Abstract
Next‐generation sequencing (NGS) enables the diagnosis of large numbers of gene aberrations during one examination, and precision medicine has been developed for patients with advanced non–small cell lung cancer (NSCLC). However, peripheral lung lesions account for the majority of advanced lung cancers, especially lung adenocarcinoma. In these cases, it is difficult to obtain tissue samples which contain sufficient tumor cells by transbronchial biopsy (TBB) with forceps. Even when the target lesions are quite small, bronchial brushing can obtain enough tumor cells by endobronchial ultrasonography using guide sheath (EBUS‐GS). In this study, we investigate the suitability of bronchial brushing cytology specimens obtained by EBUS‐GS‐TBB to evaluate the correlation between the success rate of NGS and extracted DNA/RNA yields according to biopsy method. We prospectively collected 222 tumor samples obtained from patients with advanced lung cancer. All patients were enrolled in a prospective nationwide genomic screening project for lung cancer (LC‐SCRUM‐Japan/Asia). Genomic data were obtained from the clinico‐genomic database of LC‐SCRUM‐Japan/Asia. The extraction yields of DNA/RNA from samples obtained by EBUS‐GS‐TBB were relatively low compared with tissue samples. The success rate of DNA sequencing for EBUS‐GS‐TBB was 97.9%, with no significant differences between biopsy methods. The success rate of RNA sequencing for EBUS‐GS‐TBB was 80.4%, which was relatively low compared with surgical biopsy samples (P = 0.069). However, some rare oncogenic driver aberrations were detected from these specimens. This study demonstrated that cytology samples obtained by transbronchial brushing with EBUS‐GS‐TBB were suitable for NGS analysis.
Keywords: bronchial brushing, lung cancer, next‐generation sequencing, nucleic acid yield, success rate
This study demonstrated that cytology samples obtained by transbronchial brushing with endobronchial ultrasonography using guide sheath (EBUS‐GS) were suitable for next‐generation sequencing (NGS) analysis. We believe the new knowledge of this study can contribute to the best practice and further research for patients with advanced non–small cell lung cancer (NSCLC).
1. INTRODUCTION
Recently, precision medicine has been developed for patients with advanced non–small cell lung cancer (NSCLC). Most oncogenic driver aberrations occur in less than 5% of patients. 1 , 2 To improve survival times in these patients, appropriate molecular targeted drugs are essential. 3
Currently, next‐generation sequencing (NGS) enables the diagnosis of large numbers of gene aberrations at one examination. A previous report revealed that small lung tumor biopsy tissue samples were suitable for NGS analysis. 4 However, we often see advanced‐stage NSCLC cases where only small amounts of biopsy specimen can be obtained, such as cytology samples obtained by bronchoscopy. In these patients, NGS analysis cannot be successful due to the lack of enough samples. Therefore, it can be difficult to diagnose rare oncogenic driver aberrations appropriately for all patients in clinical practice.
In advanced NSCLC patients, transbronchial biopsy (TBB) and endobronchial ultrasound sonography needle aspiration biopsy (EBUS‐TBNA) are common biopsy methods to obtain samples for pathological diagnosis. 5 Recently, there have been reports investigating the suitability of TBB materials for NGS analyses. 6 , 7 , 8 These reports investigated mainly formalin‐fixed and paraffin‐embedded (FFPE) tissue samples. Hagmeyer et al reported that the success rate of NGS was 62% using bronchoscopic brushing smear materials for central airway lung cancer in a small sample size. 9 However, peripheral lesions account for the majority of advanced lung cancer, especially lung adenocarcinoma. TBB using EBUS‐guide sheath (EBUS‐GS‐TBB) is a useful biopsy method even for smaller peripheral lung nodules. 10 We previously reported that EBUS‐GS‐TBB using virtual bronchoscopic navigation system demonstrated a high diagnostic yield (93.0%) for peripheral lung lesions. 11 However, even if we used a virtual bronchoscopic navigation system, it could be difficult to obtain sufficient tissue to confirm malignant pathological diagnosis by forceps biopsy. In these cases, bronchial brushing and washing contributed to the pathological diagnosis in peripheral lung lesions and was able to obtain enough cancer cells. 12
On the other hand, to improve the success rate of NGS analysis, the quantity of nucleic acid is an important issue. Therefore, to investigate the suitability of bronchial brushing cytology specimens obtained by EBUS‐GS‐TBB, we evaluated the correlation between success rates of NGS and extracted DNA/RNA yields pursuant to biopsy methods.
2. MATERIALS AND METHODS
2.1. Patients
We prospectively collected tumor samples which were obtained from patients with advanced lung cancer. All patients were enrolled in the prospective nationwide genomic screening project for lung cancer (LC‐SCRUM‐Japan/Asia, UMIN ID: 000010234). The study duration of this project is divided into three periods. In the first period, enrollment was conducted between March 2015 and March 2017 in Japan. The second period was undertaken from April 2017 to March 2019, and the third period started in June 2019 and has going on until present. For the third period, we have expanded the study to include East Asian countries. In March 2020, altogether 212 institutions in Japan were included.
A total of 222 patients with advanced NSCLC or SCLC were enrolled in LC‐SCRUM‐Japan/Asia between January 2016 and March 2020 from St. Marianna University School of Medicine Hospital. All patients were pathologically confirmed to have primary lung cancer, and the samples were judged to contain enough cancer cells by the pathologists. All patients provided written informed consent. Genomic data were obtained from the LC‐SCRUM‐Japan/Asia clinico‐genomic database, and clinical data including age, sex, histology, clinical stage (UICC 8th edition) were collected from electronic medical records. This study was approved by the institutional review board of St. Marianna University School of Medicine (approved no: 4455) and the National Cancer Center. Written informed consent was obtained from each of the participants.
2.2. Collection of lung cancer samples
Appropriate methods for sample collection were judged by attending physicians and the thoracic cancer board.
Cytology samples were obtained by bronchoscopy (BF‐1T260, BF‐P260, BF‐UC260FW; Olympus) or pleural effusion. For pleural effusion collection, at least 80 mL was needed by puncturing through the thoracic wall under local anesthesia. These samples were then transferred to a central laboratory (SRL, Inc) within 24 hours. Tissue samples by conventional methods were obtained by single‐port semi‐rigid (semi‐flexible) medical thoracoscopy (LTF‐260; Olympus) under local anesthesia, US guided and CT guided core needle biopsy (CNB), and surgical biopsy. These tissue samples were immediately transferred to a deep freezer and archived without DNA/RNA stabilizing solution. All tissue samples were pathologically confirmed to contain adequate tumor cells.
2.3. Bronchoscopy specimens
Transbronchial biopsy was performed for primary lung lesions with radial probe EBUS (UM‐S20‐17S; Olympus) using the guide sheath method (EBUS‐GS‐TBB) (K‐201 guide sheath kit; Olympus). We routinely performed bronchial brushing and forceps biopsy five times. 10 , 13 During every bronchial brushing, the brush was directly suspended in physiological saline solution (PSS).
EBUS‐TBNA was performed to the hilar lymph nodes and mediastinal lymph nodes. We routinely performed three punctures with a 22G needle (NA‐201SX‐4022; Olympus) using an aspiration‐back‐lock syringe. After puncture, tissue samples were expelled by needle stylet. The tissue samples were fixed in formalin, and these tissue samples were not used for this study. The needle stylet was then removed, and PSS was injected into from needle tail (EBUS‐TBNA needle washing cytology sample). This procedure was repeated for every puncture.
The suspended solutions we obtained by bronchoscopy were immediately centrifugated, and only pellets were frozen and archived without DNA/RNA‐stabilizing solution in deep freeze. To maintain the quality and stability of DNA/RNA, all freezing and archiving procedures were carried out within 10 min from the time of sample collection. All cytology samples were pathologically confirmed to contain sufficient tumor cells (Figure 1).
FIGURE 1.
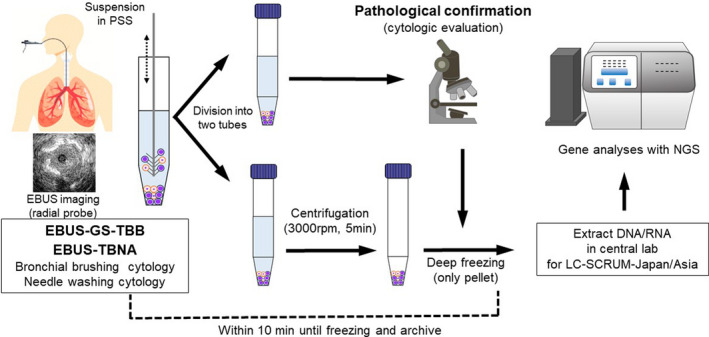
Flow chart of cytology samples obtained by bronchoscopy. EBUS, endobronchial ultrasound sonography; GS, guide sheath; NGS, next‐generation sequencing; PSS, physiological saline solution; TBB, transbronchial biopsy; TBNA, needle aspiration biopsy
2.4. Extraction of nucleic acid and library preparation
In this study, DNA/RNA were isolated from fresh frozen samples. All frozen samples were carried to a central laboratory (SRL) certified by the Clinical Laboratory Improvement Amendments (CLIA) . DNA/RNA were extracted and purified with a nucleic acid extraction kit (AllPrep DNA/RNA Mini Kit; QIAGEN) according to the manufacturer's protocol. DNA/RNA concentrations were quantified by the Qubit fluorometric assay (Thermo Fisher Scientific). The minimum concentrations were defined as 1.67 ng/μL of DNA and 2.5 ng/μL of RNA. When the concentration was under the aforementioned levels, NGS analyses were not performed. The yields of DNA/RNA were reported to each institution. After extraction of DNA/RNA, the library for NGS was prepared by the Ion AmpliSeq Library Plus Kit (Thermo Fisher Scientific).
2.5. Next‐generation sequencing
Next‐generation sequencing was performed at the central laboratory. LC‐SCRUM‐Japan/Asia prepared hot‐spot cancer panels for NGS analyses. The NGS panel was Oncomine Comprehensive Assay (OCA v1.0, 143 gene mutation and fusion; Thermo Fisher Scientific) during the first and second period. For the third period, OCA v3.0 (161 gene mutation and fusion) was available. OCA panels were required for RNA sequencing to investigate fusion genes such as ALK, ROS1, RET, NTRK, and NRG1. The quantities of input for DNA/RNA were 10 ng, which enabled the analysis of small and inferior‐quality samples, such as cytology pellets. All sequencings were performed using Ion S5 XL Systems (Thermo Fisher Scientific). In this study, we defined a complete process of sequencing with sufficient DNA/RNA as successful.
2.6. Statistical analysis
Each success rate was compared with the results of surgical biopsy by Fisher's exact test. All statistical analyses were performed with EZR (Saitama Medical Center, Jichi Medical University), which is a graphical user interface for R (The R Foundation for Statistical Computing). More precisely, it is a modified version of R commander designed to add statistical functions frequently used in biostatistics. 14 Statistical significance was indicated by P < 0.05.
3. RESULTS
3.1. Patient characteristics
In this study, a total of 222 patients were enrolled at St. Marianna University School of Medicine Hospital. The characteristics of all patients and each biopsy methods are listed in Table 1. Bronchoscopy (TBB or EBUS‐TBNA) was the dominant biopsy method. In the present cohort, the majority of patients were histologically confirmed nonsquamous (non‐Sq), NSCLC, and stage IV. The majority of biopsy sites were primary lung lesions.
TABLE 1.
Patients’ characteristics
| All | Cytology sample (N = 153) | Tissue sample (N = 69) | |||||
|---|---|---|---|---|---|---|---|
| EBUS‐GS‐TBB (bronchial brushing) | EBUS‐TBNA (needle washing) | Pleural effusion | Thoracoscopy | CNB | Surgical biopsy | ||
| Number of cases (%) | 222 (100%) | 97 (43.7%) | 45 (20.3%) | 11 (4.9%) | 16 (7.2%) | 37 (16.7%) | 16 (7.2%) |
| Median age (range) | 70 (35‐88) | 70 (46‐88) | 70 (48‐86) | 68 (49‐75) | 65 (39‐81) | 67 (35‐83) | 64 (48‐78) |
| Histology | |||||||
| Non‐squamous | 166 (74.8%) | 69 | 32 | 11 | 14 | 29 | 11 |
| Squamous | 28 (12.6%) | 15 | 4 | 0 | 0 | 6 | 3 |
| Small cell | 28 (12.6%) | 13 | 9 | 0 | 2 | 2 | 2 |
| Stage | |||||||
| IIIA/IIIB/IIIC | 40 (18.0%) | 21 | 14 | 0 | 0 | 3 | 2 |
| IVA/IVB | 170 (76.6%) | 75 | 25 | 11 | 15 | 32 | 12 |
| Postoperative recurrence | 12 (5.4%) | 1 | 6 | 0 | 1 | 2 | 2 |
| Biopsy site | |||||||
| Lung | 120 (54.1%) | 97 (100%) | 0 | 0 | 0 | 18 (48.7%) | 5 (31.3%) |
| Lymph node | 56 (25.2%) | 0 | 45 (100%) | 0 | 0 | 8 (21.6%) | 3 (18.7%) |
| Bone | 8 (3.6%) | 0 | 0 | 0 | 0 | 4 (10.8%) | 4 (25.0%) |
| Liver | 4 (1.8%) | 0 | 0 | 0 | 0 | 4 (10.8%) | 0 (0%) |
| Brain | 2 (0.9%) | 0 | 0 | 0 | 0 | 0 (0%) | 2 (12.5%) |
| Other | 32 (14.4%) | 0 | 0 | 11 (100%) | 16 (100%) | 3 (8.1%) | 2 (12.5%) |
Abbreviations: CNB, core needle biopsy; EBUS‐GS‐TBB, transbronchial biopsy using endobronchial ultrasound sonography guide sheath; EBUS‐TBNA, endobronchial ultrasound sonography–guided needle aspiration biopsy.
3.2. Extracted yield of DNA/RNA
Median yields of nucleic acids are described in Table 2. Extracted yields of RNA were consistently lower than those of DNA, regardless of biopsy method (Table 2; Figure 2). Furthermore, nucleic acid yields from cytology samples were lower than those from tissue samples. Median extracted yields of DNA from bronchoscopy samples were 70 ng/mL for TBB and 90 ng/mL for EBUS‐TBNA.
TABLE 2.
Extracted DNA/RNA and success rate of next‐generation sequencing (NGS) according to biopsy methods
| All | Cytology sample (N = 153) | Tissue sample (N = 69) | |||||
|---|---|---|---|---|---|---|---|
| EBUS‐GS‐TBB (bronchial brushing) | EBUS‐TBNA (needle washing) | Pleural effusion | Thoracoscopy | CNB | Surgical biopsy | ||
| Extracted yield of nucleic acid (ng/mL) | |||||||
| DNA median (range) | 90 (10‐400) | 70 (10‐380) | 90 (10‐400) | 270 (110‐330) | 190 (30‐350) | 70 (10‐380) | 215 (100‐350) |
| RNA median (range) | 20 (10‐790) | 10 (10‐80) | 20 (10‐290) | 50 (10‐680) | 55 (10‐340) | 10 (10‐210) | 260 (10‐790) |
| NGS success rate (95% CI) | |||||||
| DNA sequencing |
99.1% (96.8‐99.9) (220/222) |
98.7% (95.4‐98.8) (P = 0.999 a ) |
100% (95.8‐100) |
||||
|
97.9% (92.7‐99.7) (P = 0.999 b ) |
100% (93.6‐100) (P = 1.00 b ) |
100% (76.2‐100) (P = 1.00 b ) |
100% (82.9‐100) (P = 1.00b) |
100% (92.2‐100) (P = 1.00b) |
100% (82.9‐100) |
||
| RNA sequencing |
85.1% (79.8‐89.5) (189/222) |
83.0% (76.1‐88.6) (P = 0.224 a ) |
89.9% (80.2‐95.8) |
||||
|
84.4% (70.5‐93.5) (P = 0.174 b ) |
80.4% (71.1‐87.8) (P = 0.069 b ) |
100% (76.2‐100) (P = 1.00 b ) |
100% (82.9‐100) (P = 1.00 b ) |
81.1% (64.8‐92.0) (P = 0.089 b ) |
100% (82.9‐100) |
||
Abbreviations: CNB, core needle biopsy; EBUS‐GS‐TBB, transbronchial biopsy using endobronchial ultrasound sonography guide sheath; EBUS‐TBNA, endobronchial ultrasound sonography–guided needle aspiration biopsy.
Success rate of cytology sample (N = 153) was compared with tissue sample (N = 69).
Success rate of each diagnostic modality was compared with surgical biopsy.
FIGURE 2.
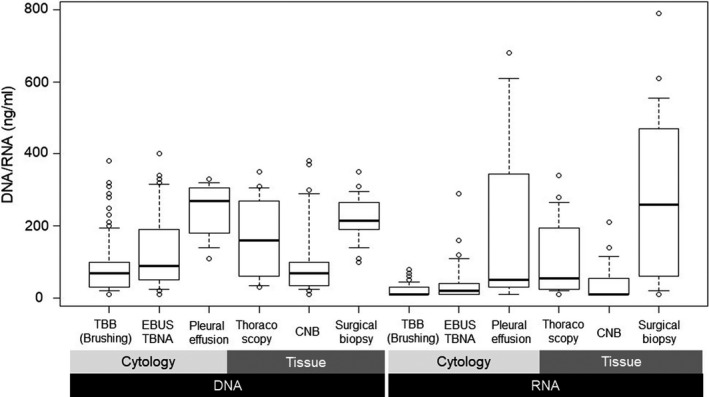
Extracted DNA/RNA yield according to biopsy methods. In the box‐whisker plot, upper and lower whiskers indicate 90 and 10 percentiles, respectively. The top and bottom of box indicate 75 and 20 percentiles, respectively. CNB, core needle biopsy; EBUS, endobronchial ultrasound sonography; TBNA, needle aspiration biopsy; TBB, transbronchial biopsy
These yields were relatively low compared with tissue samples. Median extracted yields of RNA from bronchoscopy samples were 10 ng/mL for TBB and 20 ng/mL for EBUS‐TBNA. Extracted RNA from bronchoscopy pellets were quite small compared with tissue samples. Regarding histologic classifications, the quantity of nucleic acids from non‐Sq samples was relatively low compared with other histologic classifications (Table 3).
TABLE 3.
Extracted DNA/RNA and success rate of next‐generation sequencing (NGS) according to histologic classification
| Non‐sqamous | Sqamous | Small | |
|---|---|---|---|
| Number of cases (%) | 166 (74.8%) | 28 (12.6%) | 28 (12.6%) |
| Samples for NGS | |||
| Cytology | 112 (67.5%) | 22 (78.6%) | 24 (85.7%) |
| Tissue | 54 (32.5%) | 6 (21.4%) | 4 (14.3%) |
| Extracted amount of nucleic acid (ng/mL) | |||
| DNA median (range) | 80 (10‐370) | 95 (10‐380) | 135(60‐400) |
| RNA median (range) | 20 (10‐790) | 15 (10‐210) | 20 (10‐120) |
| NGS success rate (%) | |||
| DNA sequencing | 100% (98.2‐100) | 92.9% (76.5‐99.1) | 100% (89.9‐100) |
| RNA sequencing | 86.7% (80.6‐91.5) | 60.7% (40.6‐78.5) | 100% (89.9‐100) |
3.3. Success rate and results of NGS analyses
The success rates of NGS were 99.1% (96.8%‐99.9%, 95% CI) for DNA sequencing and 85.1% (71.1%‐89.5%, 95% CI) for RNA sequencing in all patients (Table 2). Only in the case of two patients we could not perform the NGS analyses due to insufficient extraction of DNA. Therefore, there were no significant differences in the success rates between biopsy methods. On the other hand, regarding RNA sequencing, the success rate of cytology samples from bronchoscopy was relatively low compared with tissue samples. The success rates of RNA sequencing were 80.4% (71.1%‐87.8%, 95% CI) for TBB and 84.4% (70.5%‐93.5%, 95% CI) for EBUS‐TBNA.
We performed exploratory analyses according to histologic classifications. In patients with squamous cell carcinoma (Sq), the success rate of RNA sequencing was much lower than that of other histologic classifications (Table 3). We detected various rare driver oncogene aberrations (Figure 3) although we used transbronchial brushing specimens for 41.6% (69/166) of patients in the non‐Sq cohort. In the non‐Sq cohort, seven patients could be considered for investigational molecular targeted drug trials.
FIGURE 3.
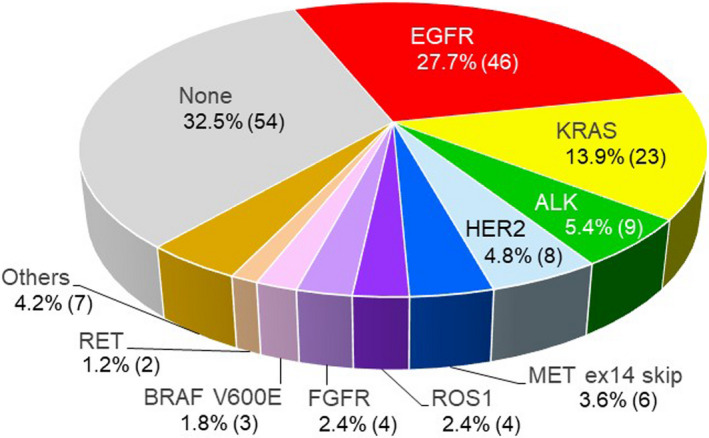
Distribution of driver oncogene aberrations in non‐Sq non–small cell lung cancer (NSCLC) (N = 166). Percentage and number of patients. Others included three cases of PIK3CA mutation, three cases of MET amplification, and one case of NRAS mutation
4. DISCUSSION
This is the first and the largest prospective cohort study to investigate the suitability of fresh frozen cytology samples obtained by transbronchial brushing for NGS analysis in patients with peripheral lung cancer using EBUS‐GS‐TBB. We demonstrated that transbronchial brushing pellets were suitable for NGS analysis even if the amount of extracted nucleic acid was relatively small compared with other biopsy methods. These findings could be useful in clinical practice.
Transbronchial brushing for peripheral lung cancer has some advantages. First, this method is less invasive and can obtain sufficient samples using the guide sheath method, without severe bleeding. 10 Second, we often see patients with small peripheral lung nodules. In these cases, it is difficult to obtain sufficient tumor cells from small lung nodules by forceps biopsy. Even if the target lesions are quite small, bronchial brushing enables sufficient tumor cells collection. 12 Third, even when fresh frozen samples are required, pathological confirmation is still possible by dividing the same sample into two tubes (Figure 1).
A few studies have reported on the feasibility of TBB for NGS analysis. 4 , 6 , 7 , 8 We selected four studies in which the number of cases was over 10 (see Table 4). When we compare these data, we need to pay attention to the number of genes in each NGS panel. Most recent studies reported on FFPE tissue samples. In the present study, we demonstrated a relatively high success rate of NGS analyses compared with previous reports (Table 4). One of the reasons for this high success rate might be that we utilized fresh frozen samples. When small FFPE biopsy samples are used for NGS analysis, there are concerns regarding DNA/RNA fragmentation and quality. In a previous report, the agreement of gene expression between fresh frozen and FFPE was not so high (63%); storage time and archival condition might have caused this low agreement. 15 Therefore, we used fresh frozen samples in LC‐SCRUM‐Japan/Asia.
TABLE 4.
Success rate of next‐generation sequencing (NGS) in previous studies by bronchoscopy
| Previous reports (reported year) | Study cohort | Biopsy method | N | Sample | NGS panel | Number of genes in NGS panel | Success rate | ||
|---|---|---|---|---|---|---|---|---|---|
| DNA seq | RNA seq | ||||||||
| Turner et al (2018) 6 | Prospective | EBUS‐TBNA | 115 | Tissue | FFPE | MSK‐IMPACT assay | DNA: 341 | 71.3% | — |
| Stoy et al (2018) 7 | Retrospective | EBUS‐TBNA | 54 | Cytology |
Smear FFPE |
OncoScreen or OncoPlus |
DNA: 50 (OncoScreen) DNA: 147 (OncoPlus) |
90.7% (49/54) a | — |
| Ku et al (2018) 8 | Retrospective |
TBB EBUS‐TBNA |
31 41 |
Tissue | FFPE |
Ion AmpliSeq Cancer Hotspot Panel v2 |
DNA: 50 | Unknown | — |
| Kage et al (2019) 4 | Retrospective |
TBB EBUS‐TBNA |
11 11 |
Tissue | FFPE | Todai OncoPanel | DNA: 464, RNA: 463 |
82% 100% |
64% 67% |
| Present study | Prospective |
EBUS‐GS‐TBB EBUS‐TBNA |
97 45 |
Cytology | Fresh frozen |
Oncomine Comprehensive Assay v1.0 or v3.0 |
DNA: 146, RNA: 51 |
97.9% 100% |
80.4% 84.4% |
Abbreviations: EBUS‐GS‐TBB, transbronchial biopsy using endobronchial ultrasound sonography guide sheath; EBUS‐TBNA, endobronchial ultrasound sonography–guided needle aspiration biopsy; FFPE, formalin‐fixed and paraffin‐embedded; TBB, transbronchial biopsy.
The data were adjusted to the definition of our study.
Interestingly, in our exploratory analyses according to histologic classifications, the success rate of RNA sequencing in Sq was much lower than that of other histologic classifications despite the larger amount of DNA/RNA extracted (Table 3). Sq is characterized by necrotic tendencies, and these necrotic samples might have caused the low success rate. Regarding this discussion point, further studies are needed.
This study has several limitations. First, this cohort was a part of LC‐SCRUM Japan/Asia, that is, it was a single‐center cohort. Second, we only collected fresh frozen samples for LC‐SCRUM Japan/Asia; however, it might be complicated to process and freeze bronchial brushing pellets after biopsy in clinical practice. When bronchial brushing materials are used, FFPE cell‐block might be considered. 16 Finally, in the present study, we focused on the difference in quantities of nucleic acids according to biopsy method. To investigate more rigidly, we should evaluate not only the quantity of DNA/RNA but also the quality by measuring DNA/RNA integrity number (DIN/RIN), ΔCt, ddCq, and DV200.
Moreover, a recent study revealed that transbronchial cryobiopsy was useful to obtain sufficient sample size and DNA/RNA with good quality for whole‐exome RNA sequencing. 17 Transbronchial techniques should be more improved to perform precision medicine for all patients with lung cancer.
In conclusion, the present study demonstrated that cytology samples obtained by transbronchial brushing using EBUS‐GS were suitable for NGS analysis. Extracted DNA/RNA yields were relatively small compared with other biopsy methods. However, we could detect some rare oncogenic driver aberrations from these specimens. These findings could contribute to precision medicine for patients diagnosed by bronchoscopy in clinical practice.
DISCLOSURE
LC‐SCRUM‐Japan/Asia was supported by Amgen, Astellas, AstraZeneca, Boehringer‐Ingelheim, Bristol‐Myers, Chugai, Daiichi‐Sankyo, Eisai, Eli Lilly, Janssen, Kyowa Kirin, Merck Serono, MBL, MSD, Novartis, Ono, Pfizer, Sumitomo Dainippon, Taiho, and Takeda.
ACKNOWLEDGEMENTS
The authors thank the staff of the Department of Respiratory Medicine of St. Marianna University School of Medicine, SCRUM‐Japan Office and SRL, especially Yuri Murata (secretary, LC‐SCRUM‐Asia), Reiko Nagata (clinical research coordinator, St. Marianna University School of Medicine), Misa Fuchioka (technical advisor of NGS, SRL) and Noriaki Kurimoto (technical advisor of bronchoscopy, Shimane University) for their advice and assistance. The authors also thank Mr Jason Tonge from St. Marianna University School of Medicine for reviewing the language of this article.
Furuya N, Matsumoto S, Kakinuma K, et al. Suitability of transbronchial brushing cytology specimens for next‐generation sequencing in peripheral lung cancer. Cancer Sci. 2021;112:380–387. 10.1111/cas.14714
REFERENCES
- 1. Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK‐RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res. 2015;4(2):156‐164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saito M, Shiraishi K, Kunitoh H, et al. Gene aberrations for precision medicine against lung adenocarcinoma. Cancer Sci. 2016;107:713‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998‐2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kage H, Kohsaka S, Shinozaki‐Ushiku A, et al. Small lung tumor biopsy samples are feasible for high quality targeted next generation sequencing. Cancer Sci. 2019;110(8):2652‐2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence‐based clinical practice guidelines. Chest. 2013;143(5 suppl):e142S‐e165S. [DOI] [PubMed] [Google Scholar]
- 6. Turner SR, Buonocore D, Desmeules P, et al. Feasibility of endobronchial ultrasound transbronchial needle aspiration for massively parallel next‐generation sequencing in thoracic cancer patients. Lung Cancer. 2018;119:85‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stoy SP, Segal JP, Mueller J, et al. Feasibility of endobronchial ultrasound‐guided transbronchial needle aspiration cytology specimens for next generation sequencing in non‐small‐cell lung cancer. Clin Lung Cancer. 2018;19(3):230‐238. [DOI] [PubMed] [Google Scholar]
- 8. Ku BM, Heo MH, Kim J‐H, et al. Molecular screening of small biopsy samples using next‐generation sequencing in Korean patients with advanced non‐small cell lung cancer: Korean Lung Cancer Consortium (KLCC‐13‐01). J Pathol Transl Med. 2018;52(3):148‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hagmeyer L, Fassunke J, Engels M, et al. Bronchoscopic brushing from central lung cancer‐next generation sequencing results are reliable. Lung. 2019;197(3):333‐337. [DOI] [PubMed] [Google Scholar]
- 10. Kurimoto N, Miyazawa T, Okimasa S, et al. Endobronchial ultrasonography using a guide sheath increases the ability to diagnose peripheral pulmonary lesions endoscopically. Chest. 2004;126(3):959‐965. [DOI] [PubMed] [Google Scholar]
- 11. Oshige M, Shirakawa T, Nakamura M, et al. Clinical application of virtual bronchoscopic navigation system for peripheral lung lesions. J Bronchology Interv Pulmonol. 2011;18(2):196‐202. [DOI] [PubMed] [Google Scholar]
- 12. Lim JH, Kim MJ, Jeon S‐H, et al. The optimal sequence of bronchial brushing and washing for diagnosing peripheral lung cancer using non‐guided flexible bronchoscopy. Sci Rep. 2020;10(1):1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamada N, Yamazaki K, Kurimoto N, et al. Factors related to diagnostic yield of transbronchial biopsy using endobronchial ultrasonography with a guide sheath in small peripheral pulmonary lesions. Chest. 2007;132(2):603‐608. [DOI] [PubMed] [Google Scholar]
- 14. Kanda Y. Investigation of the freely available easy‐to‐use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48(3):452‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lüder Ripoli F, Mohr A, Conradine Hammer S, et al. A comparison of fresh frozen vs. formalin‐fixed, paraffin‐embedded specimens of canine mammary tumors via branched‐DNA assay. Int J Mol Sci. 2016;17(5):724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young G, Wang K, He J, et al. Clinical next‐generation sequencing successfully applied to fine‐needle aspirations of pulmonary and pancreatic neoplasms. Cancer Cytopathol. 2013;121(12):688‐694. [DOI] [PubMed] [Google Scholar]
- 17. Udagawa H, Kirita K, Naito T, et al. Feasibility and utility of transbronchial cryobiopsy in precision medicine for lung cancer: prospective single‐arm study. Cancer Sci. 2020;111(7):2488‐2498. [DOI] [PMC free article] [PubMed] [Google Scholar]


