 2012;7:e37869. [DOI] [PMC free article] [PubMed]
- 223.Soga S, Kuroda D, Shirai H, Kobori M, Hirayama N. Use of amino acid composition to predict epitope residues of individual antibodies. Protein Eng Des Sel 2010;23:441–448. [DOI] [PubMed]
- 224.Brenke R, Hall DR, Chuang G-Y, Comeau SR, Bohnuud T, et al. Application of asymmetric statistical potentials to antibody–protein docking. Bioinformatics 2012;28:2608–14. [DOI] [PMC free article] [PubMed]