Interferon (IFN)‐inducible 44 like (IFI44L) is an IFN‐stimulated gene, the expression of which is induced by IFN and human immunodeficiency virus (HIV)‐1 infection. However, the mechanism has not yet been determined. In this study, we cloned the promoter of the IFI44L gene and found that interferon regulatory factor‐1 could bind directly to the IFN‐stimulated response element in the IFI44L promoter to activate IFI44L. Furthermore, we demonstrated that HIV‐1 can activate the IFI44L promoter to influence the expression of IFI44L.
Keywords: HIV‐1, IFI44L, interferon, ISRE, promoter
Abstract
Interferon (IFN)‐inducible 44 like (IFI44L) is an IFN‐stimulated gene (ISG), which is located on the same chromosome as the known antiviral ISG IFI44. Expression of IFI44L is induced by IFN and HIV‐1 infection. However, the mechanism by which IFN‐I induces IFI44L production has not yet been determined. In this study, we analyzed transcriptional regulation of IFI44L via cloning of the IFI44L promoter. We found that IFI44L has two IFN‐stimulated response elements (ISRE), which are necessary for the basal level of IFI44L transcription. IFN‐I and IFN‐II can activate the IFI44L promoter through one of the two ISREs. IFN regulatory factor (IRF)‐1 can activate transcription of IFI44L by binding to one of the ISREs. Additionally, co‐transfection of the IFI44L promoter with an HIV‐1 infectious clone or HIV‐1 infection activated IFI44L promoter transcription, but did not upregulate IFI44L expression via ISREs. These findings will help to understand the interaction between IFI44L and HIV‐1, and aid in elucidation of the role of IFI44L in the antiviral innate immune response.
Abbreviations
- IFI44L
Interferon inducible 44 like
- IFN
interferon
- IRF
interferon regulatory factor
- ISG
interferon‐stimulated gene
- ISRE
IFN‐stimulated response element
- STAT
signal transducer and activator of transcription
Interferon (IFN)‐inducible 44 like (IFI44L) is a part of the IFI44 family and is located on the same human chromosome as the previously identified antiviral interferon‐stimulated gene (ISG) IFI44, which is involved in numerous signaling pathways of the innate immune response. At the same time, IFI44L is also an ISG and can be induced by many different viruses such as influenza and the HIV‐1 [1, 2]. HIV‐1 proteins can affect the expression of IFI44L through different regulatory pathways in different cell types. For example, HIV‐1 gp120 can downregulate the expression of IFI44L by interaction with α4β7 [3]. Also, the transcription level of IFI44L is significantly upregulated in HIV‐1 gp120‐treated vaginal epithelial cells [4]. Additionally, HIV‐1 Vpr can upregulate the expression of IFI44L [5]. However, the mechanism by which HIV‐1 induces IFI44L up‐regulation has not been determined. In addition to HIV‐1, when rhesus monkeys are infected by the simian immunodeficiency virus, IFI44L is significantly upregulated up to 38‐fold on the 10th day of infection, and the up‐regulation of IFI44L is accompanied by up‐regulation of ISGs [6].
There are three types of IFNs: IFN‐I, IFN‐II, and IFN‐Ⅲ, which participate in a variety of biological activities, including antiviral response, antitumor, inflammatory response, and immunomodulatory activity [7, 8, 9]. IFNs regulate transcription of ISGs through the Janus kinase/signal transducer and activator of transcription (STAT) pathway [10]. Using the IFN‐I signaling pathway as an example, after IFN binds to its receptor, the receptor‐associated tyrosine kinases Tyk2 and Jak1 are activated, and then, STAT1 and STAT2 are phosphorylated to form a heterodimer complex and translocated to the nucleus. The complex is assembled with interferon regulatory factor (IRF)‐9 and binds to the IFN‐stimulated regulatory element (ISRE) on the ISG promoter to induce ISGs transcription [11, 12, 13, 14, 15].
Interferon regulatory factors participate in the IFN‐mediated signaling pathway. Nine members of the IRFs family have been identified in human cells. For example, IRF‐1 can bind the promoter region of IFN‐β and activate transcription. IRF‐2 can also bind to the IFN‐β promoter, but it inhibits IFN‐β expression [16]. IRF3 plays a major role in the transcription of the gene encoding IFN‐1. IRF‐5 plays a role in the immune response against viral infections. IRF‐7 can bind to MyD88 and TRIF and increases the production of IFN‐I induced by the stimulation of some TLRs [9, 17, 18, 19].
The expression of IFI44L in HIV‐1 progressor blood samples are significantly higher than that in nonprogressors, and IFI44L levels are upregulated by IFN‐α [2]. However, the mechanism by which IFN‐I induces IFI44L production is not yet determined. In this study, we cloned the promoter of IFI44L gene to better understand its transcriptional regulation. We found IRF‐1 could bind directly to the ISRE in the IFI44L promoter to activate IFI44L. Furthermore, we demonstrated that HIV‐1 can activate the IFI44L promoter to influence the expression of IFI44L.
Materials and methods
Constructs and antibodies
Plasmids pGL‐1190, pGL‐695, pGL‐390, pGL‐274, pGL‐143, pGL‐117‐1190, pGL‐237‐1190, pGL237‐390, and pGL380‐1190 were constructed by cloning the PCR‐amplified fragment of the IFI44L promoter into pGL3‐basic (Promega, Madison, WI, USA), primers are listed in Table 1. pGL‐274mISRE‐1 and pGL‐237‐1190mISRE‐2 were constructed using site‐directed mutagenesis kit (Toyobo, Osaka, Japan), and primers used are listed in Table 1. Plasmids IRF‐1, IRF‐2, IRF‐3, IRF‐5, and IRF‐7 were constructed by inserting the CDS into the pCDNA3.1 (+) (Invitrogen, Carlsbad, CA, USA) or pCMV‐Tag3B (Stratagene, La Jolla, CA, USA). Sequences of all constructs were confirmed by sequencing.
Table 1.
Primers for amplifying IFI44L promoter fragments and site‐directed mutagenesis.
| Primer | Sequence (5′–3′) |
|---|---|
| p1190 | GAGCCGCCTTAATAACTTCTC |
| p695 | GCCTCGGCAGAGACGACCCAGGAGA |
| p390 | CGACGCGTGCCTGCCTACATACATAC |
| p274 | CGACGCGTCTGCCAGCTGAGTTTTTTTGC |
| p143 | CGACGCGTCTTTCTTTCCTAGAGTCTCTG |
| p117‐1190‐R | CCCTCGAGGGCTTCAGAGACTCTAGGAAA |
| p237‐1190‐R | CCCTCGAGCCTGAAACTCAAAGCAGCAA |
| p380‐1190‐R | CCCTCGAGGGCAGGCATGAAATGATAAC |
| mISRE‐F | CTCTTCCTAGTGAGGACAAAGACAGTTAGTGGCAGTTG |
| mISRE‐R | CAACTGCCACTAACTGTCTTTGTCCTCACTAGGAAGAG |
Antibodies against Myc, IRF‐1, IRF‐2, Tubulin, IgG, and RNA Polymerase were purchased from Sigma (St. Louis, MO, USA), and antibody against IFI44L was purchased from Aviva (San Diego, CA, USA).
Cell culture and transfection
293T, HeLa, and TZM‐bl cells (maintained in our lab) were grown in Dulbecco's Modified Eagle's medium (Gibco, Gaithersburg, MD, USA) medium supplemented with 10% FBS (BI) in 5% CO2 at 37 °C. Jurkat cells were grown in RPMI 1640 (Gibco) medium supplemented with 10% FBS (BI) in 5% CO2 at 37 °C. Transfection was performed using Polyetherimide reagent (Sigma‐Aldrich) and Lipo3000.
HIV‐1 pseudovirus preparation
The HIV‐1 pseudovirus was prepared by transfecting 293T cells with NL4‐3 and pVSV‐G. To prepare cell‐free HIV‐1 stocks, culture supernatants were cleared by low‐speed centrifugation (3000 g for 10 min), filtered through a 0.22‐μm‐pore‐size filter membrane, and kept at 4 °C. HIV‐1 titers were determined by infecting TZM‐bl cells.
Chromatin immunoprecipitation (ChIP)
ChIP analysis was conducted based on manufacturer's instructions using EZ‐chip kit (Millipore, Billerica, MA, USA). PCR primers used for amplifying the IFI44L promoter are as follows: forward, 5′ CAAGGGGACCAGTGATAG‐3′ and reverse: 5′‐GATCTGTGGCTTCAGAGACTC‐3′.
Western blot
Cells were washed with PBS and lysed in lysis buffer [20 mm Tris (pH 7.4), 150 mm NaCl, 3% glycerol, 0.25% sodium deoxycholate, 1% NP‐40, complete protease inhibitor cocktail tablets (Roche, Basel, Switzerland)]. Cell lysates were applied to 10% SDS/PAGE and subsequently blotted onto Polyvinylidene fluoride membrane (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The membranes were blocked and incubated with primary antibodies and then with HRP‐conjugated secondary antibodies. Enhanced chemiluminescence detection reagents (Millipore) were used for signal detection.
Luciferase reporter assay
HeLa cells were transfected with luciferase plasmids and β‐galactosidase expression plasmids. Luciferase activity was determined with luciferase report system (Promega) and normalized to β‐galactosidase activity.
Results
Promoter activity analysis of IFI44L
To clarify the mechanism how IFI44L gene is induced by IFNs, we first studied the transcriptional regulation mechanism of IFI44L. We used the translation start site (ATG) as +1 and determined that the first exon of IFI44L is located at −16 to −183 bp through comparing the IFI44L genomic sequence in UCSU, Ensembl, and the IFI44L mRNA sequence in NCBI. Then, we cloned the 1190 bp DNA upstream of the transcription initiation site and performed sequence analysis. The results showed that it contained two ISRE components (Fig. 1A). To verify whether the cis‐acting elements are involved in the transcription, a series of deletions of the promoter were constructed (Fig. 1B). We transfected these reporter plasmids into HeLa cells and measured the luciferase activity 48 h post‐transfection. As shown in Fig. 1C, the basal transcriptional activity of truncated IFI44L promoter from +1 to −274 (with the ISRE‐2 deleted) decreased moderately, while the basal transcriptional activity of the IFI44L promoter was further reduced when the truncation spanned position −237 to −1190 (with the ISRE‐1 deleted). In addition, we also studied the basal transcriptional activity of pGL‐390 in Jurkat cells to ensure the reliability of the above results, and similar results were obtained (Fig. 1D). In order to further confirm whether the two ISREs are involved in regulating the basic transcriptional activity of the IFI44L promoter, we constructed a series of mutations on pGL‐274 and pGL‐237‐1190 (Fig. 1E). As shown in Fig. 1F,G, the basal transcriptional activity decreased by 72% (ISRE‐1 mutation) and 14% (ISRE‐2 mutation). Furthermore, when the two elements were mutated, pGL‐143 completely lost the promoter activity compared to the empty vector. Therefore, we concluded that the two ISREs are essential elements in the basal transcription of IFI44L.
Fig. 1.
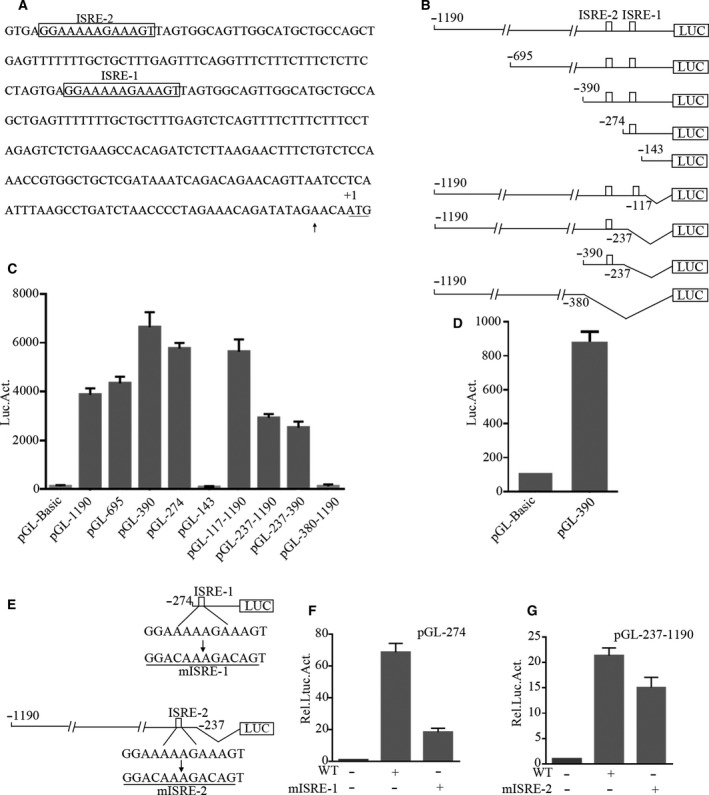
Identification and analysis of IFI44L promoter. (A) Partial sequence of IFI44L promoter. Arrow indicates the starting point of transcription and the A is designed as +1. The potential cis‐elements are boxed. (B) The 5′‐truncated plasmids of IFI44L promoter. (C) The promoter constructs (200 ng) were transfected into HeLa cells (1 × 105). Forty‐eight hours post‐transfection, luciferase assay was performed, and β‐galactosidase activity was used as a normalization control for the luciferase activity. (D) The pGL‐390 or pGL‐Basic (600 ng) was transfected into Jurkat cells (1 × 106). Forty‐eight hours post‐transfection, luciferase assay was performed, and β‐galactosidase activity was used as a normalization control for the luciferase activity. (E) Mutations of ISRE on pGL‐274 and pGL‐237‐1190, respectively. (F) The wild‐type or mutant pGL‐274 and (G) pGL‐237‐1190 or mutant pGL‐237‐1190 (200 ng) were transfected into HeLa cells (1 × 105), and luciferase was detected 48 h post‐transfection. The results shown represent the averages of the results of three independent experiments. Error bars indicate SD.
The ISRE‐1 element is essential for IFNs to activate IFI44L promoter
To verify whether the IFI44L promoter responds to IFNs, HeLa cells were transfected with pGL‐274 or pGL‐237‐1190; 36h post‐transfection, cells were treated with IFN‐α or IFN‐γ for 12 h, respectively. As shown in Fig. 2A, both IFN‐α and IFN‐γ could activate pGL‐274 (with ISRE‐1). Besides, we also transfected pGL‐274 into Jurkat cells and treated with IFNs, and got similar results as 2A (Fig. 2B). Furthermore, IFN‐α was more potent than IFN‐γ at inducing promoter response. However, IFN‐α and IFN‐γ could not activate pGL‐237‐1190 (with ISRE‐2). To further confirm whether IFNs activate pGL‐274 via the ISRE‐1, we constructed ISRE‐1 mutant plasmids (pGL274mISRE‐1). We found that the IFI44L promoter did not respond to IFNs when the ISRE‐1 was mutated (Fig. 2C,D). These results demonstrated that the ISRE‐1 is essential for IFNs to activate the IFI44L promoter.
Fig. 2.

IFNs activate IFI44L transcription via ISRE‐1. (A) pGL‐274 or pGL‐237‐1190 (200 ng) was transfected into HeLa cells (1 × 105). Thirty‐six hours post‐transfection, the cells were treated with IFN‐α or IFN‐γ (10 ng·mL−1) for 12 h. Determination of luciferase activity after 48 h transfection. (B) pGL‐274 (600 ng) was transfected into Jurkat cells (1 × 106). Thirty‐six hours post‐transfection, the cells were treated with IFN‐α or IFN‐γ (10 ng·mL−1) for 12 h. Determination of luciferase activity after 48‐h transfection. (C and D) pGL‐274, pGL‐274mISRE‐1 or pGL‐237‐1190, pGL‐237‐1190mISRE‐2 (200 ng) were transfected into HeLa cells (1 × 105), and then, cells were stimulated with IFN‐α or IFN‐γ (10 ng·mL−1) 12 h before luciferase detection. The results shown represent the averages of the results of three independent experiments. Error bars indicate SD.
IRF‐1 binds to the ISRE to increase IFI44L expression
The IRF family can participate in the transcriptional regulation of the target genes. In order to identify whether the IRF family is involved in regulating IFI44L, we co‐transfected Myc ‐IRF‐1, 3.1‐IRF‐2, Myc‐IRF‐3, Myc‐IRF‐5, Myc‐IRF‐7 encoding plasmids with pGL‐390 into HeLa cells and measured the transcription activity of pGL‐390. As shown in Fig. 3A,B, overexpression of IRF‐1 significantly activated pGL‐390 in HeLa cells, the trend measured by western blot was consistent with luciferase results, but protein levels were not as sensitive as transcriptional levels. In order to further confirm that IRF‐1 can upregulate the IFI44L promoter, we co‐transfected the IFI44L promoter and dose‐gradient Myc‐IRF‐1 into 293T cells, and found that IFI44L promoter could be upregulated by IRF‐1 in a dose‐dependent manner (Fig. 3C). We then further explored whether IRF‐1 could activate the IFI44L promoter via the ISRE element. We co‐transfected Myc‐IRF‐1 and pGL‐274mISRE‐1 or pGL‐237‐1190mISRE‐2 into HeLa cells. As shown in Fig. 3D,E, IRF‐1 did not activate the pGL‐274mISRE‐1, indicating that IRF‐1 activated the IFI44L promoter via the ISRE‐1 element. Then we examined whether endogenous IRF‐1 could also interact with ISRE when stimulated by IFN. ChIP assays were performed after treating HeLa cells with IFN‐α and followed by PCR. The results showed that endogenous IRF‐1 could interact with the ISRE of IFI44L (Fig. 3F). The results above indicate that IRF‐1 can bind to the ISRE of IFI44L promoter to regulate IFI44L expression.
Fig. 3.
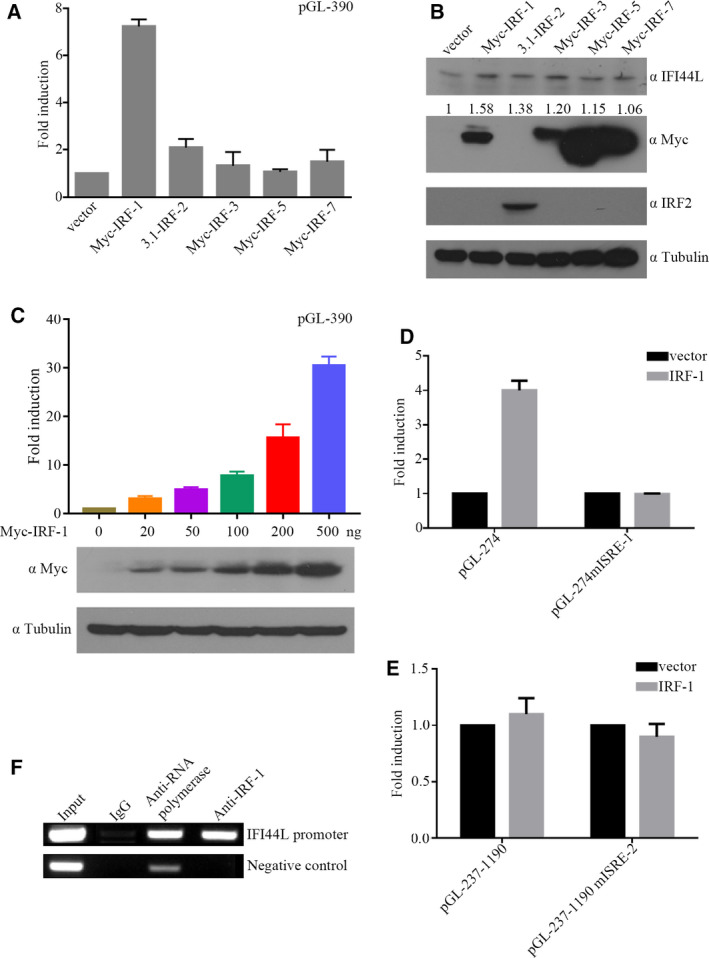
IRF‐1 binds to the ISRE‐1 to activate IFI44L promoter. (A and B) pGL‐390 (200 ng) with IRFs (400 ng) transfected into HeLa cells (1 × 105), 48 h post‐transfection, luciferase activity and western blot analysis was performed. (C) 293T cells (2 × 105) were transfected with 200 ng pGL‐390 and 20, 50, 100, 200, 500 ng IRF‐1. (D) pGL‐274, pGL‐274mISRE‐1 (200 ng) or (E) pGL‐237‐1190, pGL‐237‐1190 mISRE‐2 (200 ng) were co‐transfected with pcDNA3.1 or pcDNA3.1‐IRF‐1 (400 ng) into HeLa cells (1 × 105). Luciferase activity was measured 48 h after transfection. (F) HeLa cells (1 × 105) were treated with IFN‐α (10 ng·mL−1) for 12 h and detected with anti IRF‐1, control IgG, and anti‐RNA polymerase. Precipitated DNA was amplified by PCR with primers designed for IFI44L ISRE (up) or GAPDH promoter (down). The results shown represent the averages of the results of three independent experiments. Error bars indicate SD.
HIV‐1 can activate IFI44L promoter
To determine whether HIV‐1 can upregulate IFI44L promoter activity, we co‐transfected SG3Δenv and pGL‐390 into 293T cells. As shown in Fig. 4A,B, HIV‐1 can upregulate IFI44L promoter activity. Furthermore, co‐transfecting SG3Δenv and pGL‐274, pGL237‐1190, pGL‐274mISRE‐1, pGL‐237‐1190mISRE‐2 into 293T cells, HIV‐1 did not directly interact through ISRE elements to upregulate IFI44L promoter activity (Fig. 4C,D). To further confirm that HIV‐1 can upregulate IFI44L promoter, we co‐transfected 293T cells with dose‐gradient NL4‐3Δenv and pGL‐390, or infected 293T cells with HIV‐1 pseudovirus with dose gradient after pGL‐390 transfection. The results showed that HIV‐1 can upregulate IFI44L promoter in a dose‐dependent manner (Fig. 4E,F).
Fig. 4.
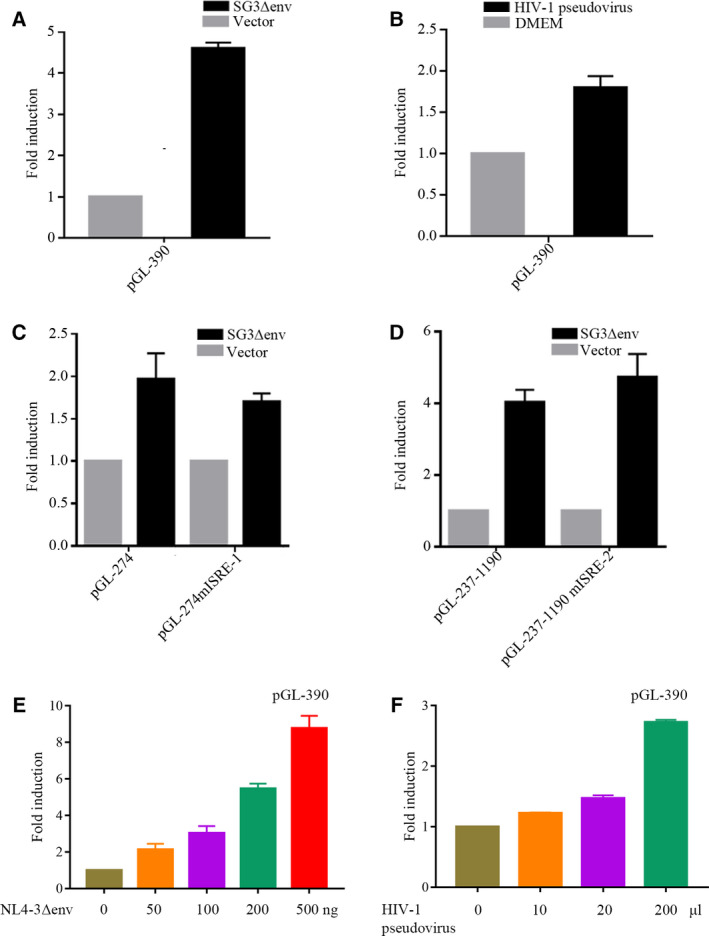
HIV‐1 activates IFI44L promoter. (A) 293T cells (2 × 105) were transfected with SG3Δenv (500 ng) and pGL‐390 (200 ng), 48 h post‐transfection, luciferase assays were performed. (B) 293T cells (2 × 105) were transfected with pGL‐390 (200 ng); after 8 h, cells were infected with HIV‐1 pseudovirus. Luciferase assays were performed. (C, D) 293T cells (2 × 105) were transfected with SG3Δenv (500 ng) and pGL‐274, pGL‐274 mISRE‐1 (200 ng) or pGL237‐1190, pGL‐237‐1190mISRE‐2 (200 ng), 48 h post‐transfection, luciferase assays were performed. (E) 293T cells (2 × 105) were transfected with 200 ng pGL‐390 and 50, 100, 200, 500 ng NL4‐3Δenv; 48 h post‐transfection, luciferase assays were performed. (F) 293T cells (2 × 105) were transfected with 200 ng pGL‐390, after 8 h, cells were infected with 10, 20, 200 μL HIV‐1 pseudovirus. Luciferase assays were performed. The results shown represent the averages of the results of three independent experiments. Error bars indicate SD.
Discussion
Interferons can activate the expression of ISGs through the JAK‐STAT pathway, which inhibit viruses at different stages of replication. HIV‐1 is able to regulate a number of ISGs to ensure its own infection [20, 21]. As an ISG, IFI44L can be induced by IFNs and HIV‐1. In this study, we explored the mechanism of how IFI44L is induced by IFNs and HIV‐1.
First, we cloned and analyzed the features of the IFI44L promoter and we found that the IFI44L promoter has two ISREs, but no TATA and GC boxes. Two ISRE components are located in the complete repeat sequence [22, 23]. We identified that the basal transcriptional activity of IFI44L is significantly decreased when the ISREs are mutated, indicating that these two ISRE elements are essential for the transcriptional regulation of IFI44L, especially ISRE‐1. This is similar to some other ISGs, which are controlled by ISRE. After IFN‐I stimulates cells, a heterodimer complex is formed by phosphorylation of STAT1 and STAT2, which is assembled with IRF‐9 and induces ISGs production by binding to ISRE [11, 12, 13, 14, 15]. Furthermore, we determined that IFN‐I can activate IFI44L promoter and that it is through ISRE‐1 rather than ISRE‐2. IFN‐Ⅱ can also induce up‐regulation of IFI44L promoter activity, but the upregulate level is lower than that of IFN‐I. We suspect that this upregulation of IFN‐II is indirect because we cannot be sure that there is a gamma‐interferon activation site element on the IFI44L promoter.
IFN regulatory factors are a class of transcriptional regulatory proteins that regulate the expression of IFNs and ISGs. Therefore, we also tested whether IRFs could activate IFI44L. We observed that IRF‐1 can significantly activate the transcription of IFI44L. This is because IRF‐1 can recognize ISRE and activate the transcription of IFN‐I and ISGs [24, 25], which indicates that IFI44L may participate in the host's innate immunity through these IRFs.
As mentioned earlier, HIV‐1 proteins can affect the expression of IFI44L through different regulatory pathways in different cell types. Furthermore, we demonstrated that HIV‐1 can affect the expression of IFI44L by upregulating the activity of the IFI44L promoter in 293T cells. Besides, Lu et al. [26] reports that high‐throughput screening experiments at the cellular level have detected a significant increase in HIV‐1 replication levels after IFI44L has been knocked out. We speculated that IFI44L plays an immunoregulatory role for HIV‐1 in host cells, one possible reason is that HIV‐1 can ensure latent infection by up‐regulating the expression of IFI44L. However, after mutation of ISRE, HIV‐1 can still upregulate the IFI44L promoter activity, therefore, HIV‐1 did not pass the ISRE to affect the IFI44L promoter activity. The mechanism by which HIV upregulates the promoter activity of IFI44L needs further study.
In summary, this study found that IFN‐I and IFN‐II can upregulate the transcription of the IFI44L promoter, and HIV‐1 can activate the IFI44L promoter to influence the expression of IFI44L. We will continue to explore the specific mechanism by which HIV‐1 upregulates IFI44L initiation activity. This will help to understand the more physiological functions of IFI44L and the interaction between IFI44L and HIV‐1, and determine the status of IFI44L in antiviral innate immune response.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
YL, JT, YL, and JZ designed the study. YL and JZ drafted the manuscript, and JT and YL helped modify the manuscript. YL, JZ, and CW performed the experiments. YL and JZ contributed equally to the work. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31870161).
Yutong Li and Junshi Zhang contributed equally to the work
Contributor Information
Yue Li, Email: yli3685@uwo.ca.
Juan Tan, Email: juantan@nankai.edu.cn.
Data Accessibility
Data will be available from the corresponding author upon reasonable request.
References
- 1. Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Nino D, Wang X, Zapata GE et al (2015) Host transcriptional response to influenza and other acute respiratory viral infections–a prospective cohort study. PLoS Pathog 11, e1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu X, Qiu C, Zhu L, Huang J, Li L, Fu W, Zhang L, Wei J, Wang Y, Geng Y et al (2014) IFN‐stimulated gene LY6E in monocytes regulates the CD14/TLR4 pathway but inadequately restrains the hyperactivation of monocytes during chronic HIV‐1 infection. J Immunol 193, 4125–4136. [DOI] [PubMed] [Google Scholar]
- 3. Jelicic K, Cimbro R, Nawaz F, da Huang W, Zheng X, Yang J, Lempicki RA, Pascuccio M, Van Ryk D, Schwing C et al (2013) The HIV‐1 envelope protein gp120 impairs B cell proliferation by inducing TGF‐beta1 production and FcRL4 expression. Nat Immunol 14, 1256–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fanibunda SE, Modi DN and Bandivdekar AH (2013) HIV gp120 induced gene expression signatures in vaginal epithelial cells. Microbes Infect 15, 806–815. [DOI] [PubMed] [Google Scholar]
- 5. Zahoor MA, Xue G, Sato H, Murakami T, Takeshima SN and Aida Y (2014) HIV‐1 Vpr induces interferon‐stimulated genes in human monocyte‐derived macrophages. PLoS One 9, e106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu W, Ma F, Churbanov A, Wan Y, Li Y, Kang G, Yuan Z, Wang D, Zhang C, Xu J et al (2014) Virus‐host mucosal interactions during early SIV rectal transmission. Virology 464–465, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taylor KE and Mossman KL (2013) Recent advances in understanding viral evasion of type I interferon. Immunology 138, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ivashkiv LB and Donlin LT (2014) Regulation of type I interferon responses. Nat Rev Immunol 14, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tamura T, Yanai H, Savitsky D and Taniguchi T (2008) The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26, 535–584. [DOI] [PubMed] [Google Scholar]
- 10. Sadler AJ and Williams BR (2008) Interferon‐inducible antiviral effectors. Nat Rev Immunol 8, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honda K, Takaoka A and Taniguchi T (2006) Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360. [DOI] [PubMed] [Google Scholar]
- 12. Lefebvre S, Berrih‐Aknin S, Adrian F, Moreau P, Poea S, Gourand L, Dausset J, Carosella ED and Paul P (2001) A specific interferon (IFN)‐stimulated response element of the distal HLA‐G promoter binds IFN‐regulatory factor 1 and mediates enhancement of this nonclassical class I gene by IFN‐beta. J Biol Chem 276, 6133–6139. [DOI] [PubMed] [Google Scholar]
- 13. Hervas‐Stubbs S, Perez‐Gracia JL, Rouzaut A, Sanmamed MF, Le Bon A and Melero I (2011) Direct effects of type I interferons on cells of the immune system. Clin Cancer Res 17, 2619–2627. [DOI] [PubMed] [Google Scholar]
- 14. Ihle JN (1996) STATs: signal transducers and activators of transcription. Cell 84, 331–334. [DOI] [PubMed] [Google Scholar]
- 15. Darnell JE Jr, Kerr IM and Stark GR (1994) Jak‐STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- 16. Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T and Taniguchi T (1989) Structurally similar but functionally distinct factors, IRF‐1 and IRF‐2, bind to the same regulatory elements of IFN and IFN‐inducible genes. Cell 58, 729–739. [DOI] [PubMed] [Google Scholar]
- 17. Richez C, Barnetche T, Miceli‐Richard C, Blanco P, Moreau JF, Rifkin I and Schaeverbeke T (2010) Role for interferon regulatory factors in autoimmunity. Joint Bone Spine 77, 525–531. [DOI] [PubMed] [Google Scholar]
- 18. Ning S, Pagano JS and Barber GN (2011) IRF7: activation, regulation, modification and function. Genes Immun 12, 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu WD, Zhang YJ, Xu K, Zhai Y, Li BZ, Pan HF and Ye DQ (2012) IRF7, a functional factor associates with systemic lupus erythematosus. Cytokine 58, 317–320. [DOI] [PubMed] [Google Scholar]
- 20. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH and Williams BR (2001) Functional classification of interferon‐stimulated genes identified using microarrays. J Leukoc Biol 69, 912–920. [PubMed] [Google Scholar]
- 21. Wie SH, Du P, Luong TQ, Rought SE, Beliakova‐Bethell N, Lozach J, Corbeil J, Kornbluth RS, Richman DD and Woelk CH (2013) HIV downregulates interferon‐stimulated genes in primary macrophages. J Interferon Cytokine Res 33, 90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Uccellini MB and Garcia‐Sastre A (2018) ISRE‐reporter mouse reveals high basal and induced type I IFN responses in inflammatory monocytes. Cell Rep 25, 2784–2796.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho HY, Lee SW, Seo SK, Choi IW, Choi I and Lee SW (2008) Interferon‐sensitive response element (ISRE) is mainly responsible for IFN‐alpha‐induced upregulation of programmed death‐1 (PD‐1) in macrophages. Biochim Biophys Acta 1779, 811–819. [DOI] [PubMed] [Google Scholar]
- 24. Saito H, Tada S, Ebinuma H, Wakabayashi K, Takagi T, Saito Y, Nakamoto N, Kurita S and Ishii H (2001) Interferon regulatory factor 1 promoter polymorphism and response to type 1 interferon. J Cell Biochem Suppl 36, 191–200. [DOI] [PubMed] [Google Scholar]
- 25. Gongora C, Degols G, Espert L, Hua TD and Mechti N (2000) A unique ISRE, in the TATA‐less human Isg20 promoter, confers IRF‐1‐mediated responsiveness to both interferon type I and type II. Nucleic Acids Res 28, 2333–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu J, Pan Q, Rong L, He W, Liu SL and Liang C (2011) The IFITM proteins inhibit HIV‐1 infection. J Virol 85, 2126–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be available from the corresponding author upon reasonable request.


