Abstract
Abstract
Streptomyces is one of the most versatile genera for biotechnological applications, widely employed as platform in the production of drugs. Although streptomycetes have a complex life cycle and metabolism that would need multidisciplinary approaches, review papers have generally reported only studies on single aspects like the isolation of new strains and metabolites, morphology investigations, and genetic or metabolic studies. Besides, even if streptomycetes are extensively used in industry, very few review papers have focused their attention on the technical aspects of biotechnological processes of drug production and bioconversion and on the key parameters that have to be set up. This mini-review extensively illustrates the most innovative developments and progresses in biotechnological production and bioconversion processes of antibiotics, immunosuppressant, anticancer, steroidal drugs, and anthelmintic agents by streptomycetes, focusing on the process development aspects, describing the different approaches and technologies used in order to improve the production yields. The influence of nutrients and oxygen on streptomycetes metabolism, new fed-batch fermentation strategies, innovative precursor supplementation approaches, and specific bioreactor design as well as biotechnological strategies coupled with metabolic engineering and genetic tools for strain improvement is described. The use of whole, free, and immobilized cells on unusual supports was also reported for bioconversion processes of drugs. The most outstanding thirty investigations published in the last 8 years are here reported while future trends and perspectives of biotechnological research in the field have been illustrated.
Key points
• Updated Streptomyces biotechnological processes for drug production are reported.
• Innovative approaches for Streptomyces-based biotransformation of drugs are reviewed.
• News about fermentation and genome systems to enhance secondary metabolite production.
Keywords: Antibiotic, Bioconversion, Biotechnological process, Secondary metabolites, Steroid, Streptomyces
Introduction
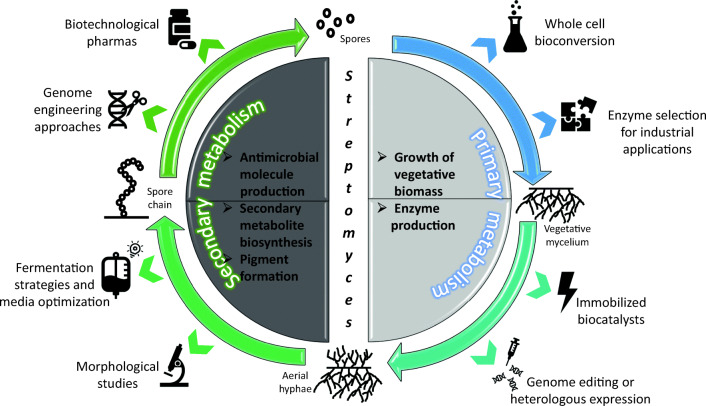
Microorganisms of the Streptomyces genus are Gram-positive and aerobic bacteria that are able to grow in various environments and to colonize a wide range of terrestrial and aquatic niches; they are saprophytes of the soil and, sometimes, plant and animal pathogens (Anderson and Wellington 2001; de Lima Procópio et al. 2012). From a morphological and microbiological point of view, these bacteria are comparable to filamentous fungi and this feature is presumably the result of adaptations to similar ecological niches (de Lima Procópio et al. 2012). During the vegetative phase, they grow by the branching of the hyphal apices and by forming a complex compactly matrix constituting the vegetative mycelium. In response to nutrient depletion or stress environmental signals, both the production of secondary metabolites and morphological differentiation initiate (de Lima Procópio et al. 2012; Chater 2016). With the age of the culture, aerial hyphae are produced that differentiate first into chains of pre-spore compartments, and then, they develop to mature spores that are released in the surrounding environment (de Lima Procópio et al. 2012; Chater 2016). When a typical Streptomyces spore encounters favorable conditions and nutrients, it germinates and grows up to form new hyphae of a vegetative mycelium, and thus, the growth process starts again. The taxonomy and the morphological, physiological, and genetic characteristics of the Streptomyces genus as well as its metabolic pathways and its enzymatic features have already been extensively described in previously reported reviews (Anderson and Wellington 2001; Chater 2016). From a biotechnological point of view, the employment of the Streptomyces strains in the diverse processes could vary according to the different growth phases in relation to the aimed applications or to the molecule types that have to be produced. During the initial vegetative growth, the streptomycetes whole cells produce vitamins and enzymes that could be interesting for commercial purposes, such as amylases. They could also perform regio-selective and stereo-specific enzymatic bioconversion reactions on externally supplied substrates that are interesting as drugs, such as steroids. The biomass in the full vegetative phase is also the most suitable for free resting and immobilized cell applications. When the streptomycetes are in the secondary metabolism phase, they produce diverse important bioactive compounds of high commercial value, such as antibiotics, anthelmintic agents, and interesting pigments (Fig. 1). In this mini-review, we have aimed to focus the attention on the processes of production and of bioconversion of drugs that could be performed by using the streptomycetes, by giving short historical outlines, by widely describing the current state of the most innovative biotechnological applications, and by discussing possible future trends and perspectives in the field.
Fig. 1.
Different biotechnological applications of streptomycetes based on their life cycle. Streptomyces life cycle includes spore chains and spores that generally differentiate on solid media while the vegetative mycelium and aerial hyphae forms are obtained after growth in liquid cultures. According to the different life cycle phases and metabolism stages, the streptomycetes could be employed in diverse biotechnological applications
Biotechnological biotransformation processes for drug production by using streptomycetes
Streptomycetes have an extraordinary capacity to bioconvert different substrates, thanks to a myriad of secreted enzymes normally involved in its complex morphological development as well as to cytochrome P450 multi-enzymatic systems (Endo et al. 2002; Tamburini et al. 2004; Roh et al. 2009; Spasic et al. 2018; Anteneh and Franco 2019). Streptomyces enzymes could perform reactions of esterification, oxidation, hydroxylation, hydrolysis, dehydrogenation, and halogenations on different substrates with great flexibility and are widely employed in industrial processes to transform numerous compounds with high regio- and stereo-selectivity as they work under milder and ecologically harmless conditions than the chemical synthetic processes (Pervaiz et al. 2013). To establish a robust and easily scalable industrial bioconversion process, conditions like pH, temperature, medium, co-factor presence, and substrate concentrations have to be investigated, especially if they are related to the complex life cycle of streptomycetes; their optimization could lead to high percentages of bioconversion with a minimum by-product formation, in a very short time, thus making the bioconversion processes economically appealing (Schmid et al. 2001; de Carvalho 2016). The first ever recorded bioconversion using Streptomyces dates back to 1952, when progesterone was obtained from pregnenolone oxidation in 24-h fermentation by whole cells of Streptomyces griseus, Streptomyces fradiae, Streptomyces aureofaciens, and Streptomyces rimosus (Perlman 1952). In the next year, the Streptomyces enzymatic pools were taken into account as tools for bioconversion and the activity of amylases from five different strains was investigated (Simpson and McCoy 1953). After these first papers, numerous studies used streptomycetes whole cells or their extracted enzymes for structural tailored-cut bioconversions of antibiotics, steroids, or other bio-active molecules to obtain new drugs with improved pharmacological characteristics (Argoudelis and Coats 1969, 1971; Walker and Skorvaga 1973; Dickens and Strohl 1996; Berrie et al. 1999; Demain and Báez-Vásquez 2000; Miranzo et al. 2010). If dated studies were more concentrated on the characterization and the discovery of the streptomycetes enzymes and/or on the different types of reactions and substrates that could be modified, in more recent years, enzyme engineering or the heterologous expression of proteins has allowed deeper operation on bioconversion processes (Chin et al. 2001; Brautaset et al. 2002; Kim et al. 2006; Hussain and Ward 2003; Molnár et al. 2006; Hayashi et al. 2008). Here, in this review, the sixteen most outstanding recent papers describing biotechnological applications of streptomycetes for bioconversion production processes of pharmaceuticals are reported: the use of both free or immobilized whole cells or of immobilized, mutated, genetically modified, or heterologous expressed enzymes is widely described (Table 1).
Table 1.
Summary of different bioconversion approaches for drug production by Streptomyces
| Streptomyces strain(s) | Free or immobilized whole cell bioconversion | Substrate(s) → product(s) | Reaction → enzyme(s) | Bioconversion conditions | Bioconversion rate/yield/productivity | References |
| Streptomyces roseochromogenes ATCC 13400 | Whole cell biocatalysis in batch and pulsed-batch fermentation | Hydrocortisone → 16α-hydroxyhydrocortisone | Hydroxylation → 16α-hydroxylase; roseoredoxin and roseoredoxin reductase | Optimization of medium composition and of the timing of substrate addition, pH and temperature in shake flask experiments; scale-up in batch and pulsed-batch processes | 84.0% | Restaino et al. 2014, 2016 |
| Streptomyces roseochromogenes ATCC 13400 plus Arthrobacter simplex ATTC31652 | Coupled whole cell biocatalysis in shake flasks | Hydrocortisone → 16α-hydroxyprednisolone | Hydroxylation → 16α-hydroxylase; roseoredoxin and roseoredoxin reductase; 1,2 dehydrogenation →Δ1-dehydrogenase | Optimization of pH and temperature of bioconversion in shake flask experiments. Bioconversion obtained at pH 6.0 and 26 °C in 120 h by coupling the two strains. | 68.8% | Restaino et al. 2020 |
| Streptomyces thermotolerans ATCC11416 | Whole cell immobilization on polyurethane foam cubes | Tylosin → acetylisovaleryltylosin | Acetylation and substitution with isovaleric acid → various enzymes | Immobilized cell activity was 1.65 times higher than free cell activity; repeated batches up to 6 cycles | 86.3% | Zhu et al. 2015 |
| Streptomyces strain(s) | Immobilized enzyme-based biocatalysis | Substrate(s) → product(s) | Reaction → enzyme(s) | Bioconversion conditions | Bioconversion rate/yield/productivity | References |
| Streptomyces sp. strain W007 | Immobilization on macroporous resins | Glycerol + long-chain n-3 polyunsaturated fatty acids → triacylglycerols | Esterification → thermostable lipase (MAS1) | Immobilized lipase was almost 7 times more stable to heat than its free form; it could be reused for up to 5 cycles | 97.5% in 24 h | Wang et al. 2017 |
| Streptomyces exfoliatus | Cross-linked enzyme aggregates (CLEAs) | Poly-(R)-3-hydroxybutyrate → chiral (R)-3-hydroxybutyric acid | Hydrolysis → poly-(R)-3-hydroxybutyrate depolymerase | Immobilized depolymerase was almost 5 times more stable to heat than its free form; it could be reused for up to 20 cycles | Not reported | Hormigo et al. 2012 |
| Streptomyces sp. | All-enzymes hydrogel | Imines → optically active amines | Reduction → NADPH-dependent-(S)-selective dimeric imine reductase | Glucose-1-dehydrogenase from Bacillus subtilis was present in the hydrogel for NADH regeneration; reaction time was 40 h; flux was 100 μL·min-1 in microfluidic bioreactor | 150 g· L-1 ·day-1 | Bitterwolf et al. 2019 |
| Streptomyces mobaraensis | Immobilization on glass microbeads | Antibodies fragments or whole antibodies → conjugated antibodies | Conjugation → microbial transglutaminase | Conjugation substrate includes fluorescent probes or metal chelator for radio-labeling | Not reported | Spycher et al. 2017 |
| Streptomyces sp. | Immobilization on silane modified iron (II, III) oxide magnetic nanoparticles (MNP) | Cholesterol → 4-cholesten-3-one | Oxidation → cholesterol oxidase | Immobilized cholesterol oxidase activity was 2 times higher than its free form; it could be reused for up to 10 cycles | Not reported | Ghosh et al. 2018 |
| Streptomyces sp. | Immobilization on silicon dioxide-coated magnetite biosensor | Cholesterol → 4-cholesten-3-one | Oxidation → cholesterol oxidase | Immobilized enzyme was more than 2 times efficient than its free form; enzyme concentration was 20 mg·mL-1, substrate concentration was 6.46 mM; it could be reused for up to 30 days | 90.0% in 180 min | Perdani et al. 2020 |
| Streptomyces strain(s) | Mutated, genetically modified, or heterologous expressed enzymes bioconversion | Substrate(s) → product(s) | Reaction → enzyme(s) | Bioconversion conditions | Bioconversion rate/yield/productivity | References |
| Streptomyces xanthochromogenes RIA 1098 | Multi-step random UV mutagenesis | Compactin → pravastatin | - | Mutagenesis was performed in order to obtain mutants resistant for high compactin concentrations; 50 h of reaction time | 91.0 % | Dzhavakhiya et al. 2015 |
| Streptomyces coelicolor CR1 | Structure-guided engineering and directed evolution | Ethyl-4-chloro-3-oxobutyrate → ethyl-(S)-4-chloro-3-hydroxylbutyrate | Reduction → carbonyl reductase | Mutagenesis was performed to improve both thermal stability and catalytic activity; substrate concentration = 100 g·L-1; bioreactor = 300 mL; reaction time = 9 h | 255 g· L−1 day-1 | Li et al. 2017 |
| Streptomyces halstedii | Heterologous expression in Pichia pastoris and displaying on its membrane surface | Phosphatidylcholine + l-serine → phosphatidylserine | Transphosphatidylation → phospholipase D | Repeated batch conditions up to 4 cycles: 8 h reaction; 200 mL of 0.2 M sodium acetate buffer; 40 °C, pH 5,5; 3 U·mL-1 biocatalyst; 1:3 phosphatidylcholine:l-serine ratio | 53.0% | Liu et al. 2019 |
| Streptomyces sp. NRRL F-4489 | Heterologous expression in E. coli BL21 (DE3) | l-tyrosine → p-coumaric acid | Non-oxidative deamination → tyrosine ammonia lyases | Bioconversion conditions: 50 °C, pH 11; 10 mM glycine-NaOH buffer; 10 OD600 cell concentration; substrate concentration 30 mM; 1 h reaction time | 58.6% | Cui et al. 2020 |
| Streptomyces toxytricini NRRL 15443; Streptomyces sp. MJ654-NF4; Streptomyces chromofuscus ATCC 49982 | Heterologous expression in E. coli BL21 (DE3) | l-glutamic acid → gamma-aminobutyric acid | Decarboxylation → glutamate decarboxylase | Best results obtained by glutamate decarboxylase from Streptomyces sp. MJ654-NF4; bioconversion conditions: water; pH 4, 6; substrate concentration 2 M; 37 °C; 20 OD600 cell concentration; 1 h reaction time; it can be reused for up to 10 cycles | 67.0% | Yuan et al. 2019 |
| Streptomyces sp. MJ654-NF4 | Heterologous expression in Saccharomyces cerevisiae | l-glutamic acid → gamma-aminobutyric acid | Decarboxylation → glutamate decarboxylase | Biocatalyst can be reused for up to 10 cycles; S. cerevisiae–based biocatalysts are GRAS | 60.8% | Yuan et al. 2020 |
| Streptomyces griseus ATCC 13273 | Heterologous expression in E. coli BL21 (DE3) | Papaverine → 6-O-demethyl-papaverine | Demethylation → CYP105D1 | Bioconversion conditions: 25 °C; pH 7, 5; 8 g of biocatalyst; 3% v/v PEG-200; substrate concentration 50 mg·L-1; 24 h reaction time | 61.1% | Shen et al. 2019 |
Bioconversion by using free or immobilized whole cells
Bioconversion by using streptomycetes whole cells as biocatalysts, either in their free or immobilized form, is a widely employed approach for the production of steroids, for antibiotic modification, or for chiral transformation of drugs. As the entire cell enzymatic machine is involved, whole cell–based bioconversions work better than immobilized enzymes because they assure an easy and natural cofactor regeneration and provide prolonged metabolic activity and extensive reusability, and could be employed in multiple reaction catalysis with low costs (el-Naggar et al. 2003; Anteneh and Franco 2019). In free whole cell bioconversion, optimal cell growth and conversion conditions, timing and concentration of substrate addition, and eventually the growth inhibiting substrate concentration are all key parameters to take into account (Dlugoński and Sedlaczek 1981; Atta and Zohri 1995). Furthermore, a wise design of the type of fermentation process (e.g., batch, pulsed batch, or fed-batch) could help to sustain the growth and to boost the conversion yields, allowing to transform substrates having initial concentrations even of g· L−1 scale. For example, in two recent studies, whole cells of Streptomyces roseochromogenes ATCC 13400 were employed in fermentation experiments to specifically perform the 16α-hydroxylation of hydrocortisone, an intermediate in the synthesis of the anti-inflammatory desfluorotriamcinolone (Restaino et al. 2014, 2016) (Table 1). In this case, the whole cells containing the P450 multi-enzymatic complex that includes a 16α-hydroxylase and two electron transport system proteins, roseoredoxin and roseoredoxin reductase, were used as microbial cell factories to establish a productive fermentation process of bioconversion. Following a wise process design, preliminary physiological studies were performed in shake flasks to test the influence of nutrients, the timing of addition, and the concentrations of substrate on both growth and bioconversion ratios. A strict correlation between the timing of the hydrocortisone addition during the microbial growth and the strain bioconversion ability was noted; the earlier the addition was, the higher was the conversion percentage obtained, thus suggesting the idea that the substrate presence ever since the beginning induced the expression of the P450 complex enzymes. The optimization of the growth medium formulation in terms of C/N ratio and type of nitrogen sources, growth temperature (26 °C), and pH (pH = 6) helped to boost the transformation percentage up to 87.0% in 120-h shake flask runs, starting with 0.1 g· L−1 of hydrocortisone, and to minimize the by-product formation under 6.0% (Restaino et al. 2014, 2016). The optimized settings were then used in batch, pulsed-batch, and fed-batch fermentations: in the latter, an 80.4% bioconversion of 1.0 g· L−1 of hydrocortisone was reached in 96 h in 22-L vessel by adding the substrate in two steps during the growth and by employing a DO-stat (dissolved oxygen-stat) feeding strategy in which glucose was added more times, during the fermentation, in small pulses as soon as the increase of the dissolved oxygen percentage indicated a carbon source lack in the medium (Table 1). The same strain was coupled, recently, with Arthrobacter simplex 31652 in whole cell experiments at pH 6.0 and 26 °C to perform also the dehydrogenation reaction and thus directly convert the hydrocortisone into the desired final product, the 16α-hydroxyprednisolone or desfluorotriamcinolone. A maximum of 68.8% of 16α-hydroxyprednisolone was obtained in a 120-h transformation starting with 0.1 g ·L−1 of hydrocortisone (Restaino et al. 2020) (Table 1; Fig. 2). In the case of immobilized whole cell bioconversions, the carrying material plays a primary role as it has to fulfil various requirements including a nontoxic nature, reusability, a high surface area for cell attachment, and a high resistance to the stressful conditions of the bioreactor environment (Zhu et al. 2015). Streptomyces whole cell immobilization has been performed on a wide variety of support matrices as calcium alginate, carrageenan, or cellulose, in the form of cell entrapment or adsorption on glass beads. The choice of the support material could be critical for the improvement of the bioconversion yields (el-Naggar et al. 2003) (Fig. 2). In a recent study, the established fermentation process for obtaining acetylisovaleryltylosin, a strongly effective antibiotic against macrolide-resistant bacterial strains, was improved by the immobilization of Streptomyces thermotolerans. Acetylisovaleryltylosin is generally obtained from S. thermotolerans ATCC11416 through the acetylation of tylosin in the 3-OH position and then by substitution with isovaleric acid in the 4-OH position. By immobilizing S. thermotolerans cells on cubes of polyurethane foam, that is, an inert material with very low commercial costs but high porosity (near 97.0%) and a large adsorption surface, a bioconversion rate 1.7 times higher (up to 33.5 mg· L−1 of acetylisovaleryltylosin) than the rate obtained with free cells was reached in batch cultures. In repeated batch processes, the activity of immobilized S. thermotolerans was stable over six cycles performing bioconversions in the range from 79.9 to 86.3%. That made this method feasible for large-scale production (Zhu et al. 2015) (Table 1). The open macrostructure of polyurethane foam was the key factor as it greatly reduced the diffusion resistance and provided excellent mass transfer. Besides being an elastic material, polyurethane foam also showed good resistance to compression deformation which is essential for its industrial applications (Zhu et al. 2015).
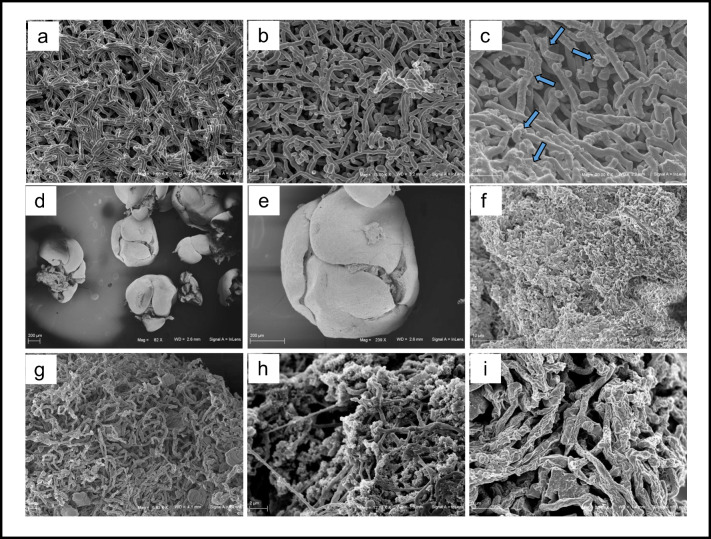
Fig. 2.
SEM pictures of Streptomyces roseochromogenes ATCC 13400 in liquid cultures during hydrocortisone bioconversion at 0, 4, and 48 h, respectively (a–c): the accumulation of material on the cell surface is due to the process of steroid bioconversion and it is indicated by the arrows (c); SEM pictures of Streptomyces roseochromogenes ATCC 13400 whole cells as immobilized in calcium alginate beads (d–f): a tight mycelium was visible in the picture of bead internal view (f); SEM pictures of Streptomyces cyanogriseus ATCC 27426 in liquid cultures during secondary metabolite production at 96, 168, and 264 h, respectively (g–i): morphology changes with increase of cell tangling, branching, and rugosity are visible. (Mag from × 82 to × 20,000, scale bar from 2 to 200 μm). (Preparation of the samples for SEM analyses: small volumes of culture (1 mL) were pelleted end suspended in 4% formalin in PBS for 18 h, dehydrated in increasing ethanol concentrations (from 30 to 100% for 5–15 min), dried in a critical point dryer, and sputtered with platinum-palladium (sputter coater Denton Vacuum Desk V). Fe-SEM Supra 40 Zeiss (5 kV, detector InLens) and Smart SEM Zeiss software were used for observation
Bioconversion by using immobilized enzymes
Immobilized enzymes from streptomycetes have been the most exploited tools in biotechnology biocatalysis in the last few years: depolymerases, laccases, and pectinases have been widely used for biomass degradation or bioremediation, while tyrosinases, lipases, or polygaracturonases were mostly employed in pharmaceutical industry (Spasic et al. 2018). Compared to their free forms, immobilized enzymes retain their specificity and activity better and show improved catalytic performances and structural stability even in a harsh industrial process reaction environment. In addition, immobilized enzymes can be used in repeated batches, thus reducing the overall operational costs, and could be easily separated from the products at the end, thus facilitating the downstream recovery protocols (Wong et al. 2020). For all these reasons, streptomycetes experimental research, as well as productive processes, has relied more and more on enzyme immobilization techniques like encapsulation in calcium alginate or in cross-linked structures or like attachment on solid support materials, such as nanoparticles, agarose beads, and celite (Spasic et al. 2018). Synthetic polymers, like macroporous resins, are widely used for enzyme immobilization as they can bind enzymes by chemical adsorption with different strengths according to the dimensions of their porous, to specific polarity and surface area. In an interesting work, five different macroporous resins were used to immobilize a thermostable lipase, named MAS1, from the marine Streptomyces sp. strain W007 (Wang et al. 2017) (Table 1). The best support resulted to be a resin named XAD1180 which gave the highest protein loading and specific esterification activity, thanks to its biggest specific surface area and pore diameter (700 m2· g−1 and 40 nm, respectively) that facilitated the enzyme immobilization both on the surface and inside the pore matrix. The immobilized lipase was not only more active but also more stable to heat than the free lipase retaining 88.0% of its initial activity after incubation at 65 °C for 30 min. This immobilized lipase was then employed in the esterification process of glycerol with a long-chain of n-3 polyunsaturated fatty acids for the synthesis of triacylglycerols, widely used as diet supplements to reduce the incidence of cardiovascular and chronic inflammatory diseases or cancer. Traditionally, triacylglycerols are obtained by chemical synthesis in organic solvent systems that create problems of environmental pollution and safety risks. Instead, this novel immobilized lipase-based process assured, in the optimized conditions, a high esterification degree (97.5%) and triacylglycerols content (92.4%) in a 24-h reaction (Wang et al. 2017) (Table 1). In the year 2000, the cross-linked enzyme aggregate technology has developed as a very simple immobilization method. It is based on enzyme precipitation with salt or organic solvent to obtain aggregates that are then cross-linked with bifunctional reactive agents. Cross-linked enzyme aggregates have great advantages like high enzyme activity, high stability, and low cost of production as no support materials for immobilization are needed. For example, the poly-(R)-3-hydroxybutyrate depolymerase from Streptomyces exfoliatus was immobilized using cross-linked enzyme aggregate technology and was tested for the conversion of poly-(R)-3-hydroxybutyrate to chiral (R)-3-hydroxybutyric acid that is used as building block for the synthesis of fine chemicals such as antibiotics, vitamins, flavors, and pheromones (Hormigo et al. 2012) (Table 1). In optimal conditions, the poly-(R)-3-hydroxybutyrate depolymerase aggregates proved to be promising biocatalysts by displaying a specific activity of 255.0 U· g−1 with high recycling capability, up to 20 consecutive batch reactions, and a strong thermal, pH, and organic solvent stress resistance (Hormigo et al. 2012) (Table 1). Progress in cross-linked enzyme aggregate technology applied to Streptomyces enzyme has also recently been reported when an all-enzyme hydrogel was developed by genetically modifying the selected enzymes with a SpyTag or SpyCatcher protein fusion domain that then spontaneously assembled in physiological conditions via covalent isopeptide bond formation (Bitterwolf et al. 2019) (Table 1). Compared to the simple cross-linked protocol, this method allows a site-specific immobilization with precise control of the enzyme stoichiometry. An all-enzyme hydrogel, constructed with a glucose-1-dehydrogenase from Bacillus subtilis and an NADPH-dependent-(S)-selective dimeric imine reductase GF3546 from Streptomyces sp., was applied, for example, in the conversion of imines to optically active amines, key structural motifs in pharmaceutical ingredients. The conversion was performed in a microfluidic reactor in a continuous flow mode for 40 h with a flux of 100 μL· min−1 obtaining a maximum space-time-yield of 150.0 g· L day−1 (Bitterwolf et al. 2019) (Table 1). Other supports like nanoparticles or beads have been reported in literature to immobilize Streptomyces enzymes that in this way are exposed on the surface of the solid support. For example, glass microbeads have been recently employed for the immobilization of Streptomyces mobaraensis transglutaminase (Spycher et al. 2017). This enzyme, displayed on the microbeads, enabled the generation of antibody fragments or whole antibodies site-specifically conjugated with various bifunctional substrates, comprising a fluorescent probe and a metal chelator for radio labeling (Spycher et al. 2017). Two recent studies also reported diverse applications of cholesterol oxidase biocatalysts from Streptomyces. One cholesterol oxidase was immobilized in silane-modified iron (II, III) oxide magnetic nanoparticles by covalent coupling methods and then used for the biotransformation of cholesterol and 7-ketocholesterol to 4-cholesten-3-one and 4-cholesten-3, 7-dione respectively, which are industrially and medically important steroid precursors (Ghosh et al. 2018). The transformation of cholesterol produced up to 0.86 mg· L-1 of 4-cholesten-3-one, thus proving to be 2 times more efficient than the same reaction performed by free enzyme. The nanobiocatalyst also showed enhanced pH and thermal stability and significant residual activity (51.0%) after 10 cycles of utilization (Ghosh et al. 2018). Recently, Streptomyces-immobilized cholesterol oxidase was also used in biomedical applications like in biosensor for cholesterol quantification on the basis of its oxidation grade (Perdani et al. 2020). In the study, cholesterol oxidase was first produced through the submerged fermentation of Streptomyces sp., and then, the crude extract was immobilized on magnetite-coated beads with a silicon dioxide layer to provide a hydrophobic surface allowing the formation of a uniform shape of the biosensor. Different oxidation conditions were tested, in terms of enzyme and substrate concentrations or incubation times. With an enzyme concentration of 20 mg ·mL−1, 6.5 mM of cholesterol were efficiently oxidated up to 90.0% in 180 min and this performance was reported to be almost unchanged up to 30 days of use (Perdani et al. 2020).
Bioconversion by using mutated, genetically modified, or heterologous expressed enzymes
Bioconversion strategies that involve preliminary protein and/or genome engineering are more and more used in Streptomyces industrial biocatalysis (Demain 2006), together with a growing trend of heterologously expressing Streptomyces enzymes in different microbial hosts that are more suitable for industrial applications as microorganisms generally recognized as safe or having a quicker time of replication (de Carvalho 2016). Several different techniques have been used for Streptomyces enzyme engineering. For example, multi-step random UV mutagenesis has been recently applied to Streptomyces xanthochromogenes strain RIA 1098 for the production of pravastatin, one of the most popular cholesterol-lowering drugs (Dzhavakhiya et al. 2015). The traditional industrial two-stage process includes the microbial synthesis of compactin by Penicillium citrinum and then its further conversion to pravastatin by S. xanthochromogenes strain RIA 1098, which is capable of hydroxylating the compactin on the C6 position (Dzhavakhiya et al. 2015). A high compactin content is required to increase the rate of conversion, but at the same time high compactin concentrations could inhibit microbial growth. Thus, improvement of the strain resistance to compactin is a desirable goal. In this recent study, S. xanthochromogenes mutants, obtained through UV irradiation, were tested for pravastatin bioconversion in the presence of a daily dose of 1.0 or 2.0 g· L−1 of compactin; with this strategy, 91.0% or 83.0% of bioconversion rates was reported, respectively, after medium optimization (Dzhavakhiya et al. 2015). A different approach was applied for the synthesis of another cholesterol-lowering drug, the Lipitor, and in particular of its key intermediate ethyl-(S)-4-chloro-3-hydroxylbutyrate from ethyl-4-chloro-3-oxobutyrate using a recombinant carbonyl reductase from Streptomyces coelicolor CR1 (Li et al. 2017). The main concerns about wild-type carbonyl reductase are the low activity and the poor stability; to solve these issues, two different tools were recently exploited like the structure-guided engineering and the directed evolution strategy. First, the crystal structure of the carbonyl reductase wild-type enzyme was complexed with NADH, generating a double mutant by structure-guided engineering. In this way, the specific activity increased from 38.8 to 168.0 U· mg−1. Then, the obtained double mutant was used as a template to generate by directed evolution a triple mutant carbonyl reductase, whose thermostability and substrate tolerance were 6.2 °C and 4.7-fold higher than those of the wild type (Li et al. 2017). Interestingly, the specific activity of the triple mutant resulted also to an improvement of up to 260.0 U· mg−1 allowing the asymmetric reduction of 100 g· L−1 of ethyl-4-chloro-3-oxobutyrate in a 300-mL scale, in 9 h, with an enantiomeric excess of 99.0% and a space-time yield of 255.0 g·(L day)−1 (Li et al. 2017). One valuable example of heterologous Streptomyces enzyme expression is the production of phosphatidylserine from bioconversion of phosphatidylcholine and l-serine by phospholipase D from Streptomyces halstedii (Liu et al. 2019). Phosphatidylserine is usually used as a supplement in diets to improve cognitive functions and prevent mental diseases and can be extracted from animal organs such as pig or turkey liver, but the low availability of these animal tissues and the risk of infectious disease transmission limit its industrial scale production. In a recent work, phosphatidylserine was obtained using a recombinant Pichia pastoris whole cell biocatalyst expressing S. halstedii phospholipase D and displaying it on its cellular surface. The use of a whole cell biocatalyst enabled also to employ a simple aqueous system to perform the phospholipase D reaction instead of the traditional biphasic system with organic solvents that could have bad environmental impact. In the optimal conditions, a 53.0% of phosphatidylserine was achieved in 4 repeated batch cycles of only 8 h, by using a 0.2 M sodium acetate buffer at 40 °C and pH 5.5 and 3 U· mL−1 of biocatalyst with 1:3 phosphatidylcholine:l-serine ratio (Liu et al. 2019). As one of the most known and exploited microorganisms both for genome engineering and for industrial scale fermentation processes, Escherichia coli has been widely used for Streptomyces protein expression. In a recent application, two new tyrosine ammonia lyases were identified from Saccharothrix sp. NRRL B-16348 and Streptomyces sp. NRRL F-4489 and heterologously expressed in E. coli BL21 (DE3) for the bioconversion of l-tyrosine to p-coumaric acid through non-oxidative deamination (Cui et al. 2020). The p-coumaric acid is a bioactive agent exhibiting protective effects against cardiovascular and inflammatory diseases and it is considered a significant building block for the production of flavonoids and polyphenols. Kinetic and genomic studies have shown a higher efficiency and expression levels of the Streptomyces-tyrosine ammonia lyase than Saccharothrix-tyrosine ammonia lyase in E. coli BL21 (DE3) strain. In the optimal temperature (50 °C) and pH conditions (pH = 11), E. coli BL21 (DE3)/Streptomyces-tyrosine ammonia lyase produced 17.6 mM (equivalent to 2.9 g· L−1) of p-coumaric acid in 1 h, with a conversion rate of 58.6%, by using a 10 mM glycine–NaOH buffer, a 10 OD600 cell concentration, and an initial l-tyrosine concentration of 30 mM. This conversion rate is the highest ever reached in a prokaryotic system for this molecule up to now (Cui et al. 2020). Another example of E. coli biocatalyst harboring Streptomyces enzymes has been proposed in a recent work for the gamma-aminobutyric acid production through the decarboxylation of monosodium glutamate or l-glutamic acid by glutamate decarboxylase (Yuan et al. 2019). Gamma-aminobutyric acid is a neurotransmission inhibitory, extensively used in functional foods and pharmaceuticals as hypotensive and anti-epilepsy, for asthma treatment, for sleep and memory improvement, and for preventing obesity (Diana et al. 2014). Three different glutamate decarboxylases from Streptomyces toxytricini NRRL 15443, Streptomyces sp. MJ654-NF4, and Streptomyces chromofuscus ATCC 49982 were identified, cloned, and expressed in E. coli BL21 (DE3). The determination of kinetics and optimization analyses showed that the best enzyme operating parameters were 37 °C temperature, 1-h reaction time, 20.0 OD600 cell concentration, and acid pH conditions. Based on these findings, a gamma-aminobutyric acid production experiment was carried out for the three biocatalysts, testing the use of either 0.1 M sodium acetate buffer or water both at pH 4.6 and 2.0 M of monosodium glutamate or l-glutamic acid as the initial substrate. In the optimal conditions, using l-glutamic acid as a substrate, the biocatalyst E. coli/Streptomyces sp. MJ654-NF4 glutamate decarboxylase showed the best performance giving a gamma-aminobutyric acid yield of 2.8 kg· L−1 and a molar conversion rate of 67.0% in 10 repeated batches (Yuan et al. 2019). To further improve this result and to establish an efficient food-grade production process of gamma-aminobutyric acid, glutamate decarboxylase from Streptomyces sp. MJ654-NF4 was then expressed also in Saccharomyces cerevisiae (Yuan et al. 2020). Although production titer and conversion rates were lower (62.6 g· L−1 and 60.8% respectively after 10 batches) than the ones obtained in E. coli, this study offers the perspective to use a microorganism generally recognized as safe as a whole cell biocatalyst in order to have a pharmaceutical-grade product (Yuan et al. 2020). A recent work reported the employment of Streptomyces griseus ATCC 13273 cytochrome-like protein, CYP105D1, for the regio-selective demethylation of papaverine to 6-O-demethyl-papaverine, when expressed in E. coli BL21 (DE3). 6-O-demethyl-papaverine is less cytotoxic than papaverine and it is generally produced using chemical methods with strong acids with poor selectivity and high costs (Shen et al. 2019). E. coli BL21 (DE3), expressing the S. griseus ATCC 13273 cytochrome-like protein, was employed as a whole cell biocatalyst in order to avoid the addition of the expensive NADH cofactor needed, in case of in vitro use of the purified CYP105D1 enzyme. In the same paper, the type and concentration of organic co-solvents to dissolve the substrate papaverine were optimized: by using PEG-200, the highest bioconversion rate and the more environmentally friendly and less toxic whole cell conditions were obtained. In the optimal conditions (25 °C, pH of 7.5, 8.0 g of biocatalyst, 3.0% v/v PEG-200), 50.0 mg· L−1 of initial papaverine was converted to 6-O-demethyl-papaverine with a rate of 61.2% in 24 h, a result never reported before for the demethylation reaction catalyzed by cytochrome P450s (Shen et al. 2019).
Biotechnological production of secondary metabolites by streptomycetes
The Streptomyces genus has an extremely important role in biotechnology as the producer of secondary metabolites with biomedical applications such as antibiotics, antifungal, antiviral, and anthelmintic agents, antitumoral drugs, anti-hypertensives, immunosuppressants, herbicides, and interesting pigments (Sanchez et al. 2012; Olmos et al. 2013). The main secondary metabolites produced by the streptomycetes are the antibiotics: the production is species specific and important for the strains in order to survive in various assorted environments, such as terrestrial soils or marine seabeds, and to compete with other microorganisms that they may come in contact with (de Lima Procópio et al. 2012). Nowadays, two-thirds of all the antibiotics present on the market for medical, veterinary, and agricultural uses are produced by Streptomyces strains, like aminoglycosides, lactams, macrolides, polyenes, and tetracyclines for more than 5000 different molecules and a global market of more than 46 billions of dollars in 2018 (Sánchez et al. 2010; Singh et al. 2014). The industrial production of secondary metabolites by streptomycetes is mainly performed by fermentation in large-scale bioreactors. In liquid submerged cultures, most of the species do not sporulate but produce mycelium whose morphology varies from filaments (Streptomyces clavuligerus) to clumps and pellet cellular aggregates (S. coelicolor or S. griseus). This morphology differentiation remains fundamental for the secondary metabolite production and even necessary to obtain high yields (Olmos et al. 2013; Sanchez et al. 2012; López-García et al. 2014). There are several issues in the streptomycetes fermentation processes for the secondary metabolite production: as these molecules are mainly produced in the late growth or stationary phase and in low concentrations, the industrial processes are usually very long and time-consuming with high costs and frequently with low yields and productivities (Olmos et al. 2013). Strain selection, medium composition, growth pH and temperature, oxygen transfer rate in the vessel, and mycelium aggregation state have been always key parameters to take into account in the development of an efficient industrial process. In recent years, the classical strategies to control these parameters have been coupled with more innovative approaches like fermentation based on mycelium immobilization, control of the metabolic pathways and antibiotic precursor supplementation, strain genetic manipulation, or metabolic engineering techniques. In this perspective, the study of the morphology, performed by scanning electron microscopy (SEM), transmission electron microscopy (TEM), or confocal-laser scanning fluorescence microscopy (CLSM), to correlate the mycelium changes with the different approaches employed and the quantity of secondary metabolites produced, has also resulted crucial in both submerged and immobilized cell cultivations (Pereira et al. 2008; López-García et al. 2014; Ng et al. 2014; Chakravarty and Kundu 2016; Scaffaro et al. 2017) (Fig. 2). Fourteen examples of the most remarkable and innovative approaches developed in the last years to increase the secondary metabolite production of Streptomyces strains are here reported (Table 2).
Table 2.
Summary of different biotechnological approaches for secondary metabolite production by Streptomyces
| Streptomyces strain | Secondary metabolite | Nutrients and metabolism | Fermentation medium (*1.0 L) | Growth conditions | Production (g·L-1) | Productivity (g·L-1 ·h-1) | Reference |
| S. griseus | Streptomycin | Optimization of pH, carbon source, and metal ions | 10.0 g Slb St, 0.3 g casein, 2.0 g K2HPO4, 2.0 g KNO3, 2.0 g NaCl, 0.2 g CaCO3, 0.01 g FeSO4, 0.05 g MgSO4·7H2O | Agar plates, 37 °C, pH = 8.5, 96 h | 4.30 | 0.045 | Singh et al. 2014 |
| S. kathirae SC-1 | Melanin | Optimization of carbon source, nitrogen source, and metal ions by response surface method | 3.27 g Amylo Dext, 37.0 g YE, 5.0 g NaCl, 0.1 g CaCl2, 50 μmol CuSO4, 0.25 mg tyrosine | Shake flask, 28 °C, pH = 6.0, 200 rpm, 128 h | 13.7 | 0.107 | Guo et al. 2014 |
| S. coelicolor A3(2) M145 | Actinorhodin and undecylprodigiosin | Optimization of phosphate and l-glutamate concentrations in the media, optimization of agitation speed and DO percentage | 55.2 g sodium-glutamate, 40.0 g Glc, 2 mmol MgSO4, 4.6 mmol phosphate, 0.028 g FeSO4∙7 H2O, 0.002 g CuSO4∙5 H2O, 0.0024 g ZnSO4∙7 H2O, 0.008 g MnSO4∙H2O, 5.6∙10−5 g Na2MoO4∙2 H2O, 11.2∙10−5 g CoCl2∙6 H2O | 3.0-L fermentor, 30 °C, pH = 7.0, 0.3–0.5 vvm, 900–1300 rpm, 70 h | - | 0.013 and 0.033 (sp. units·gcdw-1∙h-1) | Wentzel et al. 2012 |
| S. roseosporus NRRL11379 | Daptomycin | Supplementation with sodium decanoate as antibiotic precursor and optimized fed-batch feeding with dextrose | 11.0 g YE, 0.86 g Fe(NH4)2SO4·6H2O, 10.7 g dextrose, 72.0 g potato Dext, 7.2 g cane molasses | 3.6-L fermentor, 30 °C, pH = 6.5, 3.5 vvm, 450 rpm, 288 h | 0.81 | 0.003 | Ng et al. 2014 |
| Streptomyces strain | Secondary metabolite | Fermentation strategies | Fermentation medium (*1.0 L) | Growth conditions | Production (g·L-1) | Productivity (g·L-1· h-1) | Reference |
| S. cyaneogrisueus ssp. noncyanogenus | Nemadectin | WHd + RT impeller configuration, 25–55% DO by O2 supply, moderate agitation speed | 20.0 g Glc, 90.0 g CS, 25.0 g SoyF, 5.0 g YE, 4.0 g CaCO3,11.0 g MgSO4·7H2O, 0.01 g CuSO4, 0.002 g CoCl2, 0.001 g MnSO4 | 5.0-L fermentor, 28 °C, pH = 7.4, 1.3 vvm, 650 rpm,192 h | 1.37 | 0.007 | Song et al. 2018 |
| S. roseosporus NBRC 12910 | Daptomycin | Cell immobilization on ultra-porous refractory brick flakes or silk sachets, airlift bioreactor | 10.0 g Glc, 30.0 g Dext, 20.0 g SoyF, 0.6 g Fe(NH4)2SO4, 0.2 g KH2PO4 | 2.0-L fermentor, 30 °C, pH = 7.0, 200 rpm, 132 h each cycle | 4.89 on refractory bricks and 3.62 on silk sachets | 0.005 (considering 8 cycles on refractory bricks and 6 cycles on silk sachets) | Chakravarty and Kundu 2016 |
| S. coelicolor M145 | Actinorhodin | Cell encapsulation in calcium alginate beads | 20.0 g Glc, 0.2 g CSA, 10.0 g YE, 42.0 g MOPS, 0.5 g K2SO4 , 20.2 g MgCl2 | Shake flask, 30 °C, pH = 6.8, 200 rpm, 192 h | 3.17 | 0.016 | López-García et al. 2014 |
| S. coelicolor M145 | Actinorhodin and undecylprodigiosin | Cell immobilization on PLA or PLA-plasma membranes | 20.0 g Glc, 200.0 g sucrose, 0.2 g CSA, 10.0 g YE, 42.0 g MOPS, 0.5 g K2SO4, 20.2 g MgCl2 | Shake flask, 30 °C, pH = 6.8, 200 rpm, 168 h | 0.28 and 0.015 | 0.016 and 8.92*10-5 | Scaffaro et al. 2017 |
| Streptomyces strain | Secondary metabolite | Mutagenesis and genome approaches | Fermentation medium (*1.0 L) | Growth conditions | Production (g·L-1) | Productivity (g·L-1· h-1) | Reference |
| S. hygroscopicus subs. ascomyceticus SA68 | Ascomycin | Femtolaser mutation and shikimic acid addition | 24.0 g Slb St, 40.0 g Dext, 5.0 g Pep, 7.0 g YE, 2.0 g CSL, 11.0 mL SoybO, 1.5 g Sh A, 0.5 g K2HPO4·3H2O, 1.5 g (NH4)2SO4, 1.0 g MgSO4·7H2O, 1.0 g CaCO3 | Shake flask, 28 °C, pH = 6.7, 200 rpm, 144 h | 0.45 | 0.003 | Qi et al. 2014a |
| S. hygroscopicus subs. ascomyceticus FS35 | Ascomycin | Addition of resin, of n-hexadecane, valine, lysine | 24.0 g Slb St, 40.0 g Dext, 5.0 g Pep, 7.0 g YE, 2.0 g CSL, 11.0 mL Soyb O, 1.5 g Sh A, 0.5 g K2HPO4·3H2O, 1.5 g (NH4)2SO4, 1.0 g MgSO4·7H2O, 1.0 g CaCO3 | Shake flask, 28 °C, pH = 6.7, 200 rpm, 144 h | 0.46 | 0.003 | Qi et al. 2014b |
| S. tsukubaensis TJ-01 | FK506 | UV and NTG mutagenesis, with selection for disodium malonate and disodium methylmalonate resistance. Genome shuffling and FK506 resistance. Dynamic fed-batch with disodium malonate and disodium methylmalonate addition. | 60.0 g St, 2.0 g YE, 2.5 Pep, 5.0 g Soyb M, 0.5 g K2HPO4, 0.5 g MgSO4, 0.5 g CaCO3 | 7.5-L fermentor, 28 °C, pH = 6.8, 200–1000 rpm, 1.0 vvm, 144 h | 0.51 | 0.003 | Du et al. 2014 |
| S. chattanoogensis L10 | Natamycin | WhiGch gene overexpression | 0.4 g Glc, 0.03 g YE, 0.03 g ME, 0.5 g TR | Shake flask, 30 °C, pH = 7.2, 250 rpm, 120 h | wt = 1.5, LG02 mut = 2.5 | wt = 0.012, LG02 mut = 0.021 | Liu et al. 2015 |
| S. avermitilis ATCC31267 | Avermectin B1a | Enhancement of aveR and malE genes expression by treating the strain with nanosecond pulsed electric field | 70.0 g Slb St, 16.0 g YE, 0.5 g K2HPO4·3H2O, 0.5 g MgSO4·7H2O, 4.0 g KCl, 10.0 mg CoCl2·6H2O, 2.0 g CaCO3 | Shake flask, 28 °C, pH = 7.0, 220 rpm, 120 h | 0.015 | 0.0001 | Guo et al. 2016 |
| S. avermitilis DQ03 | Ivermectin B1a | Construction of a biosynthetic gene cluster | 140.0 g CS, 0.1 g α-amylase, 28.0 g SoyF, 10.0 g YE, 0.022 g Na2MoO4·2H2O, 0.0023 g MnSO4·H2O, 0.25 g (NH4)2SO4, 0.02 g CoCl2, 0.8 g CaCO3 | Shake flask, 28 °C, pH = 7.5, 220 rpm, 240 h | 1.25 | 0.005 | Deng et al. 2019 |
CSA casamino acids, CS corn starch, CSL corn step liquor, Dext dextrin, DO dissolved oxygen, Glc glucose, ME malt extract, MOPS 3-morpholinopropane-1-sulfonic acid, NTG nitrosoguanidine, Slb St soluble starch, St starch, Pep peptone, PLA polylactic acid, Sh A shikimic acid, SoyF soybean flour, Soyb M soybean meal, Soyb O soybean oil, TR tryptone, WHd + RT wide-blade hydrofoil impeller and rushton turbine; YE yeast extract
Nutrients and metabolism studies for the biotechnological production of secondary metabolites
Several factors and complex pathway-specific regulatory mechanisms influence the transition from the primary to the secondary metabolism of streptomycetes and the switch that is activated at specific developmental stages of the bacteria life cycle is responsive to environmental conditions as temperature, light, pH, carbon, nitrogen and phosphate source availability, metals and oxygen concentrations, or antifoam addition (Ruiz et al. 2010). Biotechnological approaches tried to regulate the onset and the development of the secondary metabolite biosynthesis also by wisely designing the growth media nutrient composition and by the optimization of the fermentation conditions. But these approaches have always required great efforts as the activation of the secondary metabolic pathway is species specific and tailored-cut protocols have to be developed for each Streptomyces strain. The effect of nutrients, especially of the carbon sources, on the production of secondary metabolites has been the subject of numerous and continuous studies from biochemical and fermentation process points of view (Ruiz et al. 2010), but in the last decade also from the metabolic and molecular biology points of view, thanks to the study of the different transcriptional factors involved in the metabolic balances of the streptomycetes (Martín and Liras 2020). The choice of the right carbon source could be critical for the secondary metabolism onset and the streptomycetes metabolism (Ruiz et al. 2010; Sánchez et al. 2010). At molecular levels, the use of one carbon source instead of another could influence the activation or the repression (carbon catabolite repression) of genes and of transcriptional signals; this phenomenon is strain specific and regulates the synthesis of secondary metabolite and the morphological differentiation (Ruiz et al. 2010; Sánchez et al. 2010). The synthesis of aminoglycoside, as streptomycin by S. griseus and the neomycin by S. fradiae, for example, is inhibited by glucose via repression of biosynthetic enzymes such as mannosidostreptomycinase and alkaline phosphatase, respectively. But glucose inhibits also the production of the polyketide actinorhodin by Streptomyces lividans by the repression of the synthesis of a global regulatory protein involved in the stimulation of secondary metabolite production (Ruiz et al. 2010; Sánchez et al. 2010). Recently, a transcriptomic analysis of streptomycetes has been proposed to study the carbon catabolite repression phenomena. In a recent work, a high-density micro-array approach was employed to study S. coelicolor M145 carbon catabolite repression mechanism and the role of the glucose kinase, the main glucose phosphorylating enzyme, as regulatory protein in this mechanism (Romero-Rodríguez et al. 2016). The study demonstrated that glucose and the glucose kinase influenced or modified the expression of 651 and 134 genes, respectively (Romero-Rodríguez et al. 2016). In future perspective, these studies will help to better understand how to trigger gene expression and secondary metabolite production by modulating carbon source. The use of glycerol instead of glucose could be a valid alternative, but frequently media for secondary metabolite production are formulated with more complex carbon sources like dextrin, starch, soya bean flours, and potato extract (Table 2). The use of media with complex carbon sources is still a winning classical strategy to both select and screen new high producer streptomycetes strains as well as to boost the secondary metabolite production in fermentation processes. For example, although streptomycin has been industrially produced since 1945 from S. griseus for a current global market of millions of US dollars, still nowadays literature papers report the selection of new high producer streptomycin strains from soil on the basis of the medium formulation optimization (Singh et al. 2014) (Table 2). The streptomycin is a water-soluble aminoglycoside that acts against numerous pathogenic bacteria and that is widely used in medicine for the treatment of acute infections of the respiratory and urinary tracts in humans, but it is also effective as a pesticide against pathogenic bacteria and fungi of plants and trees (Singh et al. 2014). By studying the best pH condition, in the range from 6.0 to 8.5 and by modifying a casein-based medium, by addition of Fe ions (in the form of FeSO4·7H2O), that resulted the best metal to support production compared to Ca, Zn, Fe, Co, and starch as the best carbon source instead of sucrose, maltose, lactose, and glucose, a very high concentration of streptomycin of 4.3 g· L−1 was recently obtained in 96 h of growth on agar plate by a new strain selected from soil (Singh et al. 2014) (Table 2). In the last years, statistical models have also demonstrated to be innovative and greatly helpful tools for the optimization of all the components of growth media. A response surface methodology has been applied, for example, for the optimization of the culture medium composition of a newly isolated Streptomyces kathirae SC-1 strain to boost its production of melanin (Guo et al. 2014). Melanin is a dark-black pigment and a good UV radiation and free radical protector widely used as food supplement and nutraceutical (Guo et al. 2014). The model helped to identify amylodextrin, yeast extract, and CuSO4 as key carbon, nitrogen, and metal nutrients for melanin biosynthesis and to determine their optimal concentrations to boost the melanin production up to 13.7 g· L−1 in 128 h (Guo et al. 2014). Although in literature there are many papers that have tried to better understand all the events occurring during the transition from the primary to the secondary metabolism, a complete picture of the relationship between primary and secondary metabolic networks is still lacking. A good primary metabolism is fundamental to sustain an efficient second metabolism as it supplies power and energy to build biomass but also antibiotic precursors (Coze et al. 2013). Novel biotechnological approaches are employing metabolic flux analyses and 13C-labeling to study the carbon flux in Streptomyces stains (Coze et al. 2013). In a recent paper, the primary metabolism carbon flux of two S. coelicolor A3 (2) strains, a wild type (M145) and a mutated strain, lacking the gene clusters encoding for the antibiotic actinorhodin production (M1146), was analyzed under steady-state and compared in order to better understand the influence of antibiotic biosynthesis on central carbon metabolism (Coze et al. 2013). In the mutant strain, higher growth rate, higher biomass formation, and higher flux through the pentose phosphate pathway were observed, as well as an accumulation of acetyl-CoA as precursor of actinorhodin. Thus, the metabolic differences between the two strains allowed the determination of the energetic costs of the actinorhodin synthesis. In another study, the same strain S. coelicolor A3(2) M145 was used in submerged batch fermentation to precisely determine the switching from the primary to the secondary metabolism in response to the depletion of phosphate or l-glutamate, and to correlate it to the blue-pigmented actinorhodin and red-pigmented undecylprodigiosin antibiotic production (Wentzel et al. 2012). Some nutrients, like phosphate and amino acids, result necessary for the initial primary phase of growth. When they started to be depleted and became a limiting component for growth, the stationary phase and the antibiotic production start (Wentzel et al. 2012). But their initial concentrations are also critical: with an excess of phosphate in the initial medium, for example, the growth is driven to higher biomass production and a bigger mycelium formation while the stationary phase is delayed and/or the production of a huge number of antibiotics is inhibited or repressed (Wentzel et al. 2012). A 4.6 mM phosphate concentration resulted good enough to support the growth of S. coelicolor A3(2) M145 (up to 6.0 g· L−1) and even to boost the antibiotic production, while l-glutamate resulted a good, alternative nitrogen source to the more used and complex casamino acid and better than ammonium that caused a very slow-growing mycelium. In combination with glucose, it provides sufficient energy and reducing power to maintain the growth rate high (Wentzel et al. 2012). Supplementation with precursors coupled with medium optimization has been in the last years a wise strategy to improve the yield of antibiotics too. Daptomycin is a cyclic lipopeptide antibiotic produced by Streptomyces roseosporus and approved by the FDA in 2003 as a drug for skin infections, right-sided endocarditis, and bacteremia, as it is able to act against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci strains (Chakravarty and Kundu 2016). In S. roseosporus NRRL11379 cultivations, the sodium decanoate was employed as an effective precursor of daptomycin as it seems to enhance the expression of key proteins of the biosynthetic pathways like guanosine pentaphosphate synthetase, polyribonucleotide nucleotidyltransferase phosphorylase, and tripeptidylamino peptidase (Table 2) (Ng et al. 2014). By coupling sodium decanoate supplementation (0.6 g· L−1 added at 48 h) with a culture medium made of dextrin as the major carbon source and yeast extract as the nitrogen source, whose composition was optimized with an orthogonal experimental design and with a feeding of dextrin when carbon source concentration was below 20 g· L−1, a daptomycin production of 0.81 g· L−1was reached in 288 h in a 3.6-L feed batch experiments (Table 2) (Ng et al. 2014).
Fermentation strategies for the biotechnological production of secondary metabolites
As the selection of nutrients is critical in streptomycetes cultures, oxygen is also a key factor; both have to be wisely modulated to drive the biosynthesis to the secondary metabolism and not to the biomass formation. Ensuring a sufficient oxygen transfer rate has been traditionally performed by increasing agitation speed or aeration fluxes. Studies that use combined approach of media formulation and agitation speed modulation are still frequently reported. For example, the blue-pigmented actinorhodin and red-pigmented undecylprodigiosin production from S. coelicolor A3(2) M145 changes according to the agitation speed employed (from 900 to 1300 rpm) (Wentzel et al. 2012); the higher production values were obtained by employing nonstressful agitation conditions (900-1100 rpm) while a dissolved oxygen percentage (DO) of 50.0% resulted necessary to obtain a highly reproducible process (Wentzel et al. 2012). High dissolved oxygen percentages in the vessel during the fermentation are essential for both streptomycetes mycelia and pellet differentiation and thus for the secondary metabolite production. However, reaching high oxygen rate by using elevated agitation speed could be counterproductive as it damages and stresses the mycelium itself and reduces the secondary metabolite yield. Thus, the study of new impeller configuration in the vessel or in alternative the addition of oxygen vectors in fermentation medium or the use of immobilized systems instead of liquid cultures has been taken into account during the years. In a recent interesting study, the production of nemadectin, a macrocyclic lactone, used worldwide as an efficient, broad-spectrum insecticide, acaricide, and anthelmintic agent, sourced by Streptomyces cyaneogrisueus ssp. noncyanogenus, was increased of 23.6% up to 1.3 g· L−1 by combining a new impeller configuration inside a 5.0-L vessel, made of a wide-blade hydrofoil impeller in upper stage and a rushton turbine in bottom stage (WHd+RT configuration), with insufflations of pure oxygen, thus keeping the DO (dissolved oxygen) levels between 25.0 and 55.0% during the growth phase (Song et al. 2018) (Table 2). This strategy efficiently supplied oxygen without producing a high shear rate (54.7 s−1) and that supported growth, long mycelia formation, and high nemadectin biosynthesis avoiding cell stress. Morphology is demonstrated to be critical for secondary metabolite production. The morphology of Streptomyces roseosporus, for example, has demonstrated to change according to the carbon and nitrogen source, the inoculum size, the air flow rate used, and the eventual addition of precursors (Ng et al. 2014; Chakravarty and Kundu 2016). Because of its filamentous nature, S. roseosporus free cell or pellet fermentations could drive to low daptomycin production; during fermentation, a dense biomass network could be formed and that would increase the broth viscosity and limit the volumetric oxygen transfer coefficients (Chakravarty and Kundu 2016). For these reasons, the immobilization or encapsulation of filamentous strains in submerged fermentation processes has become a more and more attractive strategy to improve the antibiotic biosynthesis in alternative to classical fermentations; in these processes, there are higher proportions of viable hyphae compared to free cell liquid cultures and higher mycelium density, aggregates, or pellets whose center has a higher proportion of dead hyphae (López-García et al. 2014; Scaffaro et al. 2017). In a recent study, very high daptomycin concentrations, between 4.89 and 3.62 g· L−1, were reached in a 2.0-L airlift bioreactor, in 6–8 repeated batches of 132 h each by using S. roseosporus NBRC 12910 cells immobilized on uncommon supports such as ultra-porous refractory brick flakes or silk sachets (Chakravarty and Kundu 2016). By encapsulating cells in calcium alginate, the actinorhodin, a blue-pigmented, polyketide, redox active antibiotic, was produced by S. coelicolor M145 up to 3.17 g· L−1 (5 mM) in 0.5-L shake flasks and in 192 h (López-García et al. 2014) (Table 2). In an another work, the same strain was employed in submerged cultivations of immobilized cells on electrospun polylactic acid (PLA) membranes, subjected or not to O2-plasma treatment, to produce both the blue-pigmented actinorhodin and red-pigmented tri-pyrrole undecylprodigiosin, another antibiotic with antimalarial, antiulcer, anticancer, and apoptotic activities (Scaffaro et al. 2017) (Table 2). The S. coelicolor M145 mycelium was able to adhere on membranes and higher biomass (up to two folds) and higher actinorhodin and undecylprodigiosin concentrations compared to the control cultures, up to 0.28 and 0.015 g· L−1 (four and ten folds higher, respectively), were obtained in 168 h (Scaffaro et al. 2017).
Mutagenesis and genome-based approaches for the biotechnological production of secondary metabolites
Innovative strategies involving mutagenesis and genetic engineering approaches have been employed in the last years to boost the secondary metabolite production coupled with new fermentation or precursor addition protocols. For example, during the years different strategies have been proposed to increase the ascomycin production by ascomyceticus and yakushimaensis subspecies of Streptomyces hygroscopicus strains. Ascomycin is a macrolide that is widely employed as an immunosuppressive agent and antifungal, anti-malaria, and antispasmodic drug (Qi et al. 2014a, b). More dated studies focused on the optimization of the fermentation parameters such as the temperature conditions (from 24 to 30 °C, optimal temperature at 24 °C) and the agitation speed from 100 to 350 rpm (Kumar et al. 2007). In a recent paper, an overproducing strain of S. hygroscopicus var. ascomyceticus SA68 was obtained by femtosecond laser mutation followed by strain selection on the basis of shikimic acid endurance. The addition of the shikimic acid (3.0 g· L−1 at 24 h) during the growth drove to an ascomycin production of 0.45 g· L−1, a 42.8% higher value than the concentration produced by the parental strain, obtained in a 24-h shorter time (Qi et al. 2014a) (Table 2). A second overproducing mutant strain, S. hygroscopicus FS35, was employed instead to produce 0.46 g· L−1 of ascomycin by combining an in situ product adsorption strategy by addition of a resin (5% m/v) in the vessel at 24 h, with an increased oxygen transfer rate approach by addition in the fermentation medium of n-hexadecane as oxygen vector and with a supplementation also of valine (Val) and lysine (Lys), key limiting antibiotic precursors (Qi et al. 2014b) (Table 2). A combination of genome shuffling and dynamic fed-batch strategies was instead recently used to increase the production of the polyketide macrolide FK506, commercially called tacrolimus, used since its approval by the Food and Drug Administration (FDA) in 1994 as an immunosuppressant drug after transplantations and an anti-inflammatory pharma for skin diseases, and whose source is the soil species Streptomyces tsukubaensis (Du et al. 2014). In one of the most recent works, four FK506 overproducer S. tsukubaensis mutants were first obtained by classical UV and N-methyl-N′-nitro-N-nitrosoguanidine (NTG) mutagenesis treatments and by screening for resistance to disodium malonate and disodium methylmalonate, two precursors of the FK506 antibiotic (Table 2). These mutants were then further subjected to five rounds of genome shuffling for protoplast fusion with an increasing evolution pressure for FK506 resistance. Thanks to this strategy a high producer recombinant strain selected was able to produce up to about 0.36 g· L−1, a 2.67-fold higher antibiotic titer compared to the concentration obtained by the wild type (Du et al. 2014). Optimization of dynamic fed-batch fermentation strategies in a 7.5-L vessel by addition of 0.05 g· L−1 h−1 of disodium malonate and of 0.03 g· L−1 h−1 of disodium methylmalonate, supplemented at 60 h and at 84 h, respectively, resulted in a further increased production of up to 0.51 g· L−1 of FK506 (Du et al. 2014) (Table 2). One of the major problems of designing processes for secondary metabolite production is our poor knowledge of the gene regulatory network. Several strategies have been used so far to manipulate the gene expression in both native and heterologous hosts. For example, both combinatorial biosynthesis and approaches based on the regulation of antibiotic signaling have become good strategies for improving the yield of production of antitumor compounds (Olano et al. 2011; Kong et al. 2019). As concerns the control of the gene, a recent study, for example, has investigated the sigma factors that regulate the secondary metabolism in Streptomyces chattanoogensis L10, an industrial strain that is the source of natamycin, a polyene macrolide widely used as an antifungal agent and whose production is associated with a yellow pigment (Liu et al. 2015). In particular, the research determined that the group 3 sigma factor, WhiGch, is not only a transcriptional regulator of the morphological differentiation of the cells but it is also a positive up-regulator of the natamycin biosynthetic gene cluster by directly binding the promoters. The over-expression of this sigma factor drove to a 26.0% increase of the antibiotic production compared to the wild type (Liu et al. 2015) (Table 2). Avermectin by Streptomyces avermitilis is an anthelmintic and insecticidal agent widely used in medicine, veterinary, and agriculture. In a recent study, the expression of two positive avermectin regulators, the aveR and malE genes, was enhanced by treating the strain with a nanosecond pulsed electric field approach; 20 pulses at 15 kV cm−1 modified the strain and caused a 42.0% increased avermectin production compared to the control, up to 0.015 g· L−1, also reducing the process time from 7 to 5 days (Table 2) (Guo et al. 2016). Combinatorial biosynthesis of gene clusters of diverse Streptomyces strains by domain swap provides a new and efficient strategy to generate structural tailored-cut secondary metabolites. Ivermectin is a broad-spectrum anti-parasitic agent and a FDA-approved drug used to fight skin infection and tuberculosis. It has recently gained great attention as it resulted to inhibit the replication of SARS-CoV-2 virus in in vitro studies (Caly et al. 2020). Ivermectin is a derivate of avermectin, traditionally produced by specific hydrogenation of the double bound at the C22 and C23 of the avermectin B1a ring by the Wilkinson catalysis (Deng et al. 2019). Streptomyces bingchenggensis is the producer of milbemycin, another avermectin derivate, used as an anthelmintic and anti-parasite agent as well, that does not show any double bound at the C22 and C23 in its 16-membered macrolide structure, as an enoylreductase is included in its biosynthetic gene clusters. By using the gene cluster combinatorial biosynthesis approach, a new S. avermitilis strain was recently developed to directly produce ivermectin B1a, up to 1.25 g· L−1 in 10 days by domain swap. In this study, polyketide synthase module of S. avermitilis, which included dehydratase and ketoreductase domains, was replaced with the one with full reductive cycle from S. bingchenggensis (Deng et al. 2019) (Table 2).
Conclusions and future perspectives
The complexity of their life cycle, gene clusters, enzymatic pools, and metabolic pathways makes the streptomycetes multi-potential microbial cell factories to be used as platforms for drug production. Because of this complexity, the study of new biotechnological processes for drug production should take into account multidisciplinary approaches. Compared to articles of 20 years ago, recent research includes morphology, genetic, and metabolic studies coupled with new technological tools. Whole cell biocatalysis is nowadays based on a wise design of physiological, metabolic, and fermentation process aspects. Heterologous expression of streptomycetes genes in microbial hosts has become a widely used practice to obtain cells more easily to grow but potentiated with versatile enzymes, while structure-guided engineering and directed evolution approaches will substitute in the future more rough mutagenesis techniques to amplify the streptomycetes catalysis ability. The employment of innovative materials is supporting new studies on immobilized biocatalysis useful for gram scale production. The never-ending research on new streptomycetes strains producing unknown antibiotics and secondary metabolites has been coupled to studies in order to better understand the morphology and metabolic changes or the signaling occurring during the switch between the primary and the secondary metabolisms. Cell immobilization in bioreactors with an innovative configuration assures nowadays gram scale production of drugs, while mutagenesis and genome approaches are giving the possibility to build biosynthetic gene clusters tailored-cut to specifically produce high-value drugs as anticancers.
Authors’ contributions
O.F.R. and S.B.F. conceived, drafted, and wrote the manuscript; C.S. obtained funds and reviewed the manuscript; M.C. performed SEM experiments. All authors read and approved the manuscript.
Funding
This work was financed by Campania Region-POR FESR 2007-2013 B25C13000290007 in the frame of project “Bio industrial processes-BIP.” S.B.F. is enrolled in a PhD program with an industrial project called “Streptomycetes as technological platform for the improvement of biotechnological productive processes of pharmaceutical active principles and/or nutraceuticals of industrial interests.”
Compliance of ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Anderson AS, Wellington EM. The taxonomy of Streptomyces and related genera. Int J Syst Evol Microbiol. 2001;51:797–814. doi: 10.1099/00207713-51-3-797. [DOI] [PubMed] [Google Scholar]
- Anteneh YS, Franco CMM (2019) Whole cell Actinobacteria as biocatalysts. Front Microbiol 10. 10.3389/fmicb.2019.00077 [DOI] [PMC free article] [PubMed]
- Argoudelis AD, Coats JH. Microbial transformation of antibiotics. II. Phosphorylation of lincomycin by Streptomyces species. J Antibiot. 1969;22:341–343. doi: 10.7164/antibiotics.22.341. [DOI] [PubMed] [Google Scholar]
- Argoudelis AD, Coats JH. Microbial transformation of antibiotics. VI. Acylation of chloramphenicol by Streptomyces coelicolor. J Antibiot. 1971;24:206–208. doi: 10.7164/antibiotics.24.206. [DOI] [PubMed] [Google Scholar]
- Atta FM, Zohri AA. Transformation reactions of progesterone by different species of Streptomyces. J Basic Microbiol. 1995;35:1–7. doi: 10.1002/jobm.3620350102. [DOI] [PubMed] [Google Scholar]
- Berrie JR, Williams RAD, Smith KE. Microbial transformations of steroids-XI. Progesterone transformation by Streptomyces roseochromogenes–purification and characterisation of the 16α-hydroxylase system. J Steroid Biochem. 1999;71:153–165. doi: 10.1016/S0960-0760(99)00132-6. [DOI] [PubMed] [Google Scholar]
- Bitterwolf P, Ott F, Rabe KS, Niemeyer CM (2019) Imine reductase based all-enzyme hydrogel with intrinsic cofactor regeneration for flow biocatalysis. Micromachines 10. 10.3390/mi10110783 [DOI] [PMC free article] [PubMed]
- Brautaset T, Bruheim P, Sletta H, Hagen L, Ellingsen TE, Strøm AR, Valla S, Zotchev SB. Hexaene derivatives of nystatin produced as a result of an induced rearrangement within the nysC polyketide synthase gene in S. noursei ATCC 11455. Chem Biol. 2002;9:367–373. doi: 10.1016/s1074-5521(02)00108-4. [DOI] [PubMed] [Google Scholar]
- Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res. 2020;178:104787. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty I, Kundu S. Improved production of daptomycin in an airlift bioreactor by morphologically modified and immobilized cells of Streptomyces roseosporus. AMB Express. 2016;6:101. doi: 10.1186/s13568-016-0274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. Recent advances in understanding Streptomyces. F1000Res. 2016;5:2795. doi: 10.12688/f1000research.9534.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin HS, Sim J, Sim TS. Mutation of N304 to leucine in Streptomyces clavuligerus deacetoxycephalosporin C synthase creates an enzyme with increased penicillin analogue conversion. Biochem Biophys Res Commun. 2001;287:507–513. doi: 10.1006/bbrc.2001.5552. [DOI] [PubMed] [Google Scholar]
- Coze F, Gilard F, Tcherkez G, Virolle M-J, Guyonvarch A. Carbon-flux distribution within Streptomyces coelicolor metabolism: a comparison between the actinorhodin-producing strain M145 and its non-producing derivative M1146. PLoS One. 2013;8:e84151. doi: 10.1371/journal.pone.0084151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P, Zhong W, Qin Y, Tao F, Wang W, Zhan J. Characterization of two new aromatic amino acid lyases from Actinomycetes for highly efficient production of p-coumaric acid. Bioprocess Biosyst Eng. 2020;43:1287–1298. doi: 10.1007/s00449-020-02325-5. [DOI] [PubMed] [Google Scholar]
- de Carvalho CCCR. Whole cell biocatalysts: essential workers from nature to the industry. Microb Biotechnol. 2016;10:250–263. doi: 10.1111/1751-7915.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lima Procópio RE, da Silva IR, Martins MK, de Azevedo JL, de Araújo JM. Antibiotics produced by Streptomyces. Braz J Infect Dis. 2012;16:466–471. doi: 10.1016/j.bjid.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Demain AL. From natural products discovery to commercialization: a success story. J Ind Microbiol Biotechnol. 2006;33:486–495. doi: 10.1007/s10295-005-0076-x. [DOI] [PubMed] [Google Scholar]
- Demain AL, Báez-Vásquez MA. Immobilized Streptomyces clavuligerus NP1 cells for biotransformation of penicillin G into deacetoxycephalosporin G. Appl Biochem Biotechnol. 2000;87:135–140. doi: 10.1385/ABAB:87:2:135. [DOI] [PubMed] [Google Scholar]
- Deng Q, Xiao L, Liu Y, Zhang L, Deng Z, Zhao C. Streptomyces avermitilis industrial strain as cell factory for Ivermectin B1a production. Synth Syst Biotechnol. 2019;4:34–39. doi: 10.1016/j.synbio.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana M, Quílez J, Rafecas M. Gamma-aminobutyric acid as a bioactive compound in foods: a review. J Funct Foods. 2014;10:407–420. doi: 10.1016/j.jff.2014.07.004. [DOI] [Google Scholar]
- Dickens ML, Strohl WR. Isolation and characterization of a gene from Streptomyces sp. strain C5 that confers the ability to convert daunomycin to doxorubicin on Streptomyces lividans TK24. J Bacteriol. 1996;178:3389–3395. doi: 10.1128/jb.178.11.3389-3395.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugoński J, Sedlaczek L. Regulation of steroid 16α-hydroxylation in Streptomyces olivoviridis. Z Allg Mikrobiol. 1981;21:499–506. doi: 10.1002/jobm.3630210703. [DOI] [PubMed] [Google Scholar]
- Du W, Huang D, Xia M, Wen J, Huang M. Improved FK506 production by the precursors and product-tolerant mutant of Streptomyces tsukubaensis based on genome shuffling and dynamic fed-batch strategies. J Ind Microbiol Biotechnol. 2014;41:1131–1143. doi: 10.1007/s10295-014-1450-3. [DOI] [PubMed] [Google Scholar]
- Dzhavakhiya VV, Voinova TM, Glagoleva EV, Petukhov DV, Ovchinnikov AI, Kartashov MI, Kuznetsov BB, Skryabin KG. Strain improvement of Streptomyces xanthochromogenes RIA 1098 for enhanced pravastatin production at high compactin concentrations. Indian J Microbiol. 2015;55:440–446. doi: 10.1007/s12088-015-0537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Naggar MY, Hassan MA, Said WYY, el-Aassar SA. Effect of support materials on antibiotic MSW2000 production by immobilized Streptomyces violatus. J Gen Appl Microbiol. 2003;49:235–243. doi: 10.2323/jgam.49.235. [DOI] [PubMed] [Google Scholar]
- Endo K, Hosono K, Beppu T, Ueda K. A novel extracytoplasmic phenol oxidase of Streptomyces: its possible involvement in the onset of morphogenesis. Microbiology (Reading) 2002;148:1767–1776. doi: 10.1099/00221287-148-6-1767. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Ahmad R, Gautam VK, Khare SK. Cholesterol-oxidase-magnetic nanobioconjugates for the production of 4-cholesten-3-one and 4-cholesten-3, 7-dione. Bioresour Technol. 2018;254:91–96. doi: 10.1016/j.biortech.2018.01.030. [DOI] [PubMed] [Google Scholar]
- Guo J, Rao Z, Yang T, Man Z, Xu M, Zhang X. High-level production of melanin by a novel isolate of Streptomyces kathirae. FEMS Microbiol Lett. 2014;357:85–91. doi: 10.1111/1574-6968.12497. [DOI] [PubMed] [Google Scholar]
- Guo J, Ma R, Su B, Li Y, Zhang J, Fang J. Raising the avermectins production in Streptomyces avermitilis by utilizing nanosecond pulsed electric fields (nsPEFs) Sci Rep. 2016;6:1–10. doi: 10.1038/srep25949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Sugimoto H, Shinkyo R, Yamada M, Ikeda S, Ikushiro S, Kamakura M, Shiro Y, Sakaki T. Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1. Biochemistry. 2008;47:11964–11972. doi: 10.1021/bi801222d. [DOI] [PubMed] [Google Scholar]
- Hormigo D, García-Hidalgo J, Acebal C, de la Mata I, Arroyo M. Preparation and characterization of cross-linked enzyme aggregates (CLEAs) of recombinant poly-3-hydroxybutyrate depolymerase from Streptomyces exfoliatus. Bioresour Technol. 2012;115:177–182. doi: 10.1016/j.biortech.2011.09.035. [DOI] [PubMed] [Google Scholar]
- Hussain HA, Ward JM. Enhanced heterologous expression of two Streptomyces griseolus cytochrome P450s and Streptomyces coelicolor ferredoxin reductase as potentially efficient hydroxylation catalysts. Appl Environ Microbiol. 2003;69:373–382. doi: 10.1128/AEM.69.1.373-382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-G, Jung B-R, Lee Y, Hur H-G, Lim Y, Ahn J-H. Regiospecific flavonoid 7-O-methylation with Streptomyces avermitilis O-methyltransferase expressed in Escherichia coli. J Agric Food Chem. 2006;54:823–828. doi: 10.1021/jf0522715. [DOI] [PubMed] [Google Scholar]
- Kong D, Wang X, Nie J, Niu G (2019) Regulation of antibiotic production by signaling molecules in Streptomyces. Front Microbiol 10. 10.3389/fmicb.2019.02927 [DOI] [PMC free article] [PubMed]
- Kumar P, Sharma S, Malviya H, Balasubramanian R, Dalal A (2007) An improved fermentation process for preparing ascomycin Patent number WO 2007/029082A2
- Li M, Zhang Z-J, Kong X-D, Yu H-L, Zhou J, Xu J-H (2017) Engineering Streptomyces coelicolor carbonyl reductase for efficient atorvastatin precursor synthesis. Appl Environ Microbiol 83. 10.1128/AEM.00603-17 [DOI] [PMC free article] [PubMed]
- Liu S-P, Yu P, Yuan P-H, Zhou Z-X, Bu Q-T, Mao X-M, Li Y-Q. Sigma factor WhiGch positively regulates natamycin production in Streptomyces chattanoogensis L10. Appl Microbiol Biotechnol. 2015;99:2715–2726. doi: 10.1007/s00253-014-6307-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Huang L, Fu Y, Zheng D, Ma J, Li Y, Xu Z, Lu F. A novel process for phosphatidylserine production using a Pichia pastoris whole-cell biocatalyst with overexpression of phospholipase D from Streptomyces halstedii in a purely aqueous system. Food Chem. 2019;274:535–542. doi: 10.1016/j.foodchem.2018.08.105. [DOI] [PubMed] [Google Scholar]
- López-García MT, Rioseras B, Yagüe P, Álvarez JR, Manteca Á. Cell immobilization of Streptomyces coelicolor: effect on differentiation and actinorhodin production. Int Microbiol. 2014;17:75–80. doi: 10.2436/20.1501.01.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín JF, Liras P (2020) The balance metabolism safety net: integration of stress signals by interacting transcriptional factors in Streptomyces and related Actinobacteria. Front Microbiol 10. 10.3389/fmicb.2019.03120 [DOI] [PMC free article] [PubMed]
- Miranzo D, Seco EM, Cuesta T, Malpartida F. Isolation and characterization of pcsB, the gene for a polyene carboxamide synthase that tailors pimaricin into AB-400. Appl Microbiol Biotechnol. 2010;85:1809–1819. doi: 10.1007/s00253-009-2195-1. [DOI] [PubMed] [Google Scholar]
- Molnár I, Jungmann V, Stege J, Trefzer A, Pachlatko JP. Biocatalytic conversion of avermectin into 4″-oxo-avermectin: discovery, characterization, heterologous expression and specificity improvement of the cytochrome P450 enzyme. Biochem Soc Trans. 2006;34:1236–1240. doi: 10.1042/BST0341236. [DOI] [PubMed] [Google Scholar]
- Ng I-S, Ye C, Zhang Z, Lu Y, Jing K. Daptomycin antibiotic production processes in fed-batch fermentation by Streptomyces roseosporus NRRL11379 with precursor effect and medium optimization. Bioprocess Biosyst Eng. 2014;37:415–423. doi: 10.1007/s00449-013-1007-2. [DOI] [PubMed] [Google Scholar]
- Olano C, Méndez C, Salas JA. Molecular insights on the biosynthesis of antitumour compounds by actinomycetes. Microb Biotechnol. 2011;4:144–164. doi: 10.1111/j.1751-7915.2010.00231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos E, Mehmood N, Haj Husein L, Goergen J-L, Fick M, Delaunay S. Effects of bioreactor hydrodynamics on the physiology of Streptomyces. Bioprocess Biosyst Eng. 2013;36:259–272. doi: 10.1007/s00449-012-0794-1. [DOI] [PubMed] [Google Scholar]
- Perdani MS, Sahlan M, Yohda M, Hermansyah H. Immobilization of cholesterol oxidase from Streptomyces sp. on magnetite silicon dioxide by crosslinking method for cholesterol oxidation. Appl Biochem Biotechnol. 2020;191:968–980. doi: 10.1007/s12010-020-03241-w. [DOI] [PubMed] [Google Scholar]
- Pereira T, Nikodinovic J, Nakazono C, Dennis GR, Barrow KD, Chuck J-A. Community structure and antibiotic production of Streptomyces nodosus bioreactors cultured in liquid environments. Microb Biotechnol. 2008;1:373–381. doi: 10.1111/j.1751-7915.2008.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D. Microbiological conversion of pregnenolone to progesterone. Science. 1952;115:529. doi: 10.1126/science.115.2993.529. [DOI] [PubMed] [Google Scholar]
- Pervaiz I, Ahmad S, Madni MA, Ahmad H, Khaliq FH. Microbial biotransformation: a tool for drug designing (Review) Prikl Biokhim Mikrobiol. 2013;49:435–449. doi: 10.7868/s0555109913050097. [DOI] [PubMed] [Google Scholar]
- Qi H, Zhao S, Fu H, Wen J, Jia X. Enhancement of ascomycin production in Streptomyces hygroscopicus var. ascomyceticus by combining resin HP20 addition and metabolic profiling analysis. J Ind Microbiol Biotechnol. 2014;41:1365–1374. doi: 10.1007/s10295-014-1473-9. [DOI] [PubMed] [Google Scholar]
- Qi H, Zhao S, Wen J, Chen Y, Jia X. Analysis of ascomycin production enhanced by shikimic acid resistance and addition in Streptomyces hygroscopicus var. ascomyceticus. Biochem Eng J. 2014;82:124–133. doi: 10.1016/j.bej.2013.11.006. [DOI] [Google Scholar]
- Restaino OF, Marseglia M, De Castro C, Diana P, Forni P, Parrilli M, De Rosa M, Schiraldi C. Biotechnological transformation of hydrocortisone to 16α-hydroxy hydrocortisone by Streptomyces roseochromogenes. Appl Microbiol Biotechnol. 2014;98:1291–1299. doi: 10.1007/s00253-013-5384-x. [DOI] [PubMed] [Google Scholar]
- Restaino OF, Marseglia M, Diana P, Borzacchiello MG, Finamore R, Vitiello M, D’Agostino A, De Rosa M, Schiraldi C. Advances in the 16α-hydroxy transformation of hydrocortisone by Streptomyces roseochromogenes. Process Biochem. 2016;51:1–8. doi: 10.1016/j.procbio.2015.11.009. [DOI] [Google Scholar]
- Restaino OF, Barbuto Ferraiuolo S, Perna A, Cammarota M, Borzacchiello MG, Fiorentino A, Schiraldi C. Biotechnological transformation of hydrocortisone into 16α-hydroxyprednisolone by coupling Arthrobacter simplex and Streptomyces roseochromogenes. Molecules. 2020;25:4912. doi: 10.3390/molecules25214912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh C, Seo S-H, Choi K-Y, Cha M, Pandey BP, Kim J-H, Park J-S, Kim DH, Chang IS, Kim B-G. Regioselective hydroxylation of isoflavones by Streptomyces avermitilis MA-4680. J Biosci Bioeng. 2009;108:41–46. doi: 10.1016/j.jbiosc.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Romero-Rodríguez A, Rocha D, Ruiz-Villafan B, Tierrafría V, Rodríguez-Sanoja R, Segura-González D, Sánchez S. Transcriptomic analysis of a classical model of carbon catabolite regulation in Streptomyces coelicolor. BMC Microbiol. 2016;16:77. doi: 10.1186/s12866-016-0690-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz B, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Rodríguez-Sanoja R, Sánchez S, Langley E. Production of microbial secondary metabolites: regulation by the carbon source. Crit Rev Microbiol. 2010;36:146–167. doi: 10.3109/10408410903489576. [DOI] [PubMed] [Google Scholar]
- Sánchez S, Chávez A, Forero A, García-Huante Y, Romero A, Sánchez M, Rocha D, Sánchez B, Ávalos M, Guzmán-Trampe S, Rodríguez-Sanoja R, Langley E, Ruiz B. Carbon source regulation of antibiotic production. J Antibiot. 2010;63:442–459. doi: 10.1038/ja.2010.78. [DOI] [PubMed] [Google Scholar]
- Sanchez J, Yague P, Manteca A (2012) New insights in Streptomyces fermentations. Ferment Technol 1. 10.4172/2167-7972.1000e105 [DOI] [PMC free article] [PubMed]
- Scaffaro R, Lopresti F, Sutera A, Botta L, Fontana RM, Gallo G. Plasma modified PLA electrospun membranes for actinorhodin production intensification in Streptomyces coelicolor immobilized-cell cultivations. Colloids Surf B: Biointerfaces. 2017;157:233–241. doi: 10.1016/j.colsurfb.2017.05.060. [DOI] [PubMed] [Google Scholar]
- Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- Shen C, Zhao W, Liu X, Liu J. Enzyme-catalyzed regio-selective demethylation of papaverine by CYP105D1. Biotechnol Lett. 2019;41:171–180. doi: 10.1007/s10529-018-2626-0. [DOI] [PubMed] [Google Scholar]
- Simpson FJ, McCoy E. The amylases of five streptomycetes. Appl Microbiol. 1953;1:228–236. doi: 10.1128/AM.1.5.228-236.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Pandey B, Mathew CY. Production, purification and optimization of streptomycin from isolated strain of Streptomyces griseus and analysis by HPLC. Indian J Sci Res. 2014;4(1):149–154. [Google Scholar]
- Song X, Zhang Y, Xue J, Li C, Wang Z, Wang Y. Enhancing nemadectin production by Streptomyces cyaneogriseus ssp. noncyanogenus through quantitative evaluation and optimization of dissolved oxygen and shear force. Bioresour Technol. 2018;255:180–188. doi: 10.1016/j.biortech.2017.09.033. [DOI] [PubMed] [Google Scholar]
- Spasic J, Mandic M, Djokic L, Nikodinovic-Runic J (2018) Streptomyces spp. in the biocatalysis toolbox. Appl Microbiol Biotechnol 102:3513–3536. 10.1007/s00253-018-8884-x [DOI] [PubMed]
- Spycher PR, Amann CA, Wehrmüller JE, Hurwitz DR, Kreis O, Messmer D, Ritler A, Küchler A, Blanc A, Béhé M, Walde P, Schibli R. Dual, site-specific modification of antibodies by using solid-phase immobilized microbial transglutaminase. Chembiochem. 2017;18:1923–1927. doi: 10.1002/cbic.201700188. [DOI] [PubMed] [Google Scholar]
- Tamburini E, Perito B, Mastromei G. Growth phase-dependent expression of an endoglucanase encoding gene (eglS) in Streptomyces rochei A2. FEMS Microbiol Lett. 2004;237:267–272. doi: 10.1111/j.1574-6968.2004.tb09706.x. [DOI] [PubMed] [Google Scholar]
- Walker JB, Skorvaga M. Phosphorylation of streptomycin and dihydrostreptomycin by Streptomyces. Enzymatic synthesis of different diphosphorylated derivatives. J Biol Chem. 1973;248:2435–2440. doi: 10.1016/S0021-9258(19)44127-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Li D, Qu M, Durrani R, Yang B, Wang Y. Immobilized MAS1 lipase showed high esterification activity in the production of triacylglycerols with n-3 polyunsaturated fatty acids. Food Chem. 2017;216:260–267. doi: 10.1016/j.foodchem.2016.08.041. [DOI] [PubMed] [Google Scholar]
- Wentzel A, Bruheim P, Øverby A, Jakobsen ØM, Sletta H, Omara WAM, Hodgson DA, Ellingsen TE. Optimized submerged batch fermentation strategy for systems scale studies of metabolic switching in Streptomyces coelicolor A3(2) BMC Syst Biol. 2012;6:59. doi: 10.1186/1752-0509-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JX, Ogura K, Chen S, Rehm BHA. Bioengineered polyhydroxyalkanoates as immobilized enzyme scaffolds for industrial applications. Front Bioeng Biotechnol. 2020;8:156. doi: 10.3389/fbioe.2020.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Wang H, Fidan O, Qin Y, Xiao G, Zhan J. Identification of new glutamate decarboxylases from Streptomyces for efficient production of γ-aminobutyric acid in engineered Escherichia coli. J Biol Eng. 2019;13:24. doi: 10.1186/s13036-019-0154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhang W, Xiao G, Zhan J. Efficient production of gamma-aminobutyric acid by engineered Saccharomyces cerevisiae with glutamate decarboxylases from Streptomyces. Biotechnol Appl Biochem. 2020;67:240–248. doi: 10.1002/bab.1840. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang W, Liu J, Caiyin Q, Qiao J. Immobilization of Streptomyces thermotolerans 11432 on polyurethane foam to improve production of acetylisovaleryltylosin. J Ind Microbiol Biotechnol. 2015;42:105–111. doi: 10.1007/s10295-014-1545-x. [DOI] [PubMed] [Google Scholar]