Abstract
Background
COVID-19, the presently prevailing global public health emergency has culminated in international instability in economy. This unprecedented pandemic outbreak pressingly necessitated the trans-disciplinary approach in developing novel/new anti-COVID-19 drugs especially, small molecule inhibitors targeting the seminal proteins of viral etiological agent, SARS-CoV-2.
Methods
Based on the traditional medicinal knowledge, we made an attempt through molecular docking analysis to explore the phytochemical constituents of three most commonly used Indian herbs in ‘steam inhalation therapy’ against well recognized viral receptor proteins.
Results
A total of 57 phytochemicals were scrutinized virtually against four structural protein targets of SARS-CoV-2 viz. 3CLpro, ACE-2, spike glycoprotein and RdRp. Providentially, two bioactives from each of the three plants i.e. apigenin-o-7-glucuronide and ellagic acid from Eucalyptus globulus; eudesmol and viridiflorene from Vitex negundo and; vasicolinone and anisotine from Justicia adhatoda were identified to be the best hit lead molecules based on interaction energies, conventional hydrogen bonding numbers and other non-covalent interactions. On comparison with the known SARS-CoV-2 protease inhibitor –lopinavir and RdRp inhibitor –remdesivir, apigenin-o-7-glucuronide was found to be a phenomenal inhibitor of both protease and polymerase, as it strongly interacts with their active sites and exhibited remarkably high binding affinity. Furthermore, in silico drug-likeness and ADMET prediction analyses clearly evidenced the usability of the identified bioactives to develop as drug against COVID-19.
Conclusion
Overall, the data of the present study exemplifies that the phytochemicals from selected traditional herbs having significance in steam inhalation therapy would be promising in combating COVID-19.
Keywords: SARS-CoV-2, COVID-19, ACE-2, 3CLpro, RdRp, Spike protein, In silico, Herbal steam inhalation therapy
Graphical abstract
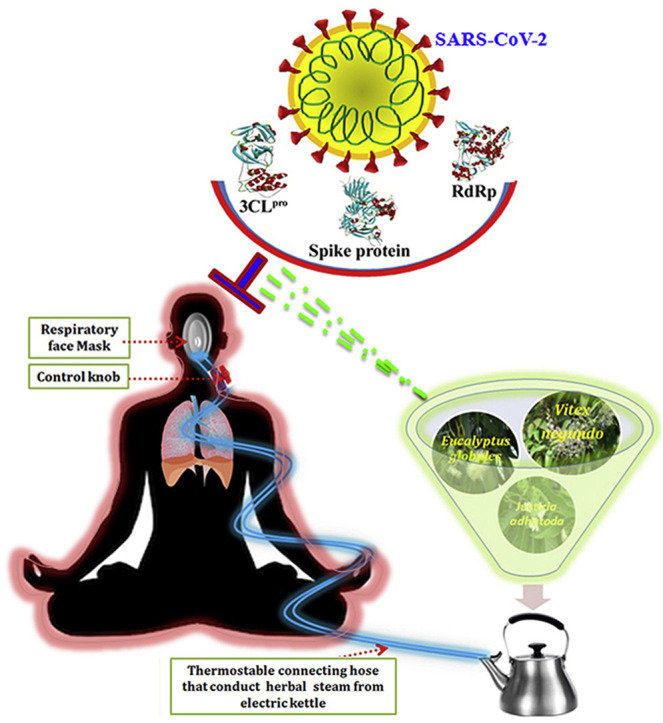
1. Introduction
Since the dawn of year 2020, a global pandemic menace prevails owing to the emergence of novel coronavirus (2019-nCoV) –the prime etiological agent of severe acute respiratory syndrome (SARS) or pneumonia denoted shortly as “coronavirus disease 2019” (COVID-19) (de Wit et al., 2016; Thuy et al., 2020; Wu et al., 2020). According to World Health Organization (WHO), SARS-CoV-2 has affected 67.5 million populations across 220 countries and the rate of mortality is 2.29% within a year (till December 09, 2020) of its outbreak. Because of the lack of early diagnosis and specific drug or vaccine to detect and treat the infection, COVID-19 has been declared as the public Health Emergency of International Concern (PHEIC) by WHO, signifying that this pandemic seeks coordinated global response in all medical aspects (de Wit et al., 2016; Wu et al., 2020).
Human coronaviruses belonging to the large family of Coronaviridae and genus beta-coronavirus has predominantly been associated with mild to major upper respiratory tract diseases, most frequently common cold (de Wit et al., 2016; Killerby et al., 2018). Other members of this genus, viz., Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) causes severe pneumonia in human, as the former caused 774 deaths in 2002 and later caused 858 in 2015 (Li et al., 2005, Li et al., 2005; de Wit et al., 2016). Similar to its antecedents, this deadly COVID-19 has an incubation period of 2 weeks to express its symptoms such as fever, cough, dyspnoea and lesions in lungs (Killerby et al., 2018; Vellingiri et al., 2020).
SARS-CoV-2, initially attaches to the nasal cavity and upon binding to the specific host receptor viz. angiotensin-converting enzyme 2 (ACE-2), makes its entry into the host cell. The active receptor binding domain (RBD) found in the viral spike protein (S-protein) is being recognized by the host ACE-2 and thereby initiate the early event of coronavirus infection. Interaction between these two proteins occur when the 394 glutamine residue of RBD in SARS-CoV-2 is recognized by the crucial lysine 31 residue of host receptor ACE-2, and establishes strong interaction by van der waals forces (Li et al., 2003; Tikellis et al., 2012; Zhou et al., 2020). The ACE-2 receptor is an integral membrane glycoprotein majorly expressed in endothelium, kidney, heart and lungs (Roberts et al., 2007; Tikellis et al., 2012). Recent reports have well demonstrated that blocking either ACE-2 host receptor or S-protein would eventually restrict the entry of coronovirus at the very beginning stage and in turn prevent the host from further infection (Paraskevis et al., 2020). Therefore, identifying phytochemicals having higher affinity towards ACE-2 receptor is critically essential to reduce the availability of host receptor for SARS-CoV-2.
Soon after the recognition by ACE-2 receptor, the SARS-CoV-2 requires main protease (Mpro; otherwise known as 3-chymotrypsin like protease (3CLpro)) for the proteolytic maturation of virus. Targeting 3CLpro could inhibit the cleavage of viral polyprotein, which ultimately prevent the viral replication in host cells (Zhang et al., 2020). The amino acids of 3CLpro viz., Thr24, Thr26 and Asn119 have previously been predicted to play an indispensable role in drug interaction (Ton et al., 2020). Thus, 3CLpro gains greater attention as attractive target for developing antiviral therapeutics (Ton et al., 2020; Alagu Lakshmi et al., 2020). In the same way, RdRp also plays an essential role in viral transcription and maturation inside the host (Lung et al., 2020). Hence, the traditional herbs with efficacy to inhibit protease (3CLpro) and polymerase (RdRp) could plausibly prevent the viral replication and arrest further spread as well as infection in respiratory tract.
Despite the immense concern of the scientific society in identifying synthetic drug(s) against SARS-CoV-2, alternative strategies that strengthen the immune status of the host would be even more promising. Following the same paradigm, of late, the management of patients chiefly focuses on the respiratory supportive care e.g., oxygenation, ventilation, and fluid management. Combination therapy of anti-virals, low-dose corticosteroids and atomization inhalation of interferon has been effective in critical COVID-19 management (Liu et al., 2020). Together, with the fact that SAR-CoV-2 enters the host through nasal cavity and infect the buccal cavity, the present study put forward the ‘herbal steam inhalation’ therapy to support and protect the COVID-19 patients. As the herbal steam abundantly wipes off the respiratory tract, it is anticipated to immunize the host by plausibly blocking viral entry, which ultimately make the virus ineffective in causing further havoc.
In Indian traditional knowledge system, the herbal steam inhalation therapy has been a well recognized home remedy for common cold. The different herbs used in the traditional in-house inhalation therapy have a wide range of proven medicinal benefits that strengthened the respiratory tracks and immune system (Wang et al., 2014; Lin et al., 2014; Ganjhu et al., 2015) Based on which, three herbal plants viz., Vitex negundo, Justicia adhatoda and Eucalyptus globules were considered for the present study. The phytochemical constituents of these plants have been well studied and documented in a plethora of earlier reports through mass spectrometry (Singh et al., 1999; Gonzalez-Burgos et al., 2018; Jha et al., 2012). In a similar fashion, several studies in the recent past have envisaged compounds from natural resources especially from plants against SARS-CoV-2 drug targets viz., ACE-2 and spike proteins (Alagu Lakshmi et al., 2020; Alexpandi et al., 2020; Gyebi et al., 2020; Joshi et al., 2020; Muthuramalingam et al., 2020; Thuy et al., 2020; Ton et al., 2020). Therefore, in the current investigation, these phyto-ligands were analysed for their inhibiting ability against Mpro, ACE-2, S-protein and RdRp through in silico approach.
2. Material and methods
2.1. Protein selection and preparation
The three dimensional crystal structure of the selected three target proteins viz. Mpro (PDB ID: 5R82), SARS-CoV-2 Spike protein (PDB ID: 6VYB), and host entry receptor ACE-2 (PDB ID: 1R42) (data of homology modelled structure of RdRp was shown as supplementary) were retrieved from the RCSB Protein Data Bank (http://www.rcsb.org/pdb/home/home.do). To minimize energy, the predicted structures were refined by removing the water molecules and co-crystal ligands of the target proteins. Finally, it was used for molecular docking simulation (Mir et al., 2016).
2.2. Ligand retrieval and preparation
The chemical structure of the phyto-ligands was retrieved from the PubChem database which is available at NCBI (http://www.pubchem.ncbi.nlm.nih.gov). The 2D conformations of the phyto-ligands were downloaded in SDF format and converted into PDB format. The structural optimization was performed using Discovery Studio visualizer. Thus obtained chemical structures were used for further docking analysis (Yadav et al., 2017).
2.3. Molecular docking simulation
The molecular docking simulation was performed with the target protein and phyto-ligands using Autodock 4.2 by employing Lamarckian genetic algorithm. In order to minimize energy, polar hydrogen and kollman charges united atomic charges were computed, and unwanted water molecules were removed in the target proteins (Han et al., 2019; Cheng et al., 2012). Ligand molecules were added with hydrogen atom and gasteiger charges were assigned. Grid box was delineated on binding pocket of target proteins and the grid points were expanded in all directions to include the binding region. The grid box dimensions considered for molecular docking of the target proteins were given in Table 1 . Finally, best orientation was determined by tethering ligands to target proteins with highest binding energy (kcal mol−1). The pose of 2D and 3D ligand-to-target interactions with high binding energy was extracted and visualized using Discovery Studio (Yousef et al., 2018; Halder et al., 2019).
Table 1.
Dimensions of grid box for selected protein targets in molecular docking analysis.
| Protein targets | Dimension of grid box | ||
| Grid points number (npts) | Center (xyz coordinates) | Grid box spacing (Å) | |
| Mpro | 80 × 80 × 85 | 30.619, 28.880, 30.200 | 0.375 |
| ACE-2 | 60 × 60 × 60 | 15.692, 37.141, 46.425 | 0.375 |
| Spike protein | 80 × 75 × 68 | 15.692, 37.141, 46.425513 | 0.375 |
2.4. Pharmacokinetic analysis
The online chemo informatics tools Molinspiration and admetSAR (http://lmmd.ecust.edu.cn:8000/) were used to analysis the ADMET profiles, pharmacokinetics, drug likeness and harmful properties of the selected phyto-ligands. The Molinspiration tool was employed to analysis the molecular descriptor and drug likeness properties of compound based on the Lipinski's rules of 5 and various pharmacokinetics properties of phyto-ligands such as blood-brain barrier penetration ability (BBBPA), human intestinal absorption ability (HIAA), cytochrome P450 (CYP450 2D6) inhibitor, renal organic cation transporter (ROCT), aqueous solubility, carcinogenicity and biodegradation were calculated using admetSAR web server (Paramashivam et al., 2015).
2.5. Molecular dynamic simulation (MDS) studies
The top three phytoligands from each medicinal plants showing good binding affinity and interaction with the Mpro and ACE2 were subjected to 25ns of MD simulation analysis using Gromacs 5.1.4 simulation package (Sepay et al., 2020). The topology parameter files of the proteins were generated with GROMOS96 all force filed and the ligand topology file was generated using PRODRG2 web server. The docked complexes were embedded into cubic box of 1.0 Å periodic boundary conditions using SPC (simple point charge) water model. Then, the overall systems were neutralized by adding the couturier ions. The processed systems were subjected to energy minimization, which was carried out by applying steepest descent algorithm and conjugate gradient minimization. The systems were equilibrated with the NPT and NVT ensembles for gradual maintenance of temperature (up to 300 K) and pressure (1 bar) for 100ps respectively, which was maintained for each system with Parrinello−Rahman barostat. Finally, the production MD simulation of each docked complexes were performed for 25 ns.
3. Results
3.1. In silico screening and identification of bioactives from Indian herbs having traditional significance towards steam inhalation therapy
Primarily, the phytochemical constituents of Indian traditional herbs used in steam inhalation therapy were taken from the earlier reports that have well characterised and documented the volatile and non-volatile biomolecules of V. negundo (Singh et al., 1999) J. adhatoda (Jha et al., 2012) and E. globules (Gonzalez-Burgos et al., 2018) using mass spectrometry technique. A total of 57 phytochemicals were chosen for the virtual screening, the 3D structures of phytochemicals were modelled and optimized, and listed on the basis of their best binding affinity towards target proteins such as Mpro, S-protein and ACE-2 (Supplementary Table 1). The crystal structures of the target proteins were used to demonstrate the molecular interactions (Meng et al., 2011). Based on the binding energy, top two bioactive ligands from each of the three selected herbs are displayed in Fig. 1, Fig. 2, Fig. 3, Fig. 4 .
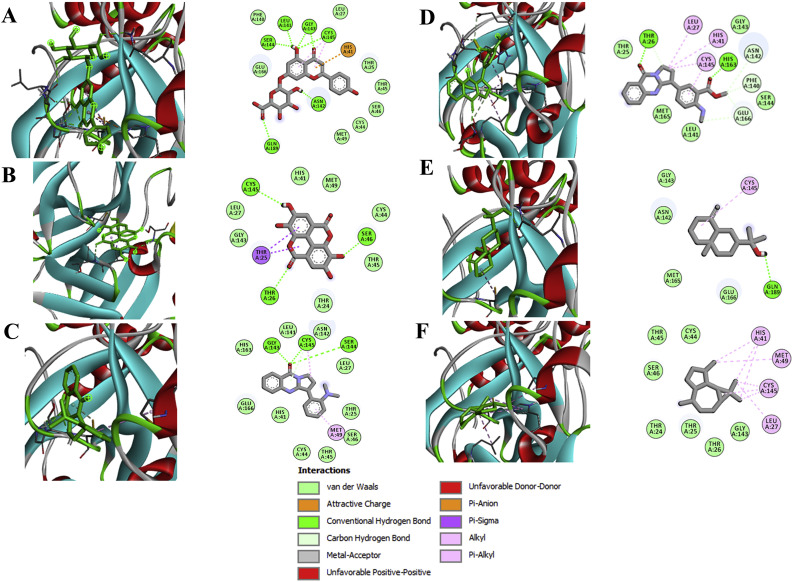
Fig. 1.
Binding interaction map of top six scoring phytoligands [(A) apigenin-o-7-glucuronide, (B) ellagic acid, (C) vasicolinone, (D) anisotine, (E) eudesmol, and (F) viridiflorene with SARS-CoV-2 drug target 3-chymotrypsin like protease (Mpro; 5R82). Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
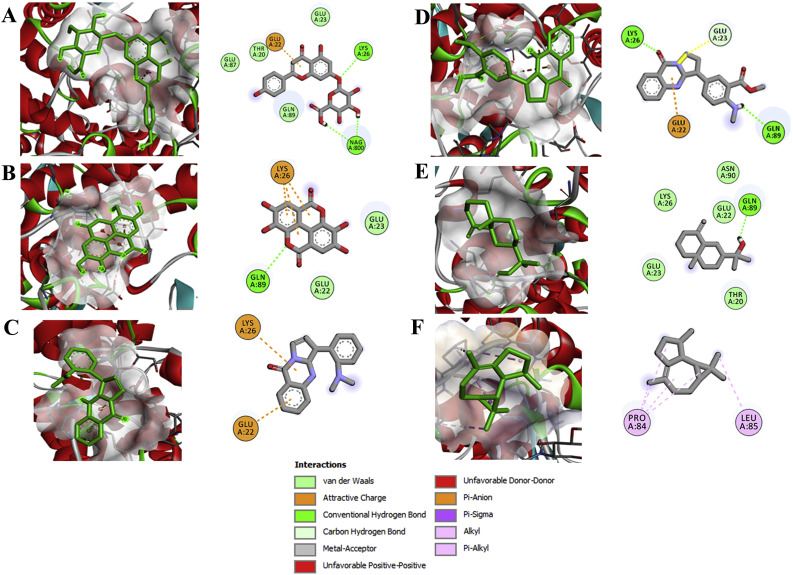
Fig. 2.
Binding interaction map of top six scoring phytoligands [(A) apigenin-o-7-glucuronide, (B) ellagic acid, (C) vasicolinone, (D) anisotine, (E) eudesmol, and (F) viridiflorene with SARS-CoV-2 drug target angiotensin-converting enzyme (ACE-2; 1R42). Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
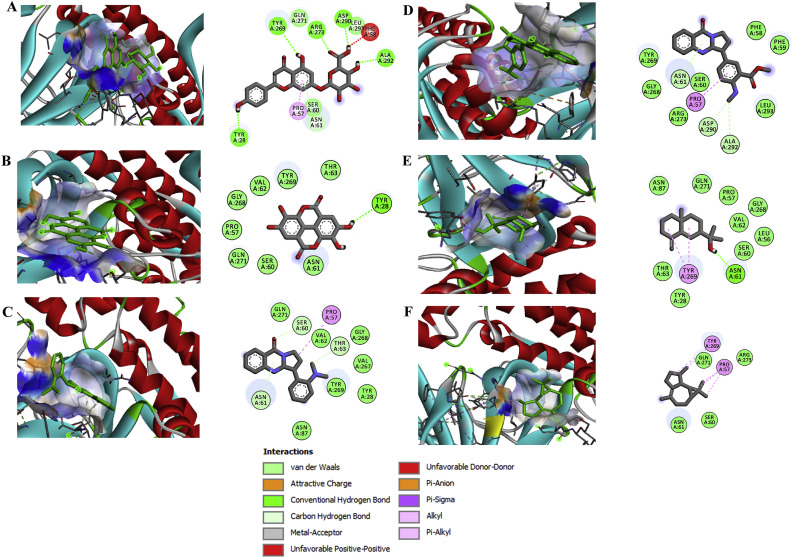
Fig. 3.
Binding interaction map of top six scoring phytoligands [(A) apigenin-o-7-glucuronide, (B) ellagic acid, (C) vasicolinone, (D) anisotine, (E) eudesmol, and (F) viridiflorene with SARS-CoV-2 drug target Spike protein (S-protein; 6VYB). Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
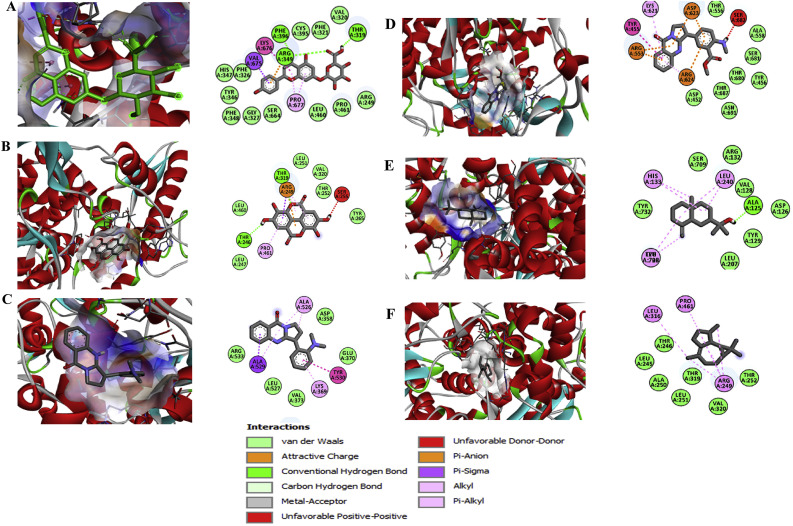
Fig. 4.
Binding interaction map of top six scoring phytoligands [(A) apigenin-o-7-glucuronide, (B) ellagic acid, (C) vasicolinone, (D) anisotine, (E) eudesmol, and (F) viridiflorene with SARS-CoV-2 drug target RNA dependent RNA polymerase (RdRp; data of homology modelled structure of RdRp was not shown). Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. Apigenin-o-7-glucuronide (AG)–a profound SARS-CoV-2 Mpro inhibitor
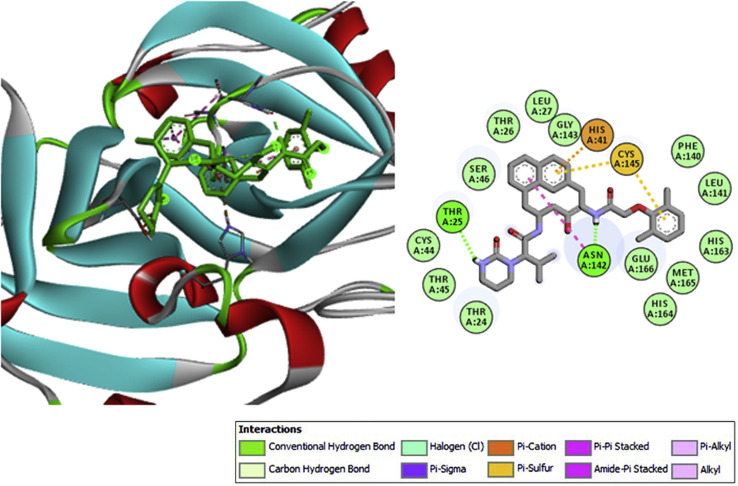
Data of the docking analysis unveiled that out of 57 phytochemicals screened from V. Negundo (n = 31), J. adhatoda (n = 8) and E. globules (n = 18), apigenin-7-o-glucuronide, a phenolic compound from E. globulus exhibited phenomenal binding affinity and interaction towards Mpro than the other phytochemicals and the positive reference lopinavir. The binding affinity of apigenin-7-o-glucuronide (AG) was observed to be −9.1 kcal mol−1, whereas −8.2 kcal mol−1 for lopinavir (Table 2 ). The highest binding energy between Mpro and AG was established through seven conventional hydrogen bonds with six residues (Cys145, Gly143, Leu141, Ser144, Asn142 and Gln189). The other significant interactions that stabilize AG and Mpro complex includes ᴫ-sulfur interaction with His41, two ᴫ-alkyl interaction with Cys145 and other non-covalent interaction such as van der Waals interaction with Met49, Cys44, Ser46, Thr45, Thr25, Leu27, Phe140 and Glu166 residues (Fig. 1).
Table 2.
List of top hit phytochemicals selected through virtual screening.
| S. No |
Name of the phytochemical |
Chemical structure |
PubChem ID |
Binding Energy toward target proteins (kcal mol−1) |
|||
|---|---|---|---|---|---|---|---|
| Mpro |
ACE-2 |
S-protein |
RdRp |
||||
| Name of the herb: Eucalyptus globulus | |||||||
| 1 | Apigenin-7-O-glucuronide | 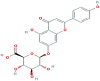 |
5319484 | −9.1 | −8.8 | −7.2 | −8.8 |
| 2 | Ellagic acid | 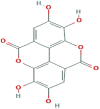 |
5281855 | −8.4 | −8.4 | −6.2 | −7.8 |
| Name of the herb: Justicia adhatoda | |||||||
| 3 | Vasicolinone | 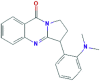 |
627712 | −8.0 | −7.5 | −6.4 | −7.6 |
| 4 | Anisotine | 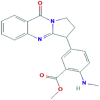 |
442884 | −7.4 | −7.8 | −6.4 | −8.2 |
| Name of the herb: Vitex negundo | |||||||
| 5 | Eudesmol | 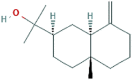 |
91457 | −8.0 | −7.1 | −6.0 | −7.2 |
| 6 | Viridiflorene | 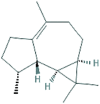 |
10910653 | −7.4 | −7.3 | −5.8 | −6.6 |
In par with the earlier reports, the active site residues (Cys145 and His41) of SARS-CoV-2 Mpro, a known CysHis catalytic dyad was found to be interactive with the known SARS-CoV-2 protease inhibitor –lopinavir (Cao et al., 2020). However, the bioactive compound AG has prominently interacted with several other crucial amino acid residues of main protease (Wu et al., 2020) alike lopinavir, which corroborate the high docking energy of the active compound AG (Supplementary Fig. 1A). These strong interactions could eventually hamper viral replication via hindering the progression of viral polyprotein. Together, the hydrophobic and non-covalent interactions were envisaged to augment the binding affinity through stabilizing the complex between protein's active site and ligand, which in turn enhanced the biological efficacy.
Vasicolinone, a quinazoline alkaloid from J. adhatoda was found to interact with the binding pocket of the target protein by building three conventional hydrogen bonds with Ser144, Cys145 and Gly143; ᴫ-alkyl interaction with Cys145 and Met49; carbon hydrogen bond interaction with Cys145; and the other non-covalent bond interactions with His41, Cys44, Thr45, Ser46, Thr25, Leu27, Asn142, Leu141, His163 and Glu166 amino acid residues with the binding affinity of −8.0 kcal mol−1 (Fig. 1). Eudesmol, a carbobicyclic compound from V. negundo exhibited one conventional hydrogen bond interaction with Gln189 residue; ᴫ-alkyl hydrophobic interaction with Cys145 and van der Waals interaction with Gly143, Asn142, Met165 and Glu166 with binding affinity of −8.0 kcal mol−1. In addition, the other bioactive ellagic acid, a polyphenol compound from E. globulus was found to profoundly interact with binding pocket of the target protein to forms three conventional hydrogen bonding with residues Cys145, Ser46 and Thr26; and two ᴫ-sigma interactions with Thr25; and other van der Waals interactions with His41, Met49, Cys44, Thr45, Thr24, Leu27 and Gly143 with a binding affinities −8.4 kcal mol−1 (Fig. 1). The positive control lopinavir explicated the binding energy of −8.1 kcal mol−1 formed two conventional hydrogen bonding with Thr25 and Asn142; two ᴫ-anionic interaction with Cys145; ᴫ-sulfur interaction with His41; amide ᴫ-stalked interaction with Asn142 and other non-covalent interaction like van der Waals interaction with Thr24, Thr45, Cys44, Ser46, Thr26, Leu27, Gly143, Phe140, Leu141, His163, Met165, His164 and Glu166 (Supplementary Fig. 1).
3.3. Binding interaction studies of drug-like molecules with ACE-2 host receptor
Data of the docking simulation and interactions between the crystal structure of angiotensin-converting enzyme-2 (ACE-2, PDB ID 1R42) and the phytochemicals revealed that AG depicted strong interactions to the binding pocket of ACE-2 as it showed a binding energy of −8.8 kcal mol−1 (Table 2). AG, through two conventional hydrogen bonding with Lys26 and Nag800 residues; ᴫ-anion interaction with Glu22 residue; and other non-covalent bond interaction viz. van der Waals interaction with residues Thr20, Glu23, Glu87 and Gln89. Next to AG, the bioactive compound ellagic acid docked complex shown a binding affinity of −8.4 kcal mol−1 building interactions via conventional hydrogen bonding with amino acid residue Gln89; ᴫ-sulfur interaction with residue Lys26; and other non-covalent bond interaction such as van der Waals interaction with residues Glu22 and Glu23 (Fig. 2). With the binding affinity of −7.8 kcal mol−1, anisotin undergoes two conventional hydrogen bonding (Lys26 and Gln89) and carbon hydrogen bond interaction (Glu23). Moreover, the other bioactive compounds ranked in the list were vasicolinone (−7.5 kcal mol−1) viridiflorene (−7.3 kcal mol−1) and eudesmol (−7.1 kcal mol−1) which were found to interact with binding pockets to form the conventional hydrogen binding with Gln89; ᴫ-sulfur interaction with residues Glu22 and Lys26; ᴫ-alkyl interaction with Pro84 and Leu85; and other non-covalent bond interaction such as van der Waals interaction with residues Thr20, Glu22, Glu23, Lys26 and Asn90 (Fig. 2).
3.4. Apigenin-o-7-glucuronide –a phenomenal SARS-CoV-2 spike protein inhibitor
As anticipated, the obtained data of docked complexes between bioactive compounds and SARS-CoV-2 spike glycoprotein unveiled that the phytochemical AG as the top bioactive expressing high binding affinity value of −7.2 kcal mol−1, which is relatively higher than the positive control lopinavir (−7.1 kcal mol−1). This top scorer AG had a very strong binding association with S-protein of SARS-CoV-2. It formed five strong conventional hydrogen bonds with crucial amino acid residues viz., Tyr28, Tyr269, Gln271, Asp290 and Ala292 (Fig. 3). Besides, it also built the ᴫ-alkyl interaction with Pro57 and unfavourable donor-donor interaction with Phe58. The additional stabilizing interaction associated with ligand and S-protein complex includes non-covalent interaction such as van der Waals interaction with Ser60, Asn61, Gln271, and Leu293. Anisotine, a quinazoline alkaloid from J. adhatoda with S-protein exhibited a binding affinity of −6.4 kcal mol−1 undergoes interaction through ᴫ-alkyl interaction with Pro57, ᴫ-sulfur interaction with Asp290, Ala292 and other non-covalent interaction like van der Waals interaction with Phe58, Phe59, Ser60, Gly268, Arg273 and Tyr269 (Fig. 3). Similarly, yet another quinazoline alkaloid –vasicolinone from the herb from J. adhatoda displayed the same binding energy of −6.4 kcal mol−1 and formed strong interactions with ᴫ-alkyl interact with Pro57, ᴫ-carbon hydrogen bonding with Ser60, Asn61 and Thr63 and other non-covalent interactions like van der Waals interaction with Tyr28, Asn87, Val267, Gly268, Tyr269 and Gln271 (Fig. 3).
Few other bioactive compounds such as ellagic acid (−6.2 kcal mol−1), endesmol (−6.0 kcal mol−1), viridiflorene (−5.8 kcal mol−1) showed the similar pattern of strong interactions with the spike glycoprotein of SARS-CoV-2 (Table 2). During all the docking orientation, hydrogen bonding, ᴫ-interaction and van der Waals interactions played major role in stabilizing the binding processes as could be observed form the relevant 2D diagrams (Fig. 1, Fig. 2, Fig. 3, Fig. 4). These results suggest that the identified bioactive compounds with the plausibly strong interactions may equally have inhibitory efficacy against SARS-CoV-2.
3.5. Virtual screening of RdRp inhibitor with remdesivir as reference
Nsp12 has been referred as the RNA-dependent RNA polymerase (RdRp), which catalyzes the process of viral transcription i.e. the synthesis of RNA from RNA template with nsp7 and nsp8 proteins as co-factors. RdRp being the key player in viral life cycle, targeting this crucial protein as drug target is considered as therapeutically potential. Remdesivir, a globally renowned antiviral compound (Wang et al., 2020) that target RdRp was used as a positive control for molecular interaction analysis in the present study. Data unveiled that the same six hit phytochemicals with profound drug efficacy against the other target proteins (Mpro, ACE-2 and S-protein) have depicted the high binding energy with the target RdRp in the order of AG, anisotine, ellagic acid, vasicolinone, eudesmol and viridiflorene (Table 2).
3.6. Apigenin-o-7-glucuronide –a promising SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) inhibitor
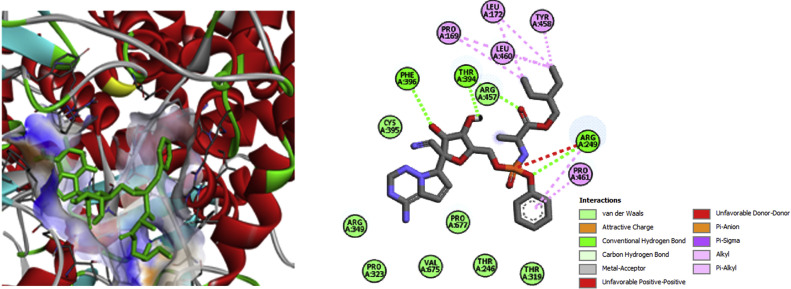
As anticipated, AG topped the list of bioactive hits with the binding energy of −8.7 kcal mol−1 in interaction with RdRp (Table 2), which is even higher than the binding energy (−7.6 kcal mol−1) exhibited by the known RdRp inhibitor –remdesivir (Supplementary Table 1). The molecular bond interactions of AG and RdRp were explicable from four conventional hydrogen bond formed with three residues of RdRp (Phe396, Thr319 and Arg349). AG-RdRp complex was stabilized by forming ᴫ-sigma interaction with Val675, ᴫ- ᴫ-stacking interaction with Lys676, ᴫ-alkyl interaction with Pro677 and other non-covalent interaction of van der Waals interaction with Cys395, Phe321, Val320, Arg249, His 347, Phe26, Tyr346, Phe348, Gly327, Ser664, Leu460 and Pro461 (Fig. 4).
The second top hit was anisotine reported a binding energy of −8.2 kcal mol−1 (Table 2). Although, the anisotine-RdRp complex formed no hydrogen bond, it built a strong interaction through the other hydrophobic, non-polar interaction with Lys621 (ᴫ-alkyl), Tyr455(ᴫ- ᴫ-stacking), Asp623, Arg553, Arg624 (ᴫ-anion), Thr556, Ala558, Ser681, Tyr456, Thr680, Thr687, Asn691 and Asp452 (Van der Waals) and also formed one unfavourable interaction with Ser682 (Fig. 4). Ellagic acid docked with RdRp reported −7.8 kcal mol−1 as binding energy (Table 2). Conformational stability of the complex (Ellagic acid-RdRp) is maintained by two hydrogen bonds with RdRp at the amino acid residues Thr319 and Thr246; and ᴫ-alkyl interaction with Arg249, ᴫ-alkyl interaction with Pro461; ᴫ-sigma interaction with Thr246; van der Waal interaction with Leu460, Thr391, Leu251, Val320, Thr252, Tyr265 and Leu247 (Fig. 4).
The known anti-viral drug –remdesivir used as positive control exhibited a binding energy of −7.6 kcal mol−1 (Supplementary Table 1) and it formed three hydrogen bond with Arg249, Thr394 and Phe396, three ᴫ-alkyl interaction with Pro169, Leu172, Leu460, Thr458, Pro461 and Arg249; van der Waals interaction with Arg457, Cys395, Arg349, Pro672, Pro323, Val675, Thr246, and Thr319 residues, and unfavourable interaction Arg249 (Supplementary Fig. 2). Other, four bioactive compounds shows the binding affinity between the range −7.6 and −6.6 kcal mol−1 interacted more strongly with the crucial amino acids residues of RdRp of SARS-CoV-2 (Fig. 4).
3.7. In silico prediction of drug-likeness properties and bioactivity score
Data of admetSAR software displayed that the molecular weight of selected active phytochemicals were under 500; LogP value was found to be under 5; the hydrogen bond acceptors (HBA) numbers were under 10 (except for AG); and the hydrogen bonds donors (HBD) were under 5 (except for AG), which indeed goes in line with the Lipinski's rule of 5. Since the screened compounds were plant derived in nature, an allowance of ±1 to both HBD and HBA would eventually be given to follow the Lipinski's rule of 5. The relative ADMET profiles of the selected bioactives are illustrated in Table 3 .
Table 3.
In silico prediction of drug-likeness properties of top hit phyto-ligands.
| Phyto-ligands | MW | HBD | HBA | Log p[<5] | nRO | nViol |
|---|---|---|---|---|---|---|
| Apigenin-7-O-glucuronide | 446.36 | 6 | 11 | 0.55 | 4 | 2 |
| Ellagic acid | 302.19 | 4 | 8 | 0.94 | 0 | 0 |
| Vasicolinone | 305.38 | 0 | 4 | 2.91 | 2 | 0 |
| Anisotine | 349.39 | 1 | 6 | 2.99 | 4 | 0 |
| Endesmol | 222.37 | 1 | 1 | 4.01 | 1 | 0 |
| Viridiflorene | 204.36 | 0 | 0 | 4.19 | 0 | 0 |
Note: MW-molecular weight, HBD-hydrogen bond donor, HBA-hydrogen bond acceptor, Log p, TPSA-Total polar surface area, nRO-Number of rotatable bond, nViol-Number of violation.
3.8. ADMET prediction analysis using admetSAR software
Furthermore, the data also unveiled the actual Absorption, Distribution, Metabolism, Excretion and Toxicity properties of the selected bioactive ligands. The aqueous solubility (Log S) of the ligand is concerned, all the six selected hit bioactives shown the Log S values between the ideal ranges i.e, 6.5 to 0.5. From the obtained results (Table 4 ), it was also obvious that the phyto-ligands were non-toxic, non-carcinogenic, absorb in the human intestine adsorption (HIA), easily permeable through CaCO2 (except the bioactive AG), mostly non-substrate and non-inhibitor to CYP enzymes, weak inhibitor for Human Ether-a-go-go- related gene (Table 4), which eventually signifies that the bioactive hits could plausibly be an efficient drug.
Table 4.
Predicted ADMET properties of top hit bioactive compounds.
| ADMET Properties |
Apigenin-7-O-glucuronide |
Ellagic acid |
Vasicolinone |
Anisotine |
Eudesmol |
Viridiflorene |
|---|---|---|---|---|---|---|
| Absorption | ||||||
| Blood brain barrier | BBB- | BBB+ | BBB+ | BBB+ | BBB+ | BBB+ |
| Human intestinal absorption | HIA+ | HIA+ | HIA+ | HIA+ | HIA+ | HIA+ |
| CaCO2 permeability | CaCO2 | CaCO2- | CaCO2+ | CaCO2- | CaCO2+ | CaCO2+ |
| Renal organic transporter | Non-inhibitor | Non-inhibitor | Inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Aqueous solubility[LogS] | −3.3684 | −3.1440 | −2.3888 | −3.4388 | −3.6183 | −5.0581 |
| Distribution | ||||||
| Subcellular localization | Mitochondria | Mitochondria | Mitochondria | Mitochondria | Lysosome | Lysosome |
| Metabolism | ||||||
| CYP450 2C9 Substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 2D6 Substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate | Non-substrate |
| CYP450 3A4 Substrate | Non-inhibitor | Non-substrate | Substrate | substrate | substrate | substrate |
| CYP450 1A2 Substrate | Non-inhibitor | Non-inhibitor | Inhibitor | Inhibitor | Non-inhibitor | Non-inhibitor |
| CYP450 2D6 Substrate | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor | Non-inhibitor |
| Toxicity | ||||||
| Human Ether-a-go-go- Related gene inhibition | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| Carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens | Non-carcinogens |
| Biodegradation | Not ready biodegradable | Not ready biodegradable | Not ready biodegradable | Not ready biodegradable | Not ready biodegradable | Not ready biodegradable |
| Acute Oral Toxicity | III | II | III | III | III | III |
| Rat Acute Toxicity [LD50][mol/kg] | 2.5393 | 0.5260 | 2.7481 | 2.6090 | 1.8911 | 1.5115 |
| Fish toxicity [pLC50,mg.L] | 0.6805 | 0.3860 | 1.3237 | 1.1395 | −0.3218 | 0.6849 |
3.9. Molecular dynamic simulation
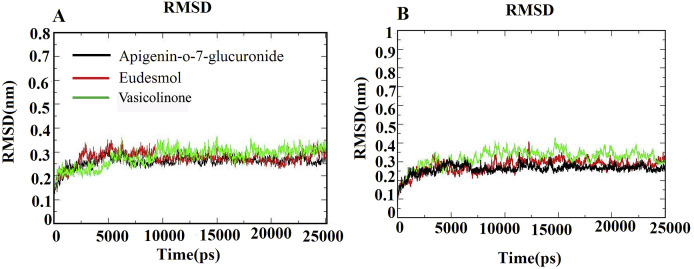
Based on the molecular docking studies, the best three phytoligands (viz., Apigenn-o-7-glucuronide, Eudesmol and Vasicolinone) from each medicinal plant showing significant interaction with most crucial amino acid residues of SARS-CoV Mpro and ACE-2 with high binding energy were selected for molecular dynamics simulation studies at 25ns using Gromacs. The stability of these docked complexes was examined using the RMSD plot, RMSF and Hydrogen bond interaction profiles. The RMSD graph (Fig. 5 A&B) showed the stable protein backbone atom of docked complexes of both Mpro and ACE-2 proteins respectively. The docked complexes of both Mpro and ACE-2 proteins displayed an average backbone RMSD of 0.3 nm. The phytochemicals Apigenn-o-7-glucuronide and Eudesmol maintained an average RMSD of 0.25 nm; however, the Eudesmol showed slight deviation up to 04 nm at 15ns with ACE-2 protein. As far as Vasicolinone is concerned, it displayed deviation in both the complexes. The Mpro -Vasicolinone complex showed deviation up to 0.3 nm at 10, 15 and 18ns. On the other hand, ACE-2- Vasicolinone complex depicted deviation up to 0.4 nm at 9 and 14ns. Such minor deviations in RMSD may be attributed to the unstructured loop region, which makes a protein structure more dynamic. Both the Mpro and ACE-2 docked complexes were quite stabilized and equilibrated with less deviation. Through the backbone RMSD plot, it was obvious that all the docked complexes of Mpro and ACE-2 remain stable throughout the simulation period.
Fig. 5.
The RMSD trajectories of all docked complexes during 25 ns simulations. (A) Mpro -Phytochemicals complexes. (B) ACE-2-Phytochemicals complexes.
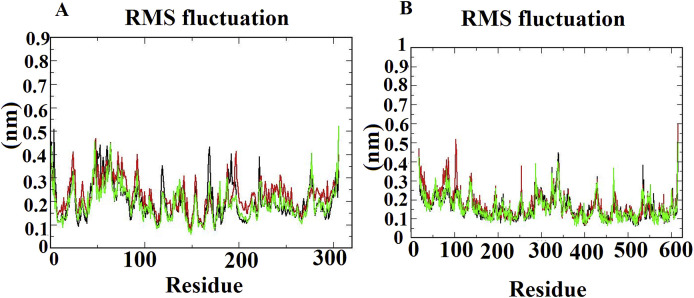
The RMSF plots demonstrated the flexibility and mobility of amino acid residues of docked complexes. RMSF plots showed higher fluctuation in some amino acid residues in the loop or disorder regions. The average RMSF is 0.3 nm and the high fluctuations did not influence any conformational change between phytoligands and proteins (Fig. 6 A&B).
Fig. 6.
The RMSF trajectories of all docked complexes during 25 ns simulations. (A) Mpro-Phytochemicals complexes. (B) ACE-2-Phytochemicals complexes.
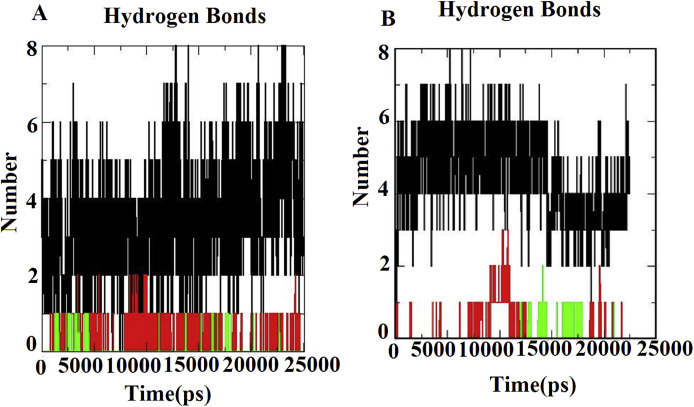
Fig. 7A&B, number of hydrogen bonds of the complexes of Mpro and ACE2 proteins. The hydrogen bonds are most important for the stability of docked complexes. It was found through H-bond analysis that the phtochemicals Eudesmol and Vasicolinone exhibit 3H-bonds and Apigenn-o-7-glucuronide forms 8H-bonds with the Mpro. Then the ACE-2 complexes revealed that the Apigenn-o-7-glucuronide exhibits 8H-bonds, Eudesmol forms 3H_bonds and Vasicolinone built 2H_bonds in the simulation period. With the overall observations of MDS, it was conferred that the docked complexes (top hit three phytochemicals and two proteins) have better structural stability (Fig. 7 A&B).
Fig. 7.
The hydrogen bond trajectories of all docked complexes during 25 ns simulations. (A) Mpro -Phytochemicals complexes. (B) ACE-2-Phytochemicals complexes.
4. Discussion
For centuries, the traditional Indian knowledge system has well demonstrated the curing effect of herbal steam inhalation therapy against common cold. Accordingly, the AYUSH, Govt. of India has listed different herbs used in the traditional in-house inhalation therapy with proven medicinal benefits that strengthened the respiratory tracks and immune system (Vellingiri et al., 2020). Most notably, an estimate by WHO signifies that around 80% of population in underdeveloped countries has been depending mostly on the traditional medicines. Together, the WHO has also enlisted nearly 21,000 therapeutically potential global plants; out of which India alone encompass around 2500 varieties (Seth and Sharma 2004).
Indian traditional system relies on proper food and exercises to maintain good health. Therefore, people believe in supplementing the medicinal herbs to get rid from severe health issues. Thus patients suffering from respiration related issues were suggested for herbal steam inhalation therapy to mitigate the symptoms (Amini et al., 2017; Singh et al., 2017). In corroboration with the present study, earlier reports by (Amini et al., 2017) had given prominence to the complementary herbal medicines viz. inhaling herbal steam and (Singh et al., 2017) signified the speedy recovery of common viral cold patients while undergoing neti treatment in supplement with antibiotics, vitamins and minerals (Amini et al., 2017; Singh et al., 2017). It is therefore likely to anticipate that these phytochemicals may put forth an inhibitory efficacy against SARS-CoV-2 either exhibiting antiviral activity or improving the immune status of an individual.
Bearing the fact that SARS-CoV-2 enters the host through nasal cavity and infect the buccal cavity, in the present study, we deliberately made an attempt to virtually authenticate the antiviral efficacy of three medicinally important herbs' (viz., E. globules, J. adhatoda and V. negundo) associated compounds against the well recognized drug targets of SARS-CoV-2 (Mpro, ACE-2, S-protein and RdRp) through in silico approach. The study was carried out with the expectation that the ‘herbal steam inhalation’ therapy involving the aforementioned herbs could combat COVID-19 infection through treating the patients' airway -the hallmark infection path of SARS-CoV-2.
Preliminarily, bioactives having potency to target the crucial protein receptors of SARS-CoV-2 were virtually screened from 57 phytochemicals. Based on the binding energy exhibited by the phytochemicals, 6 potent bioactive hits were scrutinized. Molecular interaction studies of receptor-ligand complex divulged the prevalence of hydrogen bond, non-covalent interaction (viz. van der Waals), electrostatic and non-polar interactions (Chen and Kurgan, 2009). These molecular bonding/interactions exerted by the ligand are indeed the inevitable element to be considered for biological functions and successful drug developments.
Mpro is a most commonly recognized drug target of SARS-CoV-2 belonging to Nsp family protein essential for polyprotein processing in viral life cycle (Zhang et al., 2020). A plethora of recent reports have emphasized that the inhibition of viral proteases would be a promising approach in treating the viral disease progression (Zhang et al., 2020). Similarly, SARS-CoV-2 RdRp also plays a pivotal role in viral transcription, and thus, targeting this protein would plausibly arrest the viral machinery and inactivate the virus. Alike hampering the viral replication through targeting Mpro and RdRp, blocking the host receptor ACE-2 could ultimately cause structural change, which in turn prevents the entry of virus at the initial step. Henceforth, the compounds having efficacy to bind the active sites of these crucial drug targets are reported to inhibit transcription which ultimately shuts the viral replication (Alagu Lakshmi et al., 2020; Joshi et al., 2020; Gyebi et al., 2020).
In the present study, out of 6 best high scoring hits, AG topped the list. AG is a phenolic compound found in the leaves of E. globulus. Traditionally, the E. globulus leaves are commonly used to treat asthma and bronchitis. Importantly, these leaves are employed to make herbal tea. A mounting body of recent studies on leaves extract and essential oils of E. globulus have well documented the multifaceted therapeutic efficacies such as antimicrobial, antifungal, anthelmintic and anti-diabetic (Rizeq et al., 2020; Baradaran Rahimi et al., 2020). The other bioactive compound from the same plant was ellagic acid, a naturally occurring polyphenol also predominantly found in certain fruits and nuts apart from the leaves of E. globulus. Ellagic acid being a profound antioxidant, it has been widely investigated for its anti-proliferative efficacy in few cancers, and anti-inflammatory property (Rizeq et al., 2020; Baradaran Rahimi et al., 2020). In parallel, inhaling the Eucalyptus and tulsi steam by the patients undergoing mechanical ventilation have cleared the pathogen in their airway, reduced the respiratory ailment and improved their recovery (Singh et al., 2017; Kamble et al., 2017).
Next to AG, vasicolinone – a quinazoline alkaloid from the herb from J. adhatoda was found to exhibit profound interaction with all the screened protein targets. J. adhatoda also referred as Vasaka has been used traditionally in Indian systems of medicine for nearly 2000 years. In the indigenous systems of medicine, it has been well demonstrated for beneficial effects in treating cold, cough, whooping cough, asthma and especially chronic bronchitis (Claeson et al., 2000). Although, the bronchodilatory and expectorant propensities of this plant leaves are endorsed to vasicine (Chatterjee et al., 1999), in the present study, vasicolinone is envisaged to showcase a phenomenal binding affinity towards all the significant receptors of SARS-CoV-2. Anisotine was the other quinazoline alkaloid from J. adhatoda that displayed best score in the present study. This signifies the fact that apart from vaccine and vasicinone, vasicolinone and anisotine -the under explored bioactives could plausibly play inevitable role in bronchodilatory effect of J. adhatoda leaves (Jha et al., 2012).
In the row, eudesmol is a carbobicyclic compound present in the leaves of V. negundo, found to the next high scorer targeting all the four protein targets profusely. V. negundo is a shrub commonly referred as ‘nirgundi’ in Indian ayurveda, familiar for its anti-inflammatory and hepatoprotective properties. Besides, the myriad pharmaceutical potentials of V. negundo include antioxidant, analgesic, anti-inflammatory and anti-convulsan (Singh et al., 2017) efficacies. Viridiflorene is the second bioactive compound from V. negundo, also found in many essential oils as one of the constituents. It is noteworthy to state that a report by Reichling et al. (2005), demonstrated the virucidal activity of Manuka oil consisting 4.4% of viridiflorene against HSV-1 and HSV-2 (Reichling et al., 2005). As like V. negundo, few medicinal plants belonging to the family of Leguminosae and Lamiaceae found in Himalayas have been reported to be effective against bronchitis, severe respiratory diseases that chiefly affect lungs (Amber et al., 2017). Since, remedies are prevailing in natural ways, patients those who are encountering severe side-effects by the synthetic drugs have followed the natural medicine. Furthermore, the in silico drug-likeness and ADMET prediction analyses validated that the identified bioactives are fit to the criteria of drug in comparison with known antiviral drug.
Most importantly, China treats several infectious diseases by the traditional Chinese medicines (Wang and Liu, 2014). Previously in 2003 and 2009 they have used Chinese traditional medicines to treat SARS and H1N1, respectively (Liu et al., 2004; Li et al., 2005, Li et al., 2005). Based on the experience gained in treating different viruses, they have again followed the same traditional medicine method to control and treat the COVID-19, a recent outbreak of SARS-CoV-2 and it is clinically effective. In addition, intervention of the Chinese traditional medicine during the treatment of COVID-19 has been reported to reduce the severe symptoms of patients (Zhang et al., 2020; Xu and Zhang, 2020).
Overall, the data of the present study delineated the anti-COVID-19 proficiency of phytochemicals (apigenin-o-7-glucuronide, ellagic acid, eudesmol, viridiflorene, vasicolinone and anisotine), and in turn validated the traditional medicinal significance of herbal steam inhalation therapy. Therefore, it would be pertinent to strongly emphasis the recommendation of herbal steam inhalation therapy using plants specially Eucalyptus globulus, Vitex negundo and Justicia adhatoda with myriad traditional respiratory healing potency to either arrest the viral replication or to immunize the host system against viral infection, particularly SARS-CoV-2. The overall theme of the study is portrayed as a schematic representation in Fig. 8 .
Fig. 8.
Overall schematic representation of the theme of present investigation.
CRediT authorship contribution statement
Shanmugaraj Gowrishankar: Conceived and, Conceptualization, the research idea, Writing - original draft, Data curation, Investigation, and Result interpretation. Sankar Muthumanickam: Virtual-screening, Formal analysis, In-silico result interpretation. Arumugam Kamaladevi: Writing - original draft, Critical reviewing, Writing - original draft. Chandrasekar Karthika: In-silico result interpretation and, Writing - original draft. Ravi Jothi: In-silico result interpretation and, Writing - original draft. Pandi Boomi: Formal analysis, In-silico result interpretation. Dharuman Maniazhagu: Writing - original draft. Shunmugiah Karutha Pandian: Writing - original draft.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors sincerely acknowledge UGC-SAP [Grant No. F.5-1/2018/DRS-II (SAP-II)], DST-FIST [Grant No. SR/FST/LSI-639/2015 (C)] and DST PURSE [Grant No. SR/PURSE Phase 2/38 (G)] for rendering instrumentation & infrastructure facilities. SG gratefully acknowledges UGC for Start-Up Grant (Grant No. F.30–381/2017(BSR)/F.D Diary No. 2892), Alagappa University for AURF (Ref: ALU: AURF Start-up Grant: 2018)) and RUSA 2.0 [F.24–51/2014-U, Policy (TN Multi-Gen), Department of Education, Government of India].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fct.2020.111966.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
FigS1.
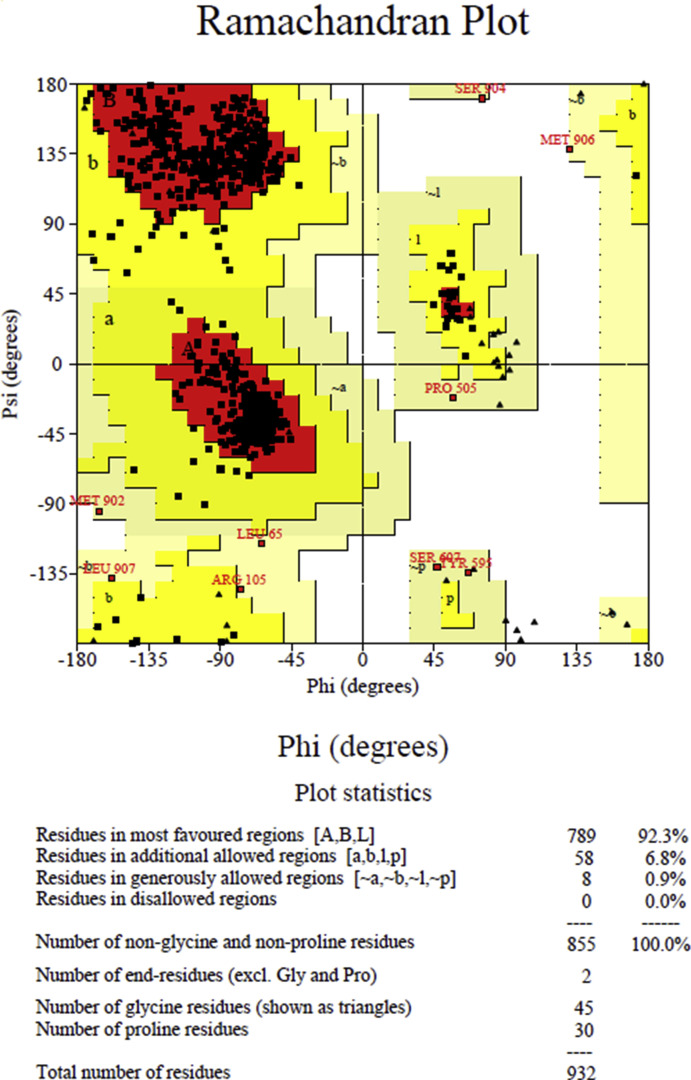
Showing the modeled structure of SARS-CoV-2 RdRp.
FigS2.
Showing the Ramachandran plot of modeled SARS-CoV-2 RdRp.
FigS3.
Binding interaction map of known SARS-CoV-2 protease inhibitor -lopinavir with SARS-CoV-2 Mpro. Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours.
FigS4.
Binding interaction map of known SARS-CoV-2 polymerase inhibitor -remdesivir with RdRp. Active site residues in binding pockets are represented in three letter amino acid code and different types of interactions are denoted in different colours.
References
- Alagu Lakshmi S., Shafreen R.M.B., Priya A., Shunmugiah K.P. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J. Biomol. Struct. Dyn. 2020;2020:1–16. doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexpandi R., De Mesquita J.F., Pandian S.K., Ravi A.V. Quinolines-based SARS-CoV-2 3CLpro and RdRp inhibitors and Spike-RBD-ACE2 inhibitor for drug-repurposing against COVID-19: an in silico analysis. Front. Microbiol. 2020;11:1796. doi: 10.3389/fmicb.2020.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amber R., Adnan M., Tariq A., Mussarat S. A review on antiviral activity of the Himalayan medicinal plants traditionally used to treat bronchitis and related symptoms. J. Pharm. Pharmacol. 2017;69(2):109–122. doi: 10.1111/jphp.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini H., Hosseini V., Schindler C., Hassankhany H., Yunesian M., Henderson S.B., Künzli N. Spatiotemporal description of BTEX volatile organic compounds in a Middle eastern megacity: tehran study of exposure prediction for environmental health research (tehran SEPEHR) Environ. Pollut. 2017;226:219–229. doi: 10.1016/j.envpol.2017.04.027. [DOI] [PubMed] [Google Scholar]
- Baradaran Rahimi V., Ghadiri M., Ramezani M., Askari V.R. Antiinflammatory and anti-cancer activities of pomegranate and its constituent, ellagic acid: evidence from cellular, animal, and clinical studies. Phytother Res. 2020;34(4) doi: 10.1002/ptr.6565. [DOI] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N. Engl. J. Med. 2020;382(19):1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S. Bronchodilatory and anti-allergic effect of PulmoFlex--a proprietary herbal formulation. Indian J. Physiol. Pharmacol. 1999;43(4):486–490. [PubMed] [Google Scholar]
- Chen K., Kurgan L. Investigation of atomic level patterns in protein--small ligand interactions. PloS One. 2009;4(2):e4473. doi: 10.1371/journal.pone.0004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012;52(11):3099–3105. doi: 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- Claeson U.P., Malmfors T., Wikman G., Bruhn J.G. Adhatoda vasica: a critical review of ethnopharmacological and toxicological data. J. Ethnopharmacol. 2000;72(1–2):1–20. doi: 10.1016/s0378-8741(00)00225-7. [DOI] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganjhu R.K., Mudgal P.P., Maity H., Dowarha D., Devadiga S., Nag S., et al. Herbal plants and plant preparations as remedial approach for viral diseases. Virus disease. 2015;26(4):225–236. doi: 10.1007/s13337-015-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos E., Liaudanskas M., Viskelis J., Zvikas V., Janulis V., Gomez-Serranillos M.P. Antioxidant activity, neuroprotective properties and bioactive constituents analysis of varying polarity extracts from Eucalyptus globulus leaves. J. Food Drug Anal. 2018;26(4):1293–1302. doi: 10.1016/j.jfda.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): an in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder S.T., Dhorajiwala T.M., Samant L.R. Multiple docking analysis and in silico absorption, distribution, metabolism, excretion, and toxicity screening of anti-leprosy phytochemicals and dapsone against dihydropteroate synthase of Mycobacterium leprae. Int. J. Mycobacteriol. 2019;8(3):229–236. doi: 10.4103/ijmy.ijmy_123_19. [DOI] [PubMed] [Google Scholar]
- Han Y., Zhang J., Hu C.Q., Zhang X., Ma B., Zhang P. In silico ADME and toxicity prediction of ceftazidime and its impurities. Front. Pharmacol. 2019;10:434. doi: 10.3389/fphar.2019.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha D.K., Panda L., Lavanya P., Ramaiah S., Anbarasu A. Detection and confirmation of alkaloids in leaves of Justicia adhatoda and bioinformatics approach to elicit its anti-tuberculosis activity. Appl. Biochem. Biotechnol. 2012;168(5):980–990. doi: 10.1007/s12010-012-9834-1. [DOI] [PubMed] [Google Scholar]
- Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., et al. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamble M., Londhe S., Rapelli P., Thakur P., Ray S.A. A comparative study to assess the effect of steam inhalation v/s Tulsi leaves inhalation on the sign and symptoms of cold and cough among adult group in selected areas of Pune city. Int. J. Med. Res . 2017;2(2):24–26. [Google Scholar]
- Killerby M.E., Biggs H.M., Haynes A., Haynes A., Dahl R.M., Mustaquim D., et al. Human coronavirus circulation in the United States 2014-2017. J. Clin. Virol. 2018;101:52–56. doi: 10.1016/j.jcv.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral. Res. Jul. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J. Tradit. Complement. Med. 2014;4(1):24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Manheimer E., Shi Y., Gluud C. Chinese herbal medicine for severe acute respiratory syndrome: a systematic review and meta-analysis. J. Alternative Compl. Med. 2004;10(6):1041–1051. doi: 10.1089/acm.2004.10.1041. [DOI] [PubMed] [Google Scholar]
- Liu Y., Li J., Feng Y. Critical care response to a hospital outbreak of the 2019-nCoV infection in Shenzhen, China. Crit. Care. 2020;24(1):56. doi: 10.1186/s13054-020-2786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A., Ismatullah H., Rauf H., Niazi U. Identification of bioflavonoid as fusion inhibitor of dengue virus using molecular docking approach. Informatics in Medicine Unlocked. 2016;3:1–6. [Google Scholar]
- Muthuramalingam P., Jeyasri R., Valliammai A., Selvaraj A., Karthika C., Gowrishankar S., Pandian S.K., Ramesh M., Chen J.T. Global multi-omics and systems pharmacological strategy unravel the multi-targeted therapeutic potential of natural bioactive molecules against COVID-19: an in silico approach. Genomics. 2020;112(6):4486–4504. doi: 10.1016/j.ygeno.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramashivam S.K., Elayaperumal K., bhagavan Natarajan B., devi Ramamoorthy M., Balasubramanian S., Dhiraviam K.N. In silico pharmacokinetic and molecular docking studies of small molecules derived from Indigofera aspalathoides Vahl targeting receptor tyrosine kinases. Bioinformation. 2015;11(2):73–84. doi: 10.6026/97320630011073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichling J., Koch C., Stahl-Biskup E., Sojka C., Schnitzler P. Virucidal activity of a beta-triketone-rich essential oil of Leptospermum scoparium (manuka oil) against HSV-1 and HSV-2 in cell culture. Planta Med. 2005;71(12):1123–1127. doi: 10.1055/s-2005-873175. [DOI] [PubMed] [Google Scholar]
- Rizeq B., Gupta I., Ilesanmi J., AlSafran M., Rahman M.M., Ouhtit A. The power of phytochemicals combination in cancer chemoprevention. J. Canc. 2020;11(15):4521–4533. doi: 10.7150/jca.34374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3(1):e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepay N., Sekar A., Halder U.C., Alarifi A., Afzal M. Anti-COVID-19 terpenoid from marine sources: a docking, admet and molecular dynamics study. J. Mol. Struct. 2020:129433. doi: 10.1016/j.molstruc.2020.129433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth S.D., Sharma B. Medicinal plants in India. Indian J. Med. Res. 2004;120(1):9–11. [PubMed] [Google Scholar]
- Singh M., Singh M., Jaiswal N., Chauhan A. Heated, humidified air for the common cold. Cochrane Database Syst. Rev. 2017;29(8) doi: 10.1002/14651858.CD001728.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Dayal R., Bartley J.P. Volatile constituents of Vitex negundo leaves. Planta Med. 1999;65(6):580–582. doi: 10.1055/s-2006-960832. [DOI] [PubMed] [Google Scholar]
- Thuy B.T.P., My T.T.A., Hai N.T.T., Hieu L.T., Hoa T.T., Thi Phuong Loan H., et al. Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega. 2020;5(14):8312–8320. doi: 10.1021/acsomega.0c00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikellis C., Thomas M.C. Angiotensin-converting enzyme 2 (ACE2) is a key modulator of the renin angiotensin system in health and disease. Int. J. Pept. 2012;2012:256294. doi: 10.1155/2012/256294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020;39:2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., Govindasamy V., Giridharan B., et al. COVID-19: a promising cure for the global panic. Sci. Total Environ. 2020;725:138277. doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu Z. Prevention and treatment of viral respiratory infections by traditional Chinese herbs. Chin. Med. J. (Engl) 2014;127(7):1344–1350. [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;6(10236):1569–1578. doi: 10.1016/S0140-6736(20)31022-9. 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang Y. Traditional Chinese medicine treatment of COVID-19. Complement. Ther. Clin. Pract. 2020;39:101165. doi: 10.1016/j.ctcp.2020.101165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Pandey S.K., Singh V.K., Goel Y., Kumar A., Singh S.M. Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: implication in designing of novel anticancer therapeutic strategies. PloS One. 2017;12(5) doi: 10.1371/journal.pone.0176403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef B.A., Dirar A.I., Elbadawi M.A.A., Awadalla M.K., Mohamed M.A. Potential deoxycytidine kinase inhibitory activity of amaryllidaceae alkaloids: an in silico approach. J. Pharm. BioAllied Sci. 2018;10(3):137–143. doi: 10.4103/jpbs.JPBS_44_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.