Abstract
Understanding of the microRNAs (miRNAs) regulatory system has become indispensable for physiological/oncological research. Tissue and organ specificities are key features of miRNAs that should be accounted for in cancer research. Further, cancer‐specific energy metabolism, referred to as the Warburg effect, has been positioned as a key cancer feature. Enhancement of the glycolysis pathway in cancer cells is what primarily characterizes the Warburg effect. Pyruvate kinase M1/2 (PKM1/2) are key molecules of the complex glycolytic system; their distribution is organ‐specific. In fact, PKM2 overexpression has been detected in various cancer cells. PKM isoforms are generated by alternative splicing by heterogeneous nuclear ribonucleoproteins. In addition, polypyrimidine tract‐binding protein 1 (PTBP1) is essential for the production of PKM2 in cancer cells. Recently, several studies focusing on non‐coding RNA elucidated PTBP1 or PKM2 regulatory mechanisms, including control by miRNAs, and their association with cancer. In this review, we discuss the strong relationship between the organ‐specific distribution of miRNAs and the expression of PKM in the context of PTBP1 gene regulation. Moreover, we focus on the impact of PTBP1‐targeting miRNA dysregulation on the Warburg effect.
Keywords: microRNA, organ‐specificity, PKM, PTBP1, Warburg effect
In this review, we discuss the miRNA‐mediated regulation of PTBP1 and PKM isoforms. In particular, we elaborate on the following points. First, under physiological conditions, the expression of PTBP1 and PKM isoforms is regulated by miRNAs that are unevenly distributed throughout the organs. Second, during carcinogenesis, dysregulation of PTBP1‐targeting miRNAs affects cancer‐specific energy metabolism in various types of cancer cells through PKM2 upregulation.
Abbreviations
- 3′UTR
3′ untranslated region
- CRC
colorectal cancer
- GBM
glioblastoma multiforme
- HCC
hepatocellular carcinoma
- hnRNP
heterogeneous nuclear ribonucleoproteins
- miRNA
microRNA
- NET
neuroendocrine tumor
- PKM
pyruvate kinase M
- PTBP1
polypyrimidine tract‐binding protein 1
- RMS
rhabdomyosarcoma
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
1. INTRODUCTION
MicroRNAs (miRNAs) are non‐coding and functional small nucleic acids. miRNAs repress gene expression at the translational level through the inhibition of translation or through induction of the degradation of target mRNAs by binding to a complementary site within the 3′UTR of target mRNAs. 1 , 2 Although the individual function of miRNAs is to fine‐tune gene expression, many miRNAs substantially orchestrally modulate major life phenomena, 3 , 4 impacting, for instance, tissue differentiation and carcinogenesis. 5 , 6 Nowadays, it is well known that dysregulation of miRNAs contributes to carcinogenesis. 7 Expression of miRNAs is frequently dysregulated as a result of epigenetic silencing (eg, via hypermethylation) 8 and the suppression of transcriptional factors (eg, hepatocyte nuclear factor). 9 miRNAs are also deeply involved in organ development and tissue differentiation. 10 , 11 , 12 Moreover, the uneven organ distribution of miRNAs implies that their expression profiles are organ‐specific. Besides, the organ distribution of miRNAs is closely associated with the biological function of the organ. 13 , 14 , 15 , 16 , 17 , 18
Recently, cancer‐specific energy metabolism (Warburg effect) has been reviewed; 19 , 20 increased glycolysis has been proposed as a cancer hallmark. 21 Although many genes regulate the glycolytic system, pyruvate kinase M1/2 (PKM1/2) are rate‐limiting glycolytic enzymes. PKM1 and PKM2 promote TCA cycle and glycolysis, respectively. 22 PKM1 is abundantly expressed in high‐energy demanding (glucose‐demanding) organs such as the brain and muscle. In contrast, PKM2 is primarily expressed in other tissues (eg, fatty tissue, lung, and kidney). 23 , 24 , 25 Notably, the dimeric form of PKM2, with low affinity to phosphoenolpyruvic acid, induces a higher nucleic acid synthesis through the pentose phosphate pathway. Furthermore, PKM2 is also expressed in various proliferating cells (eg, embryonic and tumor cells). 23 , 24 In particular, an increase in PKM2 promotes cancer progression. 26 , 27 , 28 PKM isoforms (PKM1 and PKM2) are produced through alternative splicing, 29 under the regulation of several splicing factors, such as hnRNP and serine/arginine‐rich splicing factors. 30 , 31 , 32 Of these, PTBP1, also known as hnRNPI, promotes cancer through the enhancement of PKM2 expression. 33 , 34 , 35 , 36 , 37 PTBP1 is an exonic splicing silencer, binding to optimal motifs (eg, UCUUC) in the polypyrimidine tract near the 3′ splicing site, and suppressing the downstream exon’s inclusion. 38 In the PKM mRNA, the favorable sequence for PTBP1 is located at intron 8. Therefore, PTBP1 blocks the inclusion of exon 9, resulting in the expression of PKM2 through the inclusion of exon 10. 39 Importantly, the expression of PTBP1 is promoted by transcription factors with oncogenic functions, such as MYC; 39 these transcription factors are, therefore, glycolysis enhancers in cancer cells. Moreover, based on recent findings, PTBP1 is negatively regulated by miRNAs.
In this review, we discuss the miRNA‐mediated regulation of PTBP1 and PKM isoforms. In particular, we elaborate on the following points. First, under physiological conditions, the expression of PTBP1 and PKM isoforms is regulated by miRNAs that are unevenly distributed throughout the organs. Second, during carcinogenesis, the dysregulation of PTBP1‐targeting miRNAs affects cancer‐specific energy metabolism in various types of cancer cells via PKM2 upregulation.
2. ORGAN‐SPECIFIC EXPRESSION PROFILES OF THE PTBP1‐ AND PKM‐TARGETING miRNAs
We and others have previously shown the uneven organ distribution of PTBP1‐ or PKM‐targeting miRNAs under physiological conditions, as well as their downregulation in certain cancers. 24 , 25 , 57 Notably, the introduction of PTBP1‐targeting miRNAs induced the switch of PKM isoforms from the cancer‐dominant PKM2 to PKM1. 24 , 25 , 40 , 41 , 42 , 43
Nowadays, microarray data on the distribution of miRNAs across human tissues are freely available online (https://ccb‐web.cs.uni‐saarland.de/tissueatlas/). 18 Hence, we validated the organ distribution of PTBP1‐ and PKM‐targeting miRNAs. Based on the database, we show the expression profiles of each miRNA in Figure S1. In this review, we defined these miRNAs as follows: brain‐specific (MIR9‐5p, MIR124‐3p, and MIR137), muscle‐specific (MIR1‐5p, MIR133b, and MIR206), liver‐specific (MIR122‐5p), and other PTBP1‐targeting miRNAs (MIR194‐5p and MIR340‐5p); their characteristics and tissue specificity index are shown in Tables 1 and 2.
Table 1.
Detailed information on the microRNAs regulating PTBP1 in various types of cancer
| Gene name (ID)/Chromosome location |
MIR1‐1 (406904)/20q13.33 MIR1‐2 (406905)/18q11.2 |
MIR9‐1 (407046)/1q22 MIR9‐2 (407047)/5q14.3 MIR9‐3 (407051)/15q26.1 |
MIR124‐1 (406907)/8p23.1 MIR124‐2 (406908)/8q12.3 MIR124‐3 (406909)/20q13.33 |
MIR133B (442890)/6p12.2 |
MIR137 (406928)/1p21.3 |
MIR194‐1 (406969)/1q41 MIR194‐2 (406970)/11q13.1 |
MIR206 (406989)/6p12.2 |
MIR340 (442908)/5q35.3 |
|
|---|---|---|---|---|---|---|---|---|---|
| Target gene/Species name (ID) | PTBP1 (5725)/Homo sapiens (9606) | ||||||||
| Guide strand of mature miRNA | MIR1‐3p | MIR9‐5p | MIR124‐3p | MIR133b | MIR137‐3p | MIR194‐5p | MIR206 | MIR340‐5p | |
| Sequence of the target region 5'‐3' | CATTCC | ACCAAAG |
#1: GTGCCTT #2: TGCCTT #3: TGCCTTA #4: TGCCTTA |
#1: GACCAA #2: GGACCAA |
AGCAATA | CTGTTAC a | CATTCC | TTTATA b | |
| Genomic location of MTI | 19:8810871‐810876 |
19:811859‐ 811865 |
#1:19: 811155‐811161 #2:19: 811777‐811782 #3:19: 811156‐811162 #4:19: 811777‐811783 |
#1:19: 811484‐ 811489 #2:19: 811857‐ 811863 |
19:811468‐ 814774 |
19:811607‐ 811613 |
19:8810871‐ 810876 |
19:812021‐ 812026 |
|
| Genomic location of 3'UTR | 45‐51 | 1033‐1039 |
#1:329‐336 #2:951‐957 #3:330‐337 #4:951‐957 |
#1:658‐664 #2:1031‐1038 |
642‐648 | 781‐787 | 45‐51 | 1195‐1201 | |
|
Distribution characteristics (TSI) |
Muscle‐specific (0.975) |
Brain‐specific (0.96) |
Brain‐specific (0.975) |
Muscle‐specific (0.98) |
Brain‐specific (0.94) |
Abundant in liver and colon (0.905) |
Muscle‐specific (0.99) |
Abundant in brain, but expressed in various organs (0.855) |
|
| Type of cancer |
1: RMS 2: CRC |
Glioma |
1: Glioma 2: CRC 3: CML 4: PaC |
1: RMS 2: CRC 3: GC |
1: Glioma 2: CRC |
HCC | RMS | CRC | |
|
Reference Author year/(PMID) |
1: Sugito et al 2017/(28981396) 2: Taniguchi et al 2016/(26980745) |
Zhu et al 2019/(31253583) |
1: Ferrarese et al 2014/(24865424) 2: Sun et al 2012/(22895557) Taniguchi et al 2015/(25721733) Taniguchi et al 2015/(25818238) 3: Shinohara et al 2016/(26607903) 4: Li et al 2016/(27785603) |
1: Sugito et al 2017/(28981396) 2: Taniguchi et al 2016/(26980745) 3: Sugiyama et al 2016/(27696637) |
1: Taniguchi et al 2018/(29695138) 2: Sun et al 2012/(22895557) |
Kang et al 2019/(31301177) | Taniguchi et al 2018/(29695138) | Sun et al 2012/(22895557) | |
Gene names are described according to the Gene Nomenclature Committee of Human Genome Organization (https://www.genenames.org/).
The miRNA terminology used follows the proposed miRNA nomenclature guidelines. 76
The distribution characteristics and TSI were described with reference to data from the human miRNA tissue atlas (https://ccb‐web.cs.uni‐saarland.de/tissueatlas/). 18 The actual expression values are shown in Figure S1.
The number before each reference corresponds to the number of the designated type of cancer studied.
Abbreviations: CML, chronic myelocytic leukemia; CRC, colorectal cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; MTI, microRNA‐target interaction, PaC, pancreatic cancer; PTBP1, polypyrimidine tract binding protein 1, RMS, rhabdomyosarcoma; TSI, tissue specificity index; 3'UTR, three prime untranslated region.
Poorly conserved site for microRNA families broadly conserved among vertebrates.
Poorly conserved site for microRNA families conserved among mammals. Each definition is referred to as in the TargetScan database (http://www.targetscan.org/vert_72/).
Table 2.
Implication of MIR122 in various PKM‐expressing cancers
| Gene name (ID)/Chromosome location | MIR122 (406906)/18q21.31 |
|---|---|
| Target gene/Species name (ID) | PKM (5315)/Homo sapiens (9606) |
| Guide strand of mature miRNA | MIR122‐5p |
| Sequence of the target region 5'‐3' | ACACTCC |
| Genomic location of MTI | 15:72199124‐72199130 |
| Genomic location of 3'UTR | 520‐527 |
| Distribution characteristics (TSI) | Liver‐specific (0.965) |
| Type of cancer |
1: Hepatocellular carcinoma 2: Breast cancer 3: Esophageal cancer 4: Cholangiocarcinoma 5: Renal cell carcinoma 6: Colorectal cancer |
|
Reference Author year/(PMID) |
1: Jung et al 2011/(22140464) Liu et al 2014/(24466275) Wong et al 2014/(25541689) Taniguchi et al 2018/(29695138) 2: Fong et al 2015/(25621950) 3: Zhang et al 2016/(27040384) 4: Peng et al 2019/(31115511) 5: Wang et al 2019/(31814765) 6: Wang et al 2020/(31901148) |
The miRNA terminology used follows the proposed miRNA nomenclature guidelines. 76
The distribution characteristics and TSI are described with reference to the data in the human miRNA tissue atlas (https://ccb‐web.cs.uni‐saarland.de/tissueatlas/). 18 The actual expression values are shown in Figure S1.
The number before each reference corresponds to the number of the designated type of cancer studied.
Abbreviations: MTI, microRNA‐target interaction; PKM, pyruvate kinase M1/M2; TSI, tissue specificity index; 3′UTR, three prime untranslated region.
3. THREE DISTINCT CONTEXTS OF miRNA DYSREGULATION CAUSE PTBP1/PKM2 UPREGULATION DURING CARCINOGENESIS
The organ‐specific dysregulation of miRNAs and the consequent impact on PTBP1 and PKM isoforms during carcinogenesis can lead to distinct types of cancer; in this review, we focus on three major contexts. First, in glucose‐demanding organs, dysregulation of brain‐ and muscle‐specific miRNAs directly targeting PTBP1 is associated with brain tumors and sarcomas, respectively (Section 4). Second, cooperative dysregulations of both brain‐ and muscle‐specific miRNAs are associated, especially with gastrointestinal cancers (Section 5). Third, dysregulation of liver‐specific miRNAs directly targeting PKM occurs in HCC, together with cooperative dysregulation of PTBP1‐targeting miRNAs (Section 6). The following sections describe each context in detail.
4. REGULATION OF PTBP1 BY BRAIN‐ OR MUSCLE‐SPECIFIC miRNAs
Brain‐specific MIR124‐3p is the most representative regulator of PTBP1 expression; it promotes neuronal differentiation through the repression of PTBP1 expression. 58 , 59 The relationship between MIR124‐3p and PTBP1 was discovered: PTBP1 binds to pre‐MIR124, inhibiting the expression of mature MIR124. 60 Upregulation of PTBP1 has been detected in brain tumors, such as GBM. 39 , 61 , 62 Interestingly, this upregulation is partly due to the downregulation of MIR124‐3p during carcinogenesis. 44 MIR124‐3p has the most numerous binding sites on PTBP1, which supports a secure connection between the two.
Another representative brain‐specific miRNA, MIR9‐5p, promotes differentiation of neuronal cells from retinal stem cells through downregulation of PTBP1; 63 the association of MIR9‐5p and PTBP1 was also reported in glioma. 45 A natural antisense transcript (PTB‐AS) stabilizes the expression of PTBP1, preventing the binding of MIR9‐5p to the PTBP1 3′UTR. 45 Furthermore, brain‐specific MIR137‐3p suppresses PTBP1 expression through direct binding to PTBP1 in GBM cells. 25 Similar to MIR124‐3p, a miRNA/PTBP1/PKM axis was demonstrated in these studies, suggesting that PTBP1 is strongly regulated by brain‐specific miRNAs.
Furthermore, several muscle‐specific miRNAs—MIR1‐3p, MIR133b, and MIR206—bind to the 3′UTR of PTBP1 to repress its expression. 25 , 43 As with the brain‐specific miRNAs, dysregulation of these muscle‐specific miRNAs may significantly impact carcinogenesis, especially in sarcoma of muscle origin. 25 , 43 In RMS, downregulation of MIR1‐3p and MIR133b promoted the expression of PTBP1, contributing to the Warburg effect. 43 Interestingly, the chimeric PAX3‐FOXO1 gene, a feature of alveolar RMS, was reportedly associated with PTBP1, whereas MIR133b directly regulated PAX3‐FOXO1 expression. 43 However, further research on miRNAs in the context of sarcoma (a rare tumor) is warranted.
5. IMPACT OF BRAIN‐ AND/OR MUSCLE‐SPECIFIC MICRORNA DYSREGULATION ON OTHER TYPES OF CANCER
Although the expression of brain/muscle‐specific miRNAs is unevenly distributed among organs, reportedly, the dysregulation of both miRNAs cooperatively affects carcinogenesis in various types of cancer. Impaired regulation of the PTBP1/PKM axis by MIR124‐3p has been observed in CRC, chronic myelocytic leukemia, and pancreatic cancer. 24 , 40 , 46 , 47 Increase in PTBP1 levels and a corresponding upregulation of PKM2 occurs via dysregulation of MIR124‐3p in cancer cells; MIR124‐3p overexpression induces a shift of the expression of PKM2 to PKM1 through the downregulation of PTBP1. 40 , 46 , 47 Although MIR340‐5p is abundant in the brain (Figure S1), MIR340‐5p also negatively regulates PTBP1 expression in CRC cells. 48 Furthermore, PTBP1 and PKM2 upregulation through the dysregulation of muscle‐specific MIR1‐3p and MIR133b was associated with carcinogenesis in CRC and gastric cancer. 41 , 42 Interestingly, our investigation showed that miRNAs/PTBP axis impairment was frequently detected in colorectal adenoma specimens. 40 , 41 The impairment of the miRNAs/PTBP axis may be the initial step toward carcinogenesis, especially in CRC. These findings suggest that miRNA/PTBP axis‐induced PKM2 overexpression plays a key, intrinsic mechanism of carcinogenesis.
6. miRNA‐MEDIATED REGULATION OF PKM ISOFORMS EXPRESSION IN HEPATOCELLULAR CARCINOMA
Both PKM isoforms are rarely expressed in the liver; 25 in contrast, pyruvate kinase L/R (PKLR) is specifically expressed in the liver. 23 , 25 , 64 PKL is expressed in the liver, the main gluconeogenesis‐governing organ. 23 Hence, a different perspective is required regarding PKM2 upregulation in HCC carcinogenesis. Interestingly, the 3′UTR of PKM, which is common in PKM1 and PKM2, has a binding region for liver‐specific MIR122‐5p, the only miRNA with a conserved site across most vertebrates, as determined in silico (http://www.targetscan.org/vert_72/). Notably, MIR122‐5p is a strong liver‐specific miRNA (70% of expression in the liver), 13 , 65 with potential histological and functional implications. The suppression of PKM expression may be exerted by MIR122‐5p in liver tissues.
The relationship between the dysregulation of MIR122‐5p and upregulated PKM2 in HCC has been demonstrated by many studies, counting three in the first half of the 2010s. 49 , 50 , 51 Downregulated MIR122‐5p increases the expression of both PKM1 and PKM2; of note, multiple splicing systems upregulating PKM2 may be simultaneously activated. Interestingly, the miRNA/PTBP1 axis has also been implicated in the carcinogenesis of HCC. Based on previous evidence 66 and the human miRNA tissue atlas (Figure S1), MIR194‐5p is abundant in the liver. MIR194‐5p binds to the 3′UTR of PTBP1; its dysregulation also contributes to the onset of HCC. Although we found relatively high PTBP1 expression in healthy liver compared to brain or muscle, 25 further upregulation of PTBP1 and the inverse correlation between MIR194 and PTBP1 in HCC clinical data may indicate that collapse of the miRNA/PTBP1 axis partially contributes to HCC. 52 In particular, coordinated dysregulation of MIR122‐5p and MIR194‐5p may induce a PKM2‐dominant phenotype during carcinogenesis in HCC.
Detailed information on miRNA‐PTBP1 relationships is summarized in Table 1. An important finding in these studies is the consistent organ distribution of miRNAs and PTBP1; therefore, the organ distribution of miRNA in normal conditions should always be considered. Furthermore, dysregulation of the miRNA/PTBP1 axis in multiple cancer types may suggest that this mechanism is universal and essential for the development and maintenance of the Warburg effect in cancer cells.
7. IMPACT OF LIVER‐SPECIFIC miRNA DYSREGULATION ON OTHER TYPES OF CANCER
Dysregulation of the MIR122‐5p/PKM2 axis was demonstrated in various cancer types, such as breast cancer, esophageal cancer, cholangiocarcinoma, renal cell carcinoma, and CRC. 53 , 54 , 55 , 56 , 57 These reports suggest that disruption of the MIR122‐5p‐dependent regulation of PKM is a common carcinogenic mechanism. In particular, PTBP1 upregulation due to PTBP1‐targeting miRNA downregulation may further increase the expression of PKM2. This evidence supports our viewpoint: cumulative dysregulation of different miRNAs, including PTBP1‐ and PKM‐targeting ones, cooperatively induces the upregulation of PKM2, especially in gastrointestinal cancer cells. Detailed information on PKM regulation by MIR122‐5p is summarized in Table 2; a summary of the systematic PTBP1 and PKM regulatory mechanisms by miRNAs is shown in Figure 1.
Figure 1.
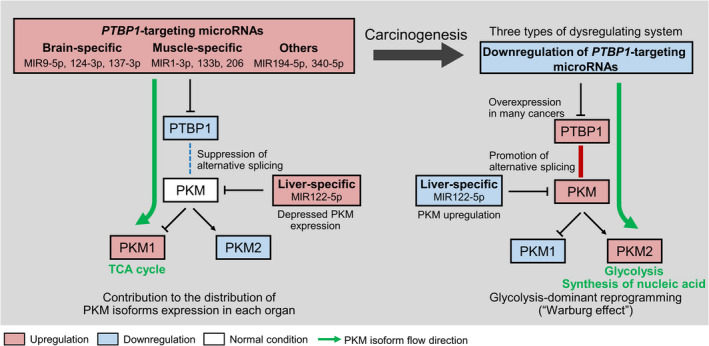
Regulation of polypyrimidine tract‐binding protein 1 (PTBP1) and pyruvate kinase M (PKM) isoforms by microRNAs: schematics. Brain and muscle‐specific miRNAs bind to the 3′ UTR of PTBP1 and downregulate PTBP1 expression. PKM1 dominance is induced through the suppression of alternative splicing in these healthy organs. PKM1 promotes the tricarboxylic acid (TCA) cycle for energy production. In the process of carcinogenesis, coordinated dysregulation of miRNAs induces PKM2 upregulation through the increment of PTBP1 expression. PKM2 promotes glycolysis and/or the synthesis of nucleic acids, especially in proliferating cells. Dysregulation of brain‐specific miRNAs such as MIR9‐5p, 124‐3p, and 137‐3p occurs in brain tumors; that of muscle‐specific miRNAs (MIR1‐3p, 133b, and 206) arises in sarcoma. In gastrointestinal cancers (eg, colorectal cancer), these miRNAs are dysregulated coordinately. In contrast, in the pyruvate kinase L (PKL) dominant normal liver, MIR122‐5p is abundant and downregulates both PKM1 and PKM2 by binding to the PKM 3′UTR. We assume that in hepatocellular carcinoma, the dominance of PKM2 is caused by harmonic dysregulation of PKM‐targeting (MIR122‐5p) and PTBP1‐targeting miRNAs (MIR194‐5p). Thus, there are three types of miRNA dysregulation behind the upregulation of PKM2 in cancer cells.
8. SIGNIFICANCE OF PTBP1 IN THE WARBURG EFFECT
Recently, we found an association between PTBP1 and PKM isoforms in the Warburg effect. 40 , 41 , 42 , 43 Many reports show upregulation of PKM2 during carcinogenesis. 26 , 27 , 28 However, this upregulation involves two different patterns. For example, in brain and muscle, the expression of PKM1 is mainly due to the suppression of PTBP1; the switching of PKM isoforms from PKM1 to PKM2 is induced by dysregulation of the miRNAs/PTBP1 axis during carcinogenesis. 24 , 25 , 43 In contrast, in gastrointestinal organs, both PKM1 and PKM2 are expressed in healthy conditions; 24 , 25 the PKM2/PKM1 ratio is increased (not switched) during carcinogenesis. 24 , 40 , 41 , 42 Perhaps, the switching of PKM isoforms causes a more significant impact on cancer‐energy metabolism. Of note, both high PKM1 or high PKM2 contexts showed that the dysregulation of PTBP1‐targeting miRNAs further contributes to the upregulation of PKM2 (Figure 2).
Figure 2.
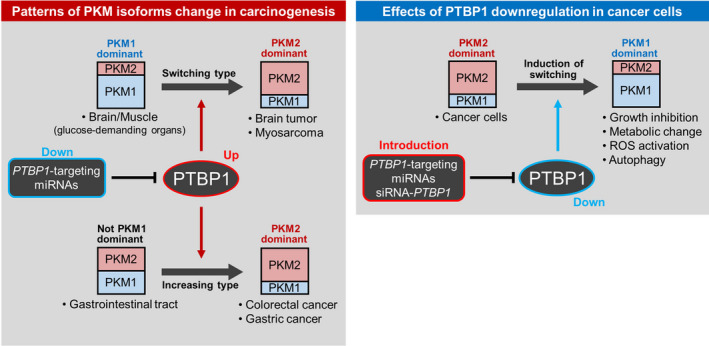
Relationship between pyruvate kinase M (PKM) isoforms, cancer development, and anticancer effects. In carcinogenesis, the establishment of PKM2 dominance follows two patterns. PKM1 to PKM2 switching occurs in PKM1‐dominant organs such as brain and muscle. Dysregulation of polypyrimidine tract‐binding protein 1 (PTBP1)‐targeting miRNAs (brain‐ and muscle‐specific) induces the switch to PKM2 dominance through PTBP1 upregulation in brain tumors and myosarcoma. This PKM2 dominant change is defined as the “switching type.” In contrast, in the gastrointestinal tract, both PKM1 and PKM2 are expressed. PKM2 expression is further upregulated through dysregulation of the PTBP1‐targeting miRNA/PTBP1 axis in carcinogenesis. This PKM2 dominant change is defined as the “increasing type.” In cancer cells, PKM2 is consistently dominant. Downregulation of PTBP1, via PTBP1‐targeting miRNAs or PTBP1 gene‐silencing of (siRNA‐PTBP1), induces growth inhibition, metabolic change, and the production of reactive oxygen species through PKM2 to PKM1 switching.
We investigated the roles of PTBP1 in cancer cells through transient PTBP1 downregulation. PTBP1 silencing induced autophagy in various cancer cells, together with a PKM2 to PKM1 switch; of note, this effect was also observed after the introduction of PTBP1‐targeting miRNAs. 40 , 41 , 42 , 43 , 46 In turn, this switch led to the production of ROS and ATP, activating the TCA cycle. N‐Acetyl‐l‐cysteine (a ROS inhibitor) also partially impacted the suppression of cell growth caused by both PTBP1 silencing and ectopic expression of PTBP1‐targeting miRNAs (Figure 2). 40 , 41 , 42 , 43 These findings suggest that PTBP1 is a central molecule in the Warburg effect of cancer cells, regulating the expression of PKM isoforms. Importantly, the PTBP1/PKM axis is strictly regulated by PTBP1‐targeting miRNAs in cancer cells.
9. FUTURE PERSPECTIVES
Our review highlights many shades of gray in the field. First, we have not discussed the organ distribution of all potential PTBP1‐binding miRNAs. In addition, several miRNAs were suggested (in silico) as PTBP1‐binding miRNAs. For example, MIR133a‐3p is a muscle‐specific miRNA, 14 , 18 and its relationship with PTBP1 has been reported in the context of human islet insulin biosynthesis and dengue virus replication. 67 , 68 miRNAs that can potentially bind to PTBP1 based on a target‐predicting database is provided in Table 3. However, further studies are needed to integrate these findings in the context of PTBP1‐targeting.
Table 3.
Detailed information of the microRNAs predicted to bind to PTBP1 based on TargetScan
| miRNA name (mature type) | MIR17‐5p/20‐5p/93‐5p/106‐5p/519‐3p | MIR133a‐3p | MIR153‐3p | MIR‐193‐3p | MIR200bc‐3p/429 | MIR216b‐5p | MIR506‐3p a |
| Features | Constitutes the MIR17 family |
Muscle‐specificity Constitutes the MIR133 family with MIR133b Three binding sites in the 3'UTR of PTBP1 |
Constitutes the MIR153 family with MIR153‐1 and ‐2 | Registered in the miRBase as hsa‐miR‐193a‐3p |
Constitutes the MIR141/200 family |
Constitutes the MIR216 family with MIR216A | Constitutes the MIR506 family with MIR507‐514 b |
| PTBP1 related references PMID and simple content | None |
20520763; Human islet insulin biosynthesis 26818704; Dengue virus replication |
None | None | None | None | None |
We searched the microRNAs with the ability to bind to PTBP1 using the TargetScan database (http://www.targetscan.org/vert_72/).
The miRNA terminology used aligns with the proposed miRNA nomenclature guidelines.76
Abbreviations: PTBP1, polypyrimidine tract‐binding protein 1; 3′UTR, three prime untranslated region.
Listed as a set of MIR124‐2 in the TargetScan database.
MIR507, 508, 509‐1, 509‐2, 509‐3, 510, 511, 512‐1, 512‐2, 513A1, 513A2, 513B, 513C, 514A1, 514A2, 514A3, and 514B are included in this family.
Second, the miRNAs regulatory mechanisms of PKM isoforms are not entirely understood. For instance, MIR369 enhances the expression of PKM2 via the stabilization of HNRNPA2B1 in cell reprogramming. 69 Various splicers and miRNAs may constitute complex PKM isoforms and impact the regulatory mechanisms, which deserve further exploration. Third, the regulatory mechanisms of PKLR remain unclear. Although a previous study showed that the expression of PKLR was not changed in HCC, 51 this finding needs to be investigated in more detail.
Fourth, the PTBP1 functions other than the regulation of PKM isoforms have not been sufficiently elucidated. PTBP1 is involved in several steps in the metabolism of mRNAs, including mRNA stability, mRNA transport, 3’‐end processing, and internal ribosome entry site‐mediated translation. 45 , 70 In cancer cells, PTBP1 was shown to impact migration, invasion, apoptosis, and cell cycle. 71 Hence, the molecular mechanisms of PTBP1 in cancer cells, with a focus on other splicing target genes or mRNA metabolism, need to be investigated.
Fifth, the roles of PKM1 are not well understood; of note, PKM1 is upregulated in various chemo‐resistant cells. 72 Moreover, PKM1 is an activator of glucose metabolism, boosting tumor cell growth. 73 Besides, in neuroendocrine lung tumors (NET), higher PKM1 expression was observed compared to non‐NET tumors. 73 Therefore, PKM1 should be considered a biomarker of chemo‐resistance and a potential therapeutic target in some types of cancer.
We should also consider organ‐specificity in the context of clinical applications. Recently, MIR34a‐5p (MRX34) was selected as a therapeutic tool in various solid tumors; a phase I study (NCT01829971) was conducted 74 and terminated due to immune‐related adverse events; the suitability of the drug delivery system was questioned. 75 Nonetheless, we suggest that the organ‐specificity of MIR34a‐5p should also be considered; the organ‐distribution of the particular miRNA in healthy conditions should also be factored in, to maximize the effectiveness of the treatment and to avoid potential side effects.
10. CONCLUSION
In summary, the regulation of PTBP1 is organ‐specific; brain‐ or muscle‐specific miRNAs partially contribute to the organ‐specific expression of PKM isoforms. Moreover, the Warburg effect in cancer cells is due to the upregulation of glycolysis‐related proteins, such as PKM2, through the dysregulation of single or multiple miRNA/PTBP1 axes. This review suggests that the organ‐specificity of miRNAs partially governs the characteristics of each tissue and that the miRNAs dysregulation profoundly contributes to carcinogenesis.
DISCLOSURE
The authors declare no conflicts of interest.
Supporting information
Figure S1
ACKNOWLEDGMENTS
We thank our collaborators, including the colleagues from the Gifu University and Osaka Medical College. We would like to thank Editage (www.editage.com) for English language editing.
Taniguchi K, Uchiyama K, Akao Y. PTBP1‐targeting microRNAs regulate cancer‐specific energy metabolism through the modulation of PKM1/M2 splicing. Cancer Sci. 2021;112:41–50. 10.1111/cas.14694
REFERENCES
- 1. Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature. 2008;455:64‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835‐840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krek A, Grün D, Poy MN, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495‐500. [DOI] [PubMed] [Google Scholar]
- 4. Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646‐3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396‐400. [DOI] [PubMed] [Google Scholar]
- 6. Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776‐780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Roosbroeck K, Calin GA. Cancer hallmarks and microRNAs: The Therapeutic Connection. Adv Cancer Res. 2017;135:119‐149. [DOI] [PubMed] [Google Scholar]
- 8. Zhou Z, Lv J, Wang J, et al. Role of microRNA‐124 as a prognostic factor in multiple neoplasms: a meta‐analysis. Dis Markers. 2019;2019:1654780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakao K, Miyaaki H, Ichikawa T. Antitumor function of microRNA‐122 against hepatocellular carcinoma. J Gastroenterol. 2014;49:589‐593. [DOI] [PubMed] [Google Scholar]
- 10. Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563‐568. [DOI] [PubMed] [Google Scholar]
- 11. Callis TE, Chen JF, Wang DZ. MicroRNAs in skeletal and cardiac muscle development. DNA Cell Biol. 2007;26:219‐225. [DOI] [PubMed] [Google Scholar]
- 12. Singh SK. miRNAs: from neurogeneration to neurodegeneration. Pharmacogenomics. 2007;8:971‐978. [DOI] [PubMed] [Google Scholar]
- 13. Lagos‐Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue‐specific microRNAs from mouse. Curr Biol. 2002;12:735‐739. [DOI] [PubMed] [Google Scholar]
- 14. Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401‐1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Weng T, Gou D, et al. Identification of rat lung‐specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genom. 2007;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hua YJ, Tang ZY, Tu K, et al. Identification and target prediction of miRNAs specifically expressed in rat neural tissue. BMC Genom. 2009;10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Minami K, Uehara T, Morikawa Y, et al. miRNA expression atlas in male rat. Sci Data. 2014;1:140005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ludwig N, Leidinger P, Becker K, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865‐3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703‐707. [DOI] [PubMed] [Google Scholar]
- 21. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646‐674. [DOI] [PubMed] [Google Scholar]
- 22. Chen M, Zhang J, Manley JL. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010;70:8977‐8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43:969‐980. [DOI] [PubMed] [Google Scholar]
- 24. Taniguchi K, Ito Y, Sugito N, et al. Organ‐specific PTB1‐associated microRNAs determine expression of pyruvate kinase isoforms. Sci Rep. 2015;5:8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taniguchi K, Sugito N, Shinohara H, et al. Organ‐specific microRNAs (MIR122, 137, and 206) contribute to tissue characteristics and carcinogenesis by regulating pyruvate kinase M1/2 (PKM) expression. Int J Mol Sci. 2018;19(5):1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wong N, De Melo J, Tang D. PKM2, a central point of regulation in cancer metabolism. Int J Cell Biol. 2013;2013:242513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zahra K, Dey T, Ashish A, et al. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol. 2020;10:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He X, Du S, Lei T, et al. PKM2 in carcinogenesis and oncotherapy. Oncotarget. 2017;8:110656‐110670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noguchi T, Inoue H, Tanaka T. The M1‐ and M2‐type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. 1986;261:13807‐13812. [PubMed] [Google Scholar]
- 30. Chen M, David CJ, Manley JL. Concentration‐dependent control of pyruvate kinase M mutually exclusive splicing by hnRNP proteins. Nat Struct Mol Biol. 2012;19:346‐354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clower CV, Chatterjee D, Wang Z, et al. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci USA. 2010;107:1894‐1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuranaga Y, Sugito N, Shinohara H, et al. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer‐specific energy metabolism in colon cancer cells. Int J Mol Sci. 2018;19(10):3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He X, Arslan AD, Ho TT, et al. Involvement of polypyrimidine tract‐binding protein (PTBP1) in maintaining breast cancer cell growth and malignant properties. Oncogenesis. 2014;3:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calabretta S, Bielli P, Passacantilli I, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031‐2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hwang SR, Murga‐Zamalloa C, Brown N, et al. Pyrimidine tract‐binding protein 1 mediates pyruvate kinase M2‐dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab Invest. 2017;97:962‐970. [DOI] [PubMed] [Google Scholar]
- 36. Jiang J, Chen X, Liu H, et al. Polypyrimidine Tract‐Binding Protein 1 promotes proliferation, migration and invasion in clear‐cell renal cell carcinoma by regulating alternative splicing of PKM. Am J Cancer Res. 2017;7:245‐259. [PMC free article] [PubMed] [Google Scholar]
- 37. Xie R, Chen X, Chen Z, et al. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett. 2019;449:31‐44. [DOI] [PubMed] [Google Scholar]
- 38. Spellman R, Smith CW. Novel modes of splicing repression by PTB. Trends Biochem Sci. 2006;31:73‐76. [DOI] [PubMed] [Google Scholar]
- 39. David CJ, Chen M, Assanah M, et al. HnRNP proteins controlled by c‐Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taniguchi K, Sugito N, Kumazaki M, et al. MicroRNA‐124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17‐27. [DOI] [PubMed] [Google Scholar]
- 41. Taniguchi K, Sakai M, Sugito N, et al. PTBP1‐associated microRNA‐1 and ‐133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7:18940‐18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugiyama T, Taniguchi K, Matsuhashi N, et al. MiR‐133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle‐splicer polypyrimidine tract‐binding protein 1. Cancer Sci. 2016;107:1767‐1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sugito N, Taniguchi K, Kuranaga Y, et al. Cancer‐specific energy metabolism in rhabdomyosarcoma cells is regulated by microRNA. Nucleic Acid Ther. 2017;27:365‐377. [DOI] [PubMed] [Google Scholar]
- 44. Ferrarese R, Harsh GR, Yadav AK, et al. Lineage‐specific splicing of a brain‐enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124:2861‐2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu L, Wei Q, Qi Y, et al. PTB‐AS, a novel natural antisense transcript, promotes glioma progression by improving PTBP1 mRNA stability with SND1. Mol Ther. 2019;27:1621‐1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shinohara H, Kumazaki M, Minami Y, et al. Perturbation of energy metabolism by fatty‐acid derivative AIC‐47 and imatinib in BCR‐ABL‐harboring leukemic cells. Cancer Lett. 2016;371:1‐11. [DOI] [PubMed] [Google Scholar]
- 47. Li C, Zhao Z, Zhou Z, et al. Linc‐ROR confers gemcitabine resistance to pancreatic cancer cells via inducing autophagy and modulating the miR‐124/PTBP1/PKM2 axis. Cancer Chemother Pharmacol. 2016;78:1199‐1207. [DOI] [PubMed] [Google Scholar]
- 48. Sun Y, Zhao X, Zhou Y, et al. miR‐124, miR‐137 and miR‐340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep. 2012;28:1346‐1352. [DOI] [PubMed] [Google Scholar]
- 49. Jung CJ, Iyengar S, Blahnik KR, et al. Epigenetic modulation of miR‐122 facilitates human embryonic stem cell self‐renewal and hepatocellular carcinoma proliferation. PLoS One. 2011;6:e27740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu AM, Xu Z, Shek FH, et al. miR‐122 targets pyruvate kinase M2 and affects metabolism of hepatocellular carcinoma. PLoS One. 2014;9:e86872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wong CC, Au SL, Tse AP, et al. Switching of pyruvate kinase isoform L to M2 promotes metabolic reprogramming in hepatocarcinogenesis. PLoS One. 2014;9:e115036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kang H, Heo S, Shin JJ, et al. A miR‐194/PTBP1/CCND3 axis regulates tumor growth in human hepatocellular carcinoma. J Pathol. 2019;249:395‐408. [DOI] [PubMed] [Google Scholar]
- 53. Fong MY, Zhou W, Liu L, et al. Breast‐cancer‐secreted miR‐122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183‐194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang HS, Zhang FJ, Li H, et al. Tanshinone ⅡA inhibits human esophageal cancer cell growth through miR‐122‐mediated PKM2 down‐regulation. Arch Biochem Biophys. 2016;598:50‐56. [DOI] [PubMed] [Google Scholar]
- 55. Peng C, Sun Z, Li O, et al. Leptin stimulates the epithelial‐mesenchymal transition and pro‐angiogenic capability of cholangiocarcinoma cells through the miR‐122/PKM2 axis. Int J Oncol. 2019;55:298‐308. [DOI] [PubMed] [Google Scholar]
- 56. Wang S, Zheng W, Ji A, et al. Overexpressed miR‐122‐5p promotes cell viability, proliferation, migration and glycolysis of renal cancer by negatively regulating PKM2. Cancer Manag Res. 2019;11:9701‐9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang X, Zhang H, Yang H, et al. Exosome‐delivered circRNA promotes glycolysis to induce chemoresistance through the miR‐122‐PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cao X, Pfaff SL, Gage FH. A functional study of miR‐124 in the developing neural tube. Genes Dev. 2007;21:531‐536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Makeyev EV, Zhang J, Carrasco MA, et al. The microRNA miR‐124 promotes neuronal differentiation by triggering brain‐specific alternative pre‐mRNA splicing. Mol Cell. 2007;27:435‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yeom KH, Mitchell S, Linares AJ, et al. Polypyrimidine tract‐binding protein blocks miRNA‐124 biogenesis to enforce its neuronal‐specific expression in the mouse. Proc Natl Acad Sci USA. 2018;115:E11061‐E11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jin W, McCutcheon IE, Fuller GN, et al. Fibroblast growth factor receptor‐1 alpha‐exon exclusion and polypyrimidine tract‐binding protein in glioblastoma multiforme tumors. Cancer Res. 2000;60:1221‐1224. [PubMed] [Google Scholar]
- 62. McCutcheon IE, Hentschel SJ, Fuller GN, et al. Expression of the splicing regulator polypyrimidine tract‐binding protein in normal and neoplastic brain. Neuro Oncol. 2004;6:9‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Qi X. The role of miR‐9 during neuron differentiation of mouse retinal stem cells. Artif Cells Nanomed Biotechnol. 2016;44:1883‐1890. [DOI] [PubMed] [Google Scholar]
- 64. Noguchi T, Yamada K, Inoue H, et al. The L‐ and R‐type isozymes of rat pyruvate kinase are produced from a single gene by use of different promoters. J Biol Chem. 1987;262:14366‐14371. [PubMed] [Google Scholar]
- 65. Jopling C. Liver‐specific microRNA‐122: biogenesis and function. RNA Biol. 2012;9:137‐142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meng Z, Fu X, Chen X, et al. miR‐194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148‐2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fred RG, Bang‐Berthelsen CH, Mandrup‐Poulsen T, et al. High glucose suppresses human islet insulin biosynthesis by inducing miR‐133a leading to decreased polypyrimidine tract binding protein‐expression. PLoS One. 2010;5:e10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Castillo JA, Castrillón JC, Diosa‐Toro M, et al. Complex interaction between dengue virus replication and expression of miRNA‐133a. BMC Infect Dis. 2016;16:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Konno M, Koseki J, Kawamoto K, et al. Embryonic microRNA‐369 controls metabolic splicing factors and urges cellular reprogramming. PLoS One. 2015;10:e0132789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sawicka K, Bushell M, Spriggs KA, et al. Polypyrimidine‐tract‐binding protein: a multifunctional RNA‐binding protein. Biochem Soc Trans. 2008;36:641‐647. [DOI] [PubMed] [Google Scholar]
- 71. Zhu W, Zhou BL, Rong LJ, et al. Roles of PTBP1 in alternative splicing, glycolysis, and oncogensis. J Zhejiang Univ Sci B. 2020;21:122‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Taniguchi K, Sakai M, Sugito N, et al. PKM1 is involved in resistance to anti‐cancer drugs. Biochem Biophys Res Commun. 2016;473:174‐180. [DOI] [PubMed] [Google Scholar]
- 73. Morita M, Sato T, Nomura M, et al. PKM1 confers metabolic advantages and promotes cell‐autonomous tumor cell growth. Cancer Cell. 2018;33:355‐367.e7. [DOI] [PubMed] [Google Scholar]
- 74. Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR‐34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180‐188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Taniguchi K, Wada SI, Ito Y, et al. alpha‐Aminoisobutyric acid‐containing amphipathic helical peptide‐cyclic RGD conjugation as a potential drug delivery system for microRNA replacement therapy in vitro. Mol Pharm. 2019;16:4542‐4550. [DOI] [PubMed] [Google Scholar]
- 76. Desvignes T, Batzel P, Berezikov E, et al. miRNA nomenclature: A view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet. 2015;31:613–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1


