Abstract
Eomesodermin (Eomes) is a T‐box transcription factor that drives the differentiation and function of cytotoxic lymphocytes. However, the underlying function and mechanism of Eomes in tumor cells remains elusive. Here, we studied the role of Eomes in human esophageal squamous cell carcinoma (ESCC). Using 2 human ESCC cell lines, we found that Eomes knockdown reduced esophageal cancer cell proliferation and that the esophageal cancer cell cycle was blocked in the G2/M phase. Mechanistically, we identified CCL20 as the main downstream target of Eomes. Furthermore, we found that CCL20 could chemoregulate regulatory T cells (Tregs) through their specific receptor CCR6, then promoting the proliferation of esophageal cancer cells. Eomes knockdown also delayed the growth of human ESCC xenografts in BALB/c nude mice. Importantly, in 133 human ESCC tissues, high Eomes levels were associated with poor clinical prognosis. Overall, our findings suggested that the Eomes‐CCL20‐CCR6 pathway plays a vital role in human ESCC progress. Therefore, targeting this pathway may represent a promising strategy for controlling human ESCC.
Keywords: CCL20, Eomes, esophageal squamous cell carcinoma, prognosis, Tregs
Lian et al report that the Eomes‐CCL20‐CCR6 pathway plays a vital role in human ESCC progress and that high Eomes levels were associated with poor clinical prognosis. They found that CCL20 could chemoregulate Tregs through their specific receptor CCR6 and then promote the proliferation of esophageal cancer cells. Eomes knockdown also delayed the growth of human ESCC xenografts in BALB/c nude mice.
Abbreviations
- CCK8
cell counting kit 8
- CFSE
carboxyfluorescein succinimidyl amino ester
- EAC
esophageal adenocarcinoma
- ELISA
enzyme‐linked immunosorbent assay
- Eomes
eomesodermin
- ESCC
esophageal squamous cell carcinoma
- IHC
immunohistochemistry
- NK
natural killer cells
- SI
staining intensity
- TCGA
The Cancer Genome Atlas
- Tregs
regulatory T cells
1. INTRODUCTION
Esophageal cancer is one of the most aggressive malignancies with high mortality and poor prognosis worldwide. 1 Esophageal cancer is generally divided into 2 subtypes, namely ESCC and EAC. 1 There are significant differences in pathogenesis, epidemiology, pathology, and geographical distribution. 2 EAC is more common in Western countries, but ESCC is the most common pathological type in Asia. In addition, about half of the total number of patients with ESCC occurs in China. 3 The overall 5‐y survival rate of most patients with ESCC is relatively poor. 4 The prognostic biomarkers of ESCC are not however well established. Therefore, effective biomarkers for diagnosis of ESCC are necessary.
Eomesodermin (Eomes) was first reported in cytotoxic T cells. This study showed that Eomes and T‐bet synergistically induced the expression of CD122 and inhibit the expression of Th17 cytokines. 5 T‐bet and Eomes were originally studied in T cells and their expression was inducible after activation and differentiation. 6 These factors are encoded by Tbx21 and Eomes genes, respectively, and are thought to be the only T‐box proteins expressed in the immune system. 6 Eomes expression is lower in CD4+ T cells compared with CD8+ T cells. Furthermore, high levels of Eomes expression was shown to rescue IFN‐γ produced by T‐bet‐deficient T cells and promote IFN‐γ production and cytotoxicity in CD4+ T cells. 7 , 8 , 9 , 10 In addition, Eomes could also promote the development and maturation of natural killer (NK) cells. 11 These studies, however, described little about the exact function of Eomes in tumor cells.
Eomes has disparate effects on different tumor types. Depending on the stage and tissue, Eomes produces the opposite effects of suppressing and promoting tumors. Eomes expression is not only related to the early recurrence of gastric cancer after surgery, but also related to poor disease‐free survival time. 12 Moreover, high expression of Eomes is associated with poor overall survival of patients with colorectal cancer, 13 however in metastatic renal cell carcinoma patients high expression of Eomes was considered to be an independent good prognostic factor for patients survival. 14 Overall, these studies have shown that Eomes exerts opposite effects in different types of tumor. Eomes methylation levels are also closely related to tumorigenesis; in patients with high‐grade bladder cancer, Eomes show tumor‐specific DNA hypermethylation. 15 The methylation level of Eomes in urine samples can be used as a diagnostic biomarker for monitoring bladder cancer recurrence. 16 Moreover, in patients with ESCC, hypermethylation of Eomes may be an effective diagnostic method for ESCC. 17 Abnormal methylation of Eomes at the promoter region leads to its downregulation and results in the occurrence and development of hepatocellular carcinoma. 18 These reports showed that Eomes plays an important role in tumor progression, however in these studies there were no insights into the detailed mechanism of Eomes in tumorigenesis. In addition, these studies did not report on the relationship between Eomes and prognosis in patients with ESCC. Finally, these studies did not explore the important role of Eomes in tumorigenesis through the esophageal tumor microenvironment.
In our study, we identified the important role of Eomes in maintaining human esophageal cancer cell proliferation through the CCL20‐CCR6 pathway. Furthermore, high levels of Eomes expression were closely related to the poor prognosis of ESCC patients. Our data also suggested that Eomes drives CCL20 secretion to promote the chemotaxis of regulatory T cells (Tregs) in the tumor microenvironment, which leads to the malignant progression of ESCC; the signaling pathway may serve as a potential therapeutic target for ESCC.
2. MATERIALS AND METHODS
2.1. Cell lines and cell cultures
Human ESCC cells lines were obtained from the Cancer Hospital of the Peking Union Medical College (Beijing, China). All cells lines were cultured in DMEM (Gibco) with 10% FBS (Gibco), 100 units/mL penicillin and 100 μg/mL streptomycin in an incubation system at 37°C with 5% CO2 in air.
2.2. RNA extraction and real‐time quantitative PCR (qRT‐PCR)
Total RNA was extracted from tumor cell lines or tissue using TRIzol reagent (TaKaRa Bio). RNA concentration and purity were determined using a NanoDrop spectrophotometer 2000 (Thermo Fisher Scientific). Here, 1 μg RNA was reverse transcribed to cDNA according to the manufacturer’s instructions for the Prime Script RT reagent kit (TaKaRa Bio). Specific primers and SYBR Green qPCR Master Mix (Roche) were used to perform qRT‐PCR experiments on an Agilent Mx3005P System (Agilent Technologies). The PCR primer sequences are as follows: Eomes forward 5′‐ACTGGTTCCCACTGGATGAG‐3′, reverse 5′‐CCACGCCATCCTCTGTAACT‐3′; CCL20 forward 5′‐TGCTGTACCAAGAGTTTGCTC‐3′, reverse 5′‐CGCACACAGACAACTTTTTCTTT‐3′; GAPDH forward 5′‐GCACCGTCAAGGCTGAGAAC‐3′, reverse 5′‐TGGTGAAGACGCCAGTGGA‐3′. Data with GAPDH as endogenous control analysis were analyzed using the 2−ΔΔCt method.
2.3. RNA interference
Thermo Fisher website software (https://rnaidesigner.thermofisher.com/rnaiexpress/) was applied to predict and design the transient knockdown sequence of Eomes as CAGGAGATTTCATTCGGGAAATTAA. siRNA produced by Genepharma was transferred to target cells using Lipofectamine 3000 (Invitrogen) and Opti‐MEM according to manufacturer’s instructions.
The lentiviral vector pSIN‐EF2‐IRES‐GFP was used to carry out stable knockdown. Eomes shRNA stably knocked down Eomes expression. DNA sequencing verified that the insertion sequences were correctly in place. Cells that expressed GFP were isolated using a flow cytometer (Beckman MoFlo XDP, USA).
2.4. Cell Counting Kit 8 (CCK8)
In total, 5 × 103 cells were placed in a 96‐well plate and 10 μL CCK8 were added to each well according to the manufacturer’s instructions for the CCK8 assay (Dojindo). Cells were cultured in DMEM (Gibco) with 10% FBS (Gibco) in an incubation system for 4 h at 37°C with 5% CO2 in air. At the same time each day, cell activity was measured using a plate reader (MULTI‐SKANMK3, Thermo Scientific) for absorbance at 450 nm.
2.5. Cell cycle analysis
Cells were obtained after trypsinization, washed with cold PBS, and fixed at 4°C. After washing, cells were treated with RNase (50 μg/mL) for 30 min at 37°C. Cell suspensions were stained with propidium iodide (50 μg/mL) in the dark at 4°C for 30 min. Detection was performed using a BD Flow cytometer (BD Pharmingen), and the cell cycle was analyzed using Modfit software.
2.6. Clone formation
Cells were counted the and seeded at 1 × 103 cells in 6‐well plates. After 2 wk of incubation, the cell colonies were stained with crystal violet dye and counted under a microscope. Results were expressed as the mean ± SEM of 3 independent experiments.
2.7. Proliferation index assay
Tumor cells were labeled with 1 mmol/L CFSE before culture; 1 × 105/well CFSE‐labeled tumor cells were cultured in 6‐well plates and then collected at different time points and the proliferation index measured using flow cytometry.
2.8. Apoptosis detection
Cells were collected and washed twice with pre‐chilled PBS. The cells were suspended in 200 μL annexin V binding buffer (Biolegend) to a final cell concentration of 106 cells/mL, then 1 μL of Alexa Fluor 647 Annexin V (Biolegend) was added to the cell suspension in the dark, which was incubated at 4°C for 15 min, and then propidium iodide was added (Sigma). Flow cytometry was used to analyze apoptosis.
2.9. Transwell migration assays
Transwell migration assays were carried out in 24‐well plates with insertions (5 μm, Corning) according to the manufacturer’s instructions. Cell suspensions (2 × 105 cells/well) were added to the upper chamber and 600 μL RPMI‐1640 medium containing no FBS was added to the lower chambers. After 24 h, cells that attached to the lower surface were stained with 0.1% crystal violet for 15 min.
2.10. Human cytokine array
Here, 5 × 103/cells per well were seeded into 24‐well plates and cultured overnight in an incubator. The medium was changed for fresh medium the next day and cells were incubated for 48 h, then the supernatant was collected. Human LEGENDplex array kits (BioLegend, San Diego, California, USA) were used to detect the secretion of various chemokines in accordance with the manufacturer’s instructions.
2.11. ELISA
An ELISA kit (BioLegend) was used to determine the CCL20 level in the cell culture supernatants following the manufacturer’s instructions.
2.12. Tumor xenografts
Institutional guidelines for the care and use of laboratory animals were applied to all animal experiments using the committee of the First Affiliated Hospital of Zhengzhou University. Here, 7‐wk‐old female BALB/c nude mice (Vital River Laboratory Animal Technology) were divided randomly into 2 groups (n = 5/group) and injected with designated tumor cells (5 × 106). Tumor size was measured and tumor volume was calculated: (length × width2)/2. At the end of the experiment, all animals were sacrificed, and tumors were collected for further experiments.
2.13. Collection of ESCC patients’ samples
Human ESCC tissue samples were from the First Affiliated Hospital of Zhengzhou University in China. In total, 133 fresh ESCC tissues and pairs of adjacent normal tissues were collected with the full knowledge and consent of patients from the First Affiliated Hospital of Zhengzhou University. Pathology testing confirmed that these specimens could be used. Eomes expression was checked at the mRNA level; 45 formalin‐fixed paraffin‐embedded ESCC tissue samples were collected to check Eomes expression at the protein level. Patient age, gender, tumor stage, lymph nodes metastasis, and histological grade were assessed and listed in Table 1.
TABLE 1.
Association between Eomes expression and clinicopathological parameters in patients with ESCC
| Variables | Number | Eomes expression | χ2 | P‐value | |
|---|---|---|---|---|---|
| High group | Low group | ||||
| Age (y) | |||||
| ≤60 | 47 | 37 | 10 | 1.064 | .302 |
| >60 | 81 | 57 | 24 | ||
| Gender | |||||
| Male | 79 | 39 | 40 | 0.165 | .684 |
| Female | 49 | 26 | 23 | ||
| Depth of invasion | |||||
| T1‐T2 | 78 | 61 | 17 | 3.958 | .046 |
| T3‐T4 | 50 | 31 | 19 | ||
| Lymph node metastasis | |||||
| Positive | 47 | 30 | 17 | 4.189 | .040 |
| Negative | 81 | 65 | 16 | ||
| Differentiation | |||||
| Well vs moderately | 60 | 36 | 24 | 9.106 | .002 |
| Poorly | 68 | 57 | 11 | ||
| Stage | |||||
| I, II, III, IV | 70 | 52 | 18 | 4.248 | .039 |
| 58 | 40 | 18 | |||
| Survival condition | |||||
| Living | 45 | 36 | 9 | 5.749 | .016 |
| Dead | 83 | 49 | 34 | ||
2.14. Immunohistochemistry
Immunohistochemistry was performed; anti‐Eomes antibody (1:40, R&D, #644730) was used according to the manufacturer’s protocol for this experiment. The results were analyzed and scored by 2 individuals. The percentages of tumor cells expressing the target protein were classified as follows: 0 (no positive tumor cells), 1 (<10% positive tumor cells), 2 (10%‐50% positive tumor cells), and 3 (>50% positive tumor cells). Based on the SI, cells were divided into the following grades: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). The staining index was calculated as the intensity of staining × proportion of positive tumor cells, with scores of 0, 1, 2, 3, 4, 6, and 9, respectively. A staining index score ≥4 was considered as high expression, and a score ≤3 was considered as low expression.
2.15. Western blotting
Cells were placed in pre‐chilled lysis buffer (150 mmol/L NaCl, 5 mmol/L EDTA, 50 mmol/L Tris, 1% NP‐40, 0.5% sodium deoxycholate, 0.1% SDS) and protease inhibitor cocaine (Sigma‐Aldrich). An equal amount of total protein (50 μg) from each sample was separated on a 10% SDS‐PAGE gel and transferred to nitrocellulose blotting membranes (GE Healthcare Life science). Membranes were blocked with 5% BSA and then incubated with a primary antibody to detect Eomes (1:1000, R&D, #644730) and β‐actin (Cell Signaling Technology). Membranes were incubated with the corresponding secondary antibody (Cell Signaling Technology); protein expression was detected and photographed using a chemiluminescence imaging system (Bio‐Rad Laboratories).
2.16. TCGA database analysis
Clinical data obtained from ESCC cases were analyzed together with gene expression using TCGA; https://cancergenome.nih.gov/). All patients had ESCC and had not undergone any treatment.
2.17. Statistical analysis
SPSS statistical software or GraphPad Prism 6 (Graph Pad Software) was used to analyze data by log‐rank test, Student t test, chi‐squared test, log‐rank test, or one‐way ANOVA P < .05. Statistically significant difference was identified, *P < .05, ** P < .01, ***P < .001. Data were presented as the mean ± SD from at least 2 or 3 independent experiments.
3. RESULTS
3.1. Eomes is highly expressed in ESCC tissues and correlates with poor prognosis in ESCC patients
Eomes mRNA was found to be highly expressed in cancer tumor tissues from 133 patients with ESCC compared with their adjacent normal tissues (Figure 1A). Eomes mRNA was also related to the clinical tumor stage of patients with ESCC. Eomes expression levels were higher in patients with stage III/IV disease compared with stage I/II (Figure 1B). Eomes was highly expressed in patients with metastatic ESCC (Figure 1C). There was also a strong correlation between Eomes expression and tumor differentiation status of patients with ESCC (Figure 1D). In addition, as shown in Table 1, Eomes expression was related to clinical histopathology of ESCC. Eomes mRNA expression had no significant correlation with patient age or gender. IHC was used to investigate Eomes expression at the protein level in formalin‐fixed paraffin‐embedded tumor tissues compared with the adjacent normal tissues. Figure 1E shows the protein expression levels of Eomes in tumor tissues compared with the adjacent normal tissues of patients with ESCC. Among ESCC patients for whom overall survival data were available, patients with higher Eomes protein expression had a shorter overall survival time compared with patients with lower Eomes protein levels (Figure 1F). Furthermore, we used TCGA database to verify the correlation between Eomes expression and the clinical tumor stages of patients with ESCC (Figure 1G). TCGA database revealed that the high expression Eomes group had a poor prognosis compared with the low expression group (Figure 1H). These data indicated that Eomes expression was significantly upregulated in patients with ESCC and correlated with overall survival.
FIGURE 1.
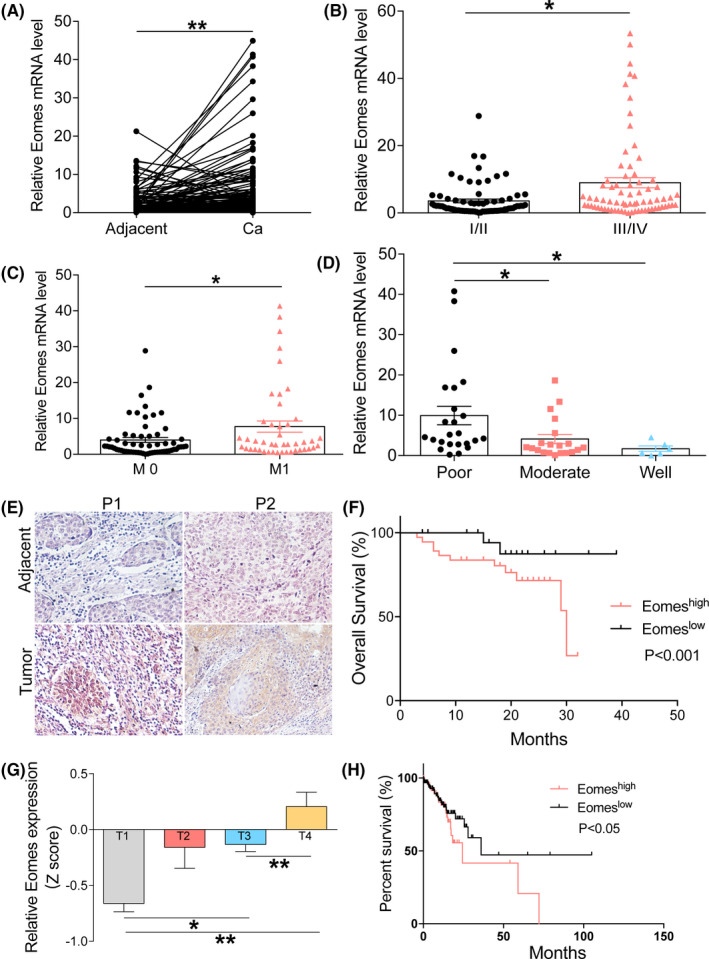
Eomes was highly expressed in ESCC tissues and correlates with the prognosis of ESCC patients. A, Eomes mRNA expression was detected in matched cancer and adjacent cancer patients (n = 133). B‐D, Correlation between Eomes expression mRNA and tumor clinical stage, metastasis, and tumor differentiation status of ESCC patients. E, IHC was used to detect Eomes expression protein in matched cancer and adjacent of ESCC patients. F, Correlation between Eomes expression protein and overall survival in ESCC patients. G, TCGA database analyzed the expression level of Eomes in ESCC patients with different tumor stages. H, Correlation between Eomes expression level and prognosis of ESCC patients by TCGA database. Data are presented as mean ± SD. *P < .05, **P < .01, ***P < .001
3.2. Eomes promotes the proliferation of esophageal cancer cells in vitro
To investigate the effect of Eomes on the function of esophageal cancer cells, we used qRT‐PCR to detect the expression levels of Eomes in different esophageal cancer cell lines. We found that Eomes expression was higher in KYSE450 and TE7 cell lines compared with in other cell lines (Figure 2A). Therefore, we used siRNA interference to establish 2 esophageal cancer cell lines that transiently knocked down Eomes. We detected Eomes knockdown efficiency at the mRNA level (Figure 2B). We found that the colony‐forming ability of esophageal cancer cells was reduced when Eomes was knocked down (Figure 2C,D). We analyzed the effects of Eomes on the cell cycle and found that the cell cycle was blocked in the G2/M phase after knocking down Eomes in KYSE450 and TE7 cell lines (Figure 2E). We used CCK8 to test the effect of Eomes knockdown on the proliferation of esophageal cancer cells and found that proliferation decreased after Eomes knockdown (Figure 2F). Proliferation index of esophageal cancer cells after knocking down Eomes was assessed by CFSE assay, which verified that knocking down Eomes affected the proliferation index (Figure 2G). Flow cytometry analysis found that Eomes had no effect on apoptosis of esophageal cancer cells (Figure 2H). Moreover, we also found that after knocking down Eomes, the expression levels of genes related to proliferation were reduced, but gene expression related to apoptosis was not changed (Figure S1). These findings suggested a potential role for Eomes in the proliferative capacity of ESCC.
FIGURE 2.
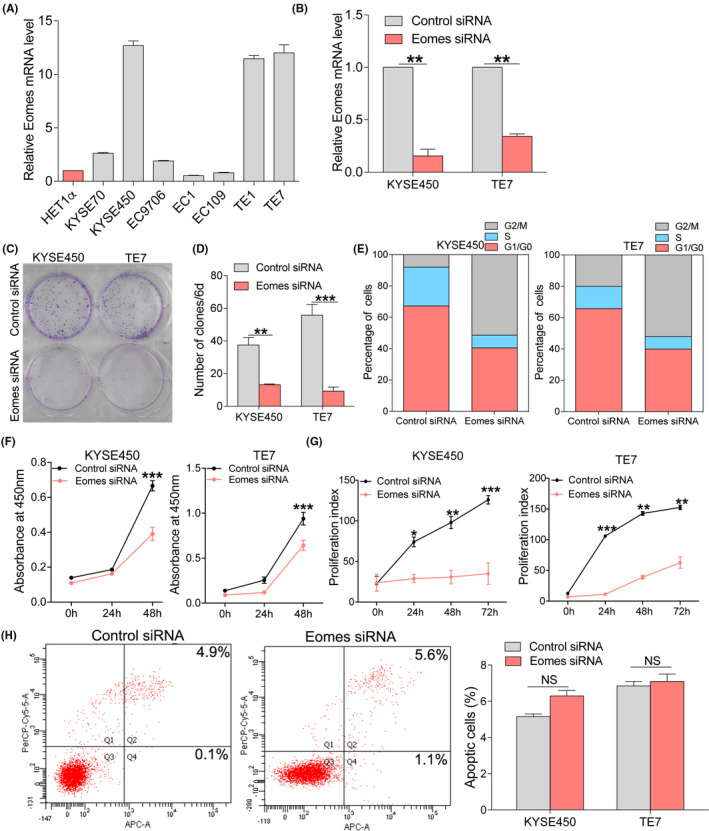
Eomes knockdown led to reduced proliferation of esophageal cancer cells in vitro. A, Different expression levels of Eomes mRNA in different esophageal cancer cell lines. B, The knockdown efficiency of Eomes was detected by qRT‐PCR in KYSE450 and TE7 cell lines. C, D, Effects of Eomes knockdown on colony formation in esophageal cancer cells. E, Effects of Eomes knockdown on the esophageal cancer cells cycle by flow cytometry. F, Effects of Eomes knockdown on the proliferation of esophageal cancer cells by CCK8. G, CFSE staining to detect the effect of Eomes knockdown on the proliferation of esophageal cancer cells. H, Effects of Eomes knockdown on apoptosis of esophageal cancer cells by flow cytometry. Mean ± SD of relative fold changes from triplicate experiments was plotted. *P < .05, **P < .01, ***P < .001
3.3. Eomes knockdown resulted in decreased CCL20 expression
In order to investigate the mechanism by which Eomes regulated the proliferation of esophageal cancer cells, we used the KYSE450 line to establish an esophageal cancer cell line that stably knocked down Eomes. We tested Eomes knockdown efficiency at the mRNA (Figure 3A) and protein levels (Figure 3B). Using multiplex ELISA assays, we found that the Eomes stable knockdown had markedly blocked CCL20 secretion (Figure 3C). We further found by qRT‐PCR that Eomes knockdown affected chemokine expression and confirmed that Eomes regulated CCL20 (Figure 3D). By analyzing TCGA database, we found that there was a strong positive correlation between Eomes and CCL20 in patients with ESCC (Figure 3E). We validated the regulation of CCL20 by Eomes knockdown models that were constructed in KYSE450 and TE7 cell lines. When knocking down Eomes, the CCL20 level was seen to decrease in qRT‐PCR (Figure 3F) and ELISA (Figure 3G). We used a CCL20 recombinant protein to find if Eomes affected the proliferation of esophageal cancer cells through CCL20 (Figure 3H). Using TCGA database, we found that ESCC patients with high CCL20 expression had a poor prognosis (Figure 3I). These data indicated that CCL20 was a downstream target of the Eomes signaling pathway.
FIGURE 3.
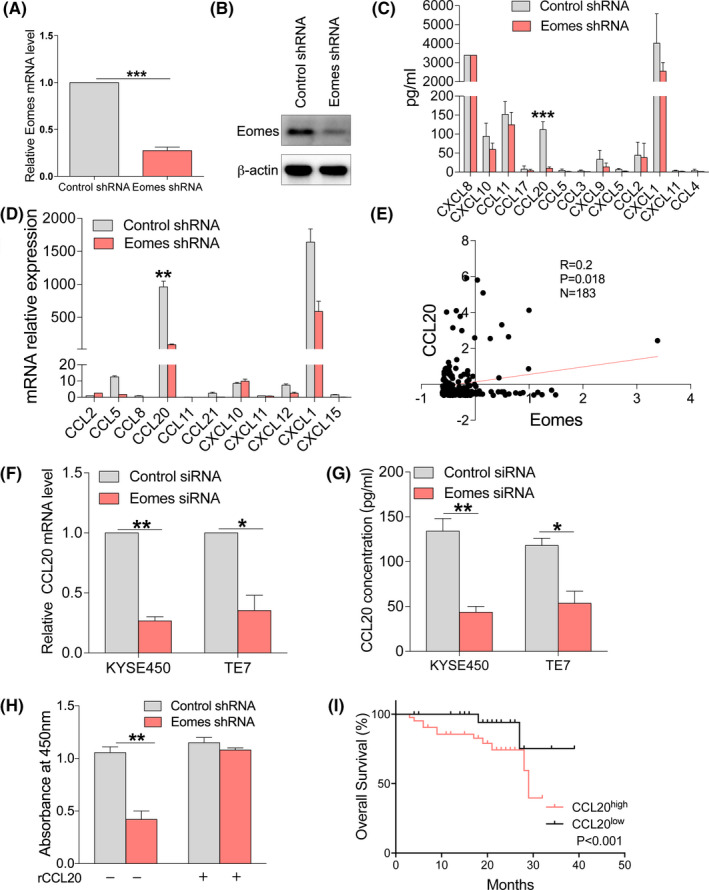
Eomes knockdown resulted in decreased expression of CCL20. A, Analysis of efficiency after stable knockdown of Eomes in the KYSE450 cell line using qRT‐PCR. B, Analysis of efficiency after stable knockdown of Eomes in the KYSE450 cell line using western blotting. C, Supernatants were collected from KYSE450 cells that had stably knocked down Eomes and then screened by human chemokine assay. D, Effects of Eomes knockdown on expression of chemokines by qRT‐PCR. E, TCGA database was used to analyze the correlation between Eomes and CCL20 in patients with ESCC. F, Expression of CCL20 after knockdown of Eomes was detected by qRT‐PCR. G, Expression of CCL20 after knockdown of Eomes was detected by ELISA. H, Effects of Eomes on the proliferation of esophageal cancer cells after addition of recombinant protein CCL20. I, TCGA database was used to analyze the differences in the overall survival of patients with ESCC with different expression levels of CCL20. Data are presented as mean ± SD. *P < .05, **P < .01, ***P < .001
3.4. The Eomes‐CCL20 pathway regulates migration of Tregs
Chemokines regulate tumor progression in the tumor microenvironment by chemotactic immune cells or immunosuppressive cells. 19 CCL20 has been reported to affect tumor progression through chemotactic Tregs in the tumor microenvironment. 20 Therefore, we next wanted to explore whether Eomes affected chemotaxis of Tregs through CCL20 and thus affected tumor cell proliferation in ESCC. To obtain PBMC, we collected peripheral blood from esophageal cancer patients, and found that in esophageal cancer patients the cell supernatant after knocking down Eomes had a reduced ability for chemotactic Tregs (Figure 4A). Figure 4B shows the statistical analysis. Through TCGA database analysis we found that the Eomes and Foxp3 expression were correlated in esophageal cancer patients (Figure 4C). TCGA database analysis also showed high Foxp3 expression in esophageal cancer compared with adjacent cancer tissue (Figure S3A). Furthermore, we found high Foxp3 protein expression in tumor tissue (Figure S3B).
FIGURE 4.
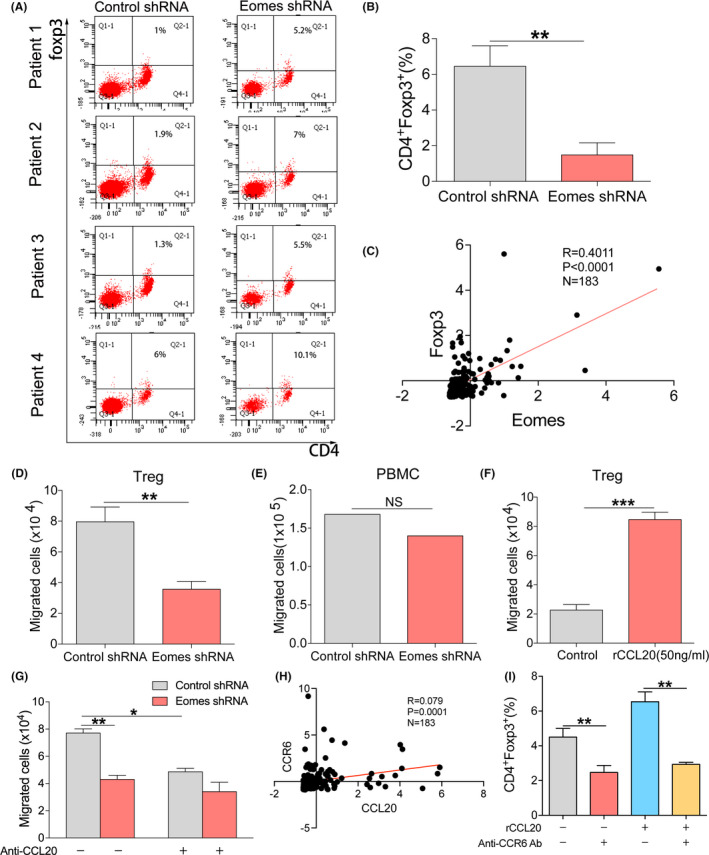
Eomes‐CCL20‐CCR6 pathway regulates migration of Tregs. A, The effect of Eomes knockdown cell line supernatant on Tregs chemotaxis in patients with ESCC by flow cytometry. B, Statistical analysis of (A). C, TCGA database analysis of the correlation between Eomes and Foxp3 expression in ESCC patients’ tissues. D, The effect of Eomes knockdown cell line supernatant on the chemotactic capacity for Tregs. E, Effect of Eomes knockdown cell line supernatant on chemotaxis of PBMC. F, Transwell assay detects the chemotactic ability of Tregs after adding CCL20 recombinant protein in vitro. G, Effect of Eomes on Tregs chemotaxis was detected after neutralizing CCL20. H, TCGA database analysis of the correlation between CCL20 and CCR6 expression in ESCC tissues. I, CCL20‐CCR6 pathway affects the chemotaxis of Tregs as shown using CCL20 recombinant protein and CCR6 neutralizing antibody. Data are presented as mean ± SD. **P < .01, ***P < .001
To further verify that knocking down Eomes led to a reduction in CCL20 expression and thus reduced the chemotactic ability of Tregs, we collected peripheral blood from healthy donors and isolated PBMC. We found that the ability to chemotactically attract Tregs was reduced in the supernatants of Eomes knockdown cells (Figure 4D), but there was no effect on the chemotaxis of PBMC (Figure 4E). In addition, we added CCL20 recombinant protein in vitro to verify the chemotactic effect of CCL20 on Tregs (Figure 4F). We used a neutralizing antibody for CCL20, indicating that Eomes chemotactically attracted Treg cells through CCL20 (Figure 4G). As CCR6 is a specific receptor for CCL20, we speculated that chemotaxis of Treg cells could be due to binding of CCL20 to CCR6. By analyzing TCGA database, we found that there was a strong positive correlation between CCL20 and CCR6 levels in ESCC patients (Figure 4H). As seen in Figure 4I, using a CCR6 neutralizing antibody we showed that Eomes affected the chemotaxis of Tregs by regulating the CCL20‐CCR6 pathway. From this result we speculated that the Eomes‐CCL20‐CCR6 signaling pathway affected chemotaxis of Tregs in the esophageal cancer microenvironment and then affected the progression of esophageal cancer cells.
3.5. Effects of Eomes on the progression of esophageal cancer in vivo
To further verify the proliferation effect of Eomes on esophageal cancer cells, we used in vivo experiments. We subcutaneously inoculated BALB/c nude mice with an esophageal cancer cell line KYSE450 that had stably knocked down Eomes. We found that tumor proliferation was suppressed in the knockdown Eomes group and tumor proliferation rate became slower (Figure 5A). There was also a significant difference in tumor tissue weight after Eomes was knocked down (Figure 5B). We used qRT‐PCR to analyze Eomes expression in tumor tissues and found that Eomes expression was reduced in the knockdown group (Figure 5C). Correspondingly, we also found that CCL20 expression was also reduced in knockdown Eomes tumor tissues (Figure 5D). Taken together, these findings indicated that Eomes inhibition reduced esophageal cancer progression in vivo. A schematic diagram of the effect of Eomes on ESCC progress is given in Figure 5E.
FIGURE 5.
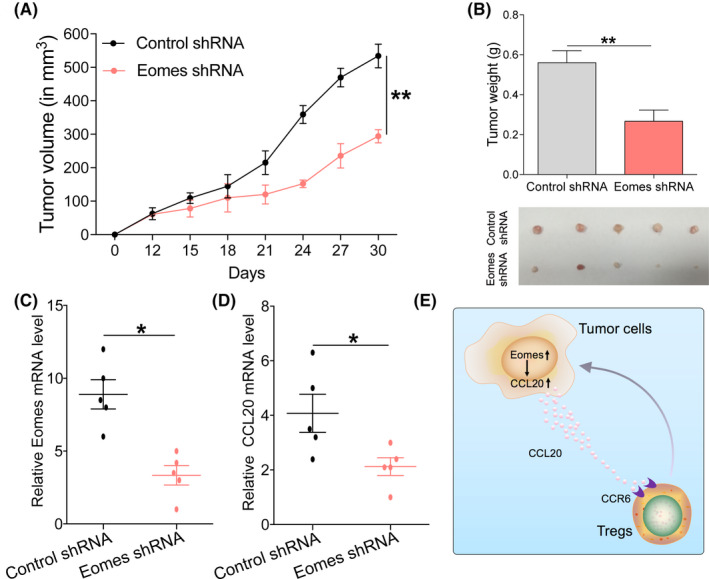
Effects of Eomes on the progression of esophageal cancer in vivo. A, Tumor proliferation rate of subcutaneous vaccination with KYSE450 with Eomes knockdown. B, Tumor tissue weight of xenografts harvested after tumor challenge. C, Detection of Eomes expression in xenograft tumor tissues by qRT‐PCR. D, Detection of CCL20 expression in xenograft tumor tissues by qRT‐PCR. E, Research summary: Eomes through the CCL20‐CCR6 pathway and chemotaxis of Tregs in the ESCC tumor microenvironment affects the progression of esophageal cancer. Data are presented as mean ± SD. *P < .05, **P < .01
4. DISCUSSION
Eomes is a T‐box family transcription factor and is closely related to T‐bet. 11 It is highly expressed in NK cells and various subsets of effector and memory CD8+ T cells. 21 , 22 Eomes plays an important role in the differentiation of CD8+ T cells and NK cells. 23 Eomes expression can be found in almost all Th subtypes 24 and can induce IFN‐γ expression, 25 however the role of Eomes in tumors has been rarely reported and results in existing reports on tumor progression are not consistent. Our study found that Eomes was overexpressed in ESCC and recruited Tregs through CCL20 to promote ESCC progression. We also found that Eomes overexpression was associated with poor survival in ESCC patients. These data provide the theoretical basis for use of the Eomes‐CCL20‐CCR6 signaling pathway as a new strategy for esophageal cancer treatment.
The effects of Eomes on tumor cell function have not been reported consistently. Overexpression of Eomes in colorectal cancer resulted in a poor prognosis for colorectal cancer patients, and Eomes knockdown caused a reduction in colorectal cancer cell proliferation. 13 This result was consistent with our findings, however in contrast Eomes expression in metastatic renal cell carcinoma patients was associated with a good prognosis. 14 Therefore, in our study, we first explored Eomes expression in ESCC. We observed that both Eomes mRNA and protein levels were upregulated in esophageal cancer tissues and that Eomes expression was related to the poor prognosis of ESCC patients and to clinicopathology. This difference in research results may be due to the different types of tumor pathology.
In addition to the important role of Eomes for T cells and NK cells, it also functions as a regulator in activating various downstream signals. In the current study, we found that Eomes knockdown could significantly inhibit CCL20 expression in ESCC, suggesting that Eomes plays a vital role in regulating the CCL20‐CCR6 pathway. CCR6 is the only specific receptor for CCL20 and it has been reported that CCL20 could be used as a potential predictor of survival in ESCC patients. 20 Our study raised the possibility that CCL20 is a downstream target of Eomes, in a related pathway for Eomes‐mediated tumorigenesis, however in this research the mechanism by which Eomes regulated the CCL20‐CCR6 pathway remained unknown. We initially screened using TCGA database and found a significant positive correlation between Eomes and NFκB expression (Figure S2). We speculated that Eomes may regulate the CCL20‐CCR6 pathway through phosphorylation of NFκB, this molecular mechanism will be explored in depth in future research.
Esophageal tumor xenograft experiments showed that Eomes had a profound effect on tumor growth. In vitro experiments also showed the effect of Eomes on esophageal cancer cell proliferation and that Eomes knockdown affected CCL20 secretion. However, it should be noted that CCL20 is an inflammatory chemokine and could also recruit suppressive immune cells to downregulate the antitumor response. 26 CCL20 is a chemokine that promotes tumor progression by recruiting Tregs. 27 The CCL20‐CCR6 interaction mediates migration of Tregs to the tumor microenvironment, which in turn leads to tumor progression and poor prognosis in hepatocellular carcinoma patients. 28 CCL20 secreted by colorectal cancer cells can recruit Tregs through FOXO1/CEBPB/NFκB signaling to enhance chemoresistance. 29 Human oral squamous cell carcinoma is rich in the chemokine CCL20, which is conducive to the recruitment and retention of CCR6+ Treg cells, thereby promoting cancer cell invasion and tumor progression, 30 therefore Tregs may be involved in esophageal cancer progression. In our research, we found that in the esophageal cancer microenvironment, the supernatant from knocked down Eomes lines significantly reduced chemotaxis of Tregs. We found that Eomes affected chemotaxis of Tregs by regulating CCL20, however by which pathway Tregs participate in Eomes‐induced esophageal cancer progression still needs to be determined. In the next step, we will explore the related mechanisms of the Eomes‐CCL20‐Treg pathway induced esophageal cancer cell proliferation and promotion of esophageal cancer progression.
In conclusion, our findings suggest that overexpression of Eomes may affect the esophageal tumor microenvironment through the CCL20‐CCR6 pathway and thus promote the progression of ESCC. Given the clinical relevance, this signaling pathway may become a potential therapeutic target for esophageal cancer treatment.
DISCLOSURE
Authors declare no conflicts of interest for this article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Our study was approved by the Ethics Committee of First Hospital of Zhengzhou University (ethical approval number: 2018‐KY‐92), and informed consent was obtained from all participants included in the study, in agreement with institutional guidelines.
Supporting information
Figure S1
Figure S2
Figure S3
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (grant no. 9194230059) and the State’s Key Project of Research and Development Plan (grant no. 2016YFC1303500).
Lian J, Liu S, Yue Y, et al. Eomes promotes esophageal carcinoma progression by recruiting Treg cells through the CCL20‐CCR6 pathway. Cancer Sci. 2021;112:144–154. 10.1111/cas.14712
These authors contributed equally to this work.
Contributor Information
Shengli Yang, Email: slyang@sibs.ac.cn.
Yi Zhang, Email: yizhang@zzu.edu.cn.
REFERENCES
- 1. Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400‐412. [DOI] [PubMed] [Google Scholar]
- 2. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241‐2252. [DOI] [PubMed] [Google Scholar]
- 3. Wei W‐Q, Hao C‐Q, Guan C‐T, et al. Esophageal histological precursor lesions and subsequent 8.5‐year cancer risk in a population‐based prospective study in China. Am J Gastroenterol. 2020;115(7):1036‐1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Besharat S, Jabbari A, Semnani S, Keshtkar A, Marjani J. Inoperable esophageal cancer and outcome of palliative care. World J Gastroenterol. 2008;14:3725‐3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Intlekofer AM, Takemoto N, Wherry EJ, et al. Effector and memory CD8+ T cell fate coupled by T‐bet and eomesodermin. Nat Immunol. 2005;6:1236‐1244. [DOI] [PubMed] [Google Scholar]
- 6. Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T‐bet, directs Th1 lineage commitment. Cell. 2000;100:655‐669. [DOI] [PubMed] [Google Scholar]
- 7. Yang Y, Xu J, Niu Y, Bromberg JS, Ding Y. T‐bet and eomesodermin play critical roles in directing T cell differentiation to Th1 versus Th17. J Immunol. 1950;2008(181):8700‐8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steiner DF, Thomas MF, Hu JK, et al. MicroRNA‐29 regulates T‐box transcription factors and interferon‐γ production in helper T cells. Immunity. 2011;35:169‐181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curran MA, Geiger TL, Montalvo W, et al. Systemic 4–1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J Exp Med. 2013;210:743‐755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suto A, Wurster AL, Reiner SL, Grusby MJ. IL‐21 inhibits IFN‐gamma production in developing Th1 cells through the repression of Eomesodermin expression. J Immunol. 1950;2006(177):3721‐3727. [DOI] [PubMed] [Google Scholar]
- 11. Zhang J, Marotel M, Fauteux‐Daniel S, et al. T‐bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur J Immunol. 2018;48:738–750. [DOI] [PubMed] [Google Scholar]
- 12. Cui Y, Yu S, Zhu M, et al. Identifying predictive factors of recurrence after radical resection in gastric cancer by RNA Immune‐oncology Panel. J Cancer. 2020;11:638‐647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang R, Kang Y, Löhr CV, et al. Reciprocal regulation of BMF and BIRC5 (Survivin) linked to Eomes overexpression in colorectal cancer. Cancer Lett. 2016;381:341‐348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dielmann A, Letsch A, Nonnenmacher A, Miller K, Keilholz U, Busse A. Favorable prognostic influence of T‐box transcription factor Eomesodermin in metastatic renal cell cancer patients. Cancer Immunol Immunother. 2016;65:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olkhov‐Mitsel E, Savio AJ, Kron KJ, et al. Epigenome‐wide DNA methylation profiling identifies differential methylation biomarkers in high‐grade bladder cancer. Transl Oncol. 2017;10:168‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reinert T, Borre M, Christiansen A, Hermann GG, Ørntoft TF, Dyrskjøt L. Diagnosis of bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PLoS One. 2012;7:e46297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang C, Pu W, Zhao D, et al. Identification of hyper‐methylated tumor suppressor genes‐based diagnostic panel for esophageal squamous cell carcinoma (ESCC) in a Chinese Han Population. Front Genet. 2018;9:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao F, Xia Y, Wang J, et al. Integrated analyses of DNA methylation and hydroxymethylation reveal tumor suppressive roles of ECM1, ATF5, and EOMES in human hepatocellular carcinoma. Genome Biol. 2014;15:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17:559‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu JY, Li F, Wang LP, et al. CTL‐ vs Treg lymphocyte‐attracting chemokines, CCL4 and CCL20, are strong reciprocal predictive markers for survival of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2015;113:747‐755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller CH, Klawon DEJ, Zeng S, Lee V, Socci ND, Savage PA. Eomes identifies thymic precursors of self‐specific memory‐phenotype CD8 T cells. Nat Immunol. 2020;21:567‐577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Held W, Jeevan‐Raj B, Charmoy M. Transcriptional regulation of murine natural killer cell development, differentiation and maturation. Cell Mol Life Sci. 2018;75:3371‐3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simonetta F, Pradier A, Roosnek E. T‐bet and eomesodermin in NK cell development, maturation, and function. Front Immunol. 2016;7:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dejean AS, Joulia E, Walzer T. The role of Eomes in human CD4 T cell differentiation: a question of context. Eur J Immunol. 2019;49:38‐41. [DOI] [PubMed] [Google Scholar]
- 25. Stienne C, Michieletto MF, Benamar M, et al. Foxo3 transcription factor drives pathogenic T helper 1 differentiation by inducing the expression of Eomes. Immunity. 2016;45:774‐787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen W, Qin Y, Liu S. CCL20 signaling in the tumor microenvironment. Adv Exp Med Biol. 2020;1231:53‐65. [DOI] [PubMed] [Google Scholar]
- 27. Yamazaki T, Yang XO, Chung Y, et al. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 1950;2008(181):8391‐8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li WM, Liu HR. CCL20‐CCR6 cytokine network facilitate Treg activity in advanced grades and metastatic variants of hepatocellular carcinoma. Scand J Immunol. 2016;83:33‐37. [DOI] [PubMed] [Google Scholar]
- 29. Wang D, Yang L, Yu W, et al. Colorectal cancer cell‐derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF‐κB signaling. J Immunother Cancer. 2019;7:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J‐J, Kao K‐C, Chiu Y‐L, et al. Enrichment of human CCR6 regulatory T cells with superior suppressive activity in oral cancer. J Immunol. 1950;2017(199):467‐476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3


