Arbutin, a natural polyphenol, inhibits OS cell viability, invasion and migration by down‐regulating MTHFD1L expression. miR‐338‐3p directly targets MTHFD1L. This regulatory action is associated with the inactivation of the AKT/mTOR signaling pathway.
Keywords: AKT/mTOR, Arbutin, miR‐338‐3p, MTHFD1L, osteosarcoma, proliferation
Abstract
Arbutin, a glycoside extracted from the plant Arctostaphylos uva‐ursi, has been previously reported to possess antioxidant, anti‐inflammatory and anticancer effects. Here, we investigated whether arbutin affects the proliferation of the cells of the osteosarcoma (OS) cell lines MG‐63 and SW1353. Arbutin suppressed OS cell viability in a dose‐ and time‐dependent manner, as shown by Cell Counting Kit‐8 assay. Furthermore, arbutin exposure decreased the protein levels of MTHFD1L, CCND1 and phosphorylated‐protein kinase B (AKT)/phosphorylated‐mammalian target of rapamycin (mTOR). Potential upstream miRNAs of MTHFD1L were predicted using TargetScan, PICTAR5, miRanda and miRWalk. We performed luciferase activity assays to show that miR‐338‐3p directly targets and negatively regulates the expression of MTHFD1L. Knockdown of miR‐338‐3p promoted cell invasion, migration and proliferation in arbutin‐treated OS cells via MTHFD1L. In summary, our data suggest that arbutin inhibits OS cell proliferation, migration and invasion via miR‐338‐3p/MTHFD1L and by inactivating the AKT/mTOR pathway.
Abbreviations
- AKT
protein kinase B
- CCK‐8
Cell Counting Kit‐8
- CTDbase
Comparative Toxicogenomics Database
- GEO
Gene Expression Omnibus
- miRNA
microRNA
- mTOR
mammalian target of rapamycin
- mut
mutant
- NC
negative control
- OS
osteosarcoma
- p
phosphorylated
- qRT‐PCR
quantitative real‐time PCR
- SD
standard deviation
- si‐
small interfering
- wt
wild‐type
Osteosarcoma (OS) is a malignant neoplasm of the bone, which originates from the distal end of long bone and is most common in young adolescents [1]. Despite that prodigious progress has been achieved in OS treatment, the long‐term overall survival rate of patients with OS was still unfavorable, attributed to its sharp progression and early metastasis [2, 3]. The current main therapy is surgical resection with systemic multidrug chemotherapy, which eventually leads to serious resistance, aggravates metastasis and increases recurrence rate [4].
Increasing publications indicate that the wide utility of natural agents opens up a novel avenue for the management of cancers [5]. Arbutin, also namely as 4‐hydroxyphenyl‐β‐d‐glucopyranoside, is a natural polyphenol extracted from the bearberry plant Arctostaphylos uva‐ursi (molecular mass: 272 Da) [6]. It exhibits a range of biological activities. Traditionally, Arbutin is used as a skin‐whitening or depigmenting agent [7]. The antioxidant, anti‐inflammatory, and even anticancer capabilities of Arbutin have been fully focused [8]. Arbutin is able to attenuate the melanin content, as well as tyrosinase activity, in melanoma cells [9]. Previous study has indicated that Arbutin could facilitate apoptosis of human bladder cancer and melanoma A375 cells [10, 11]. Nawarak et al. [11] have confirmed that the repression of Arbutin to aggressive melanoma development is associated with the dysregulation of p53, ENOA and VIME. Based on our prior study, MTHFD1L has been concluded as a prospective therapeutic target for OS [12]. The Comparative Toxicogenomics Database (CTDbase) website analysis revealed that Arbutin treatment can decrease the expression of MTHFD1L. However, the effects of Arbutin in OS progression are largely unknown.
Cumulating investigations manifest that the progression of malignant OS is related to the aberrant microRNAs (miRNAs) expression. miR‐338‐3p serves as a tumor repressor in different human cancers, such as breast cancer [13], colorectal cancer [14], non‐small cell lung cancer [15] and prostate cancer [16]. More than that, miR‐338‐3p has been documented to hinder proliferation, invasion and other malignant behaviors in OS cells via targeting ATPase homolog 1 or RUNX2/CDK4 [17, 18]. Bioinformatics analysis of our work predicted that miR‐338‐3p may be an upstream of MTHFD1L in OS. It is not known, however; there are still rare investigations on its association between Arbutin and MTHFD1L in OS.
Hence we detected the pharmacological impacts of Arbutin on cell viability, invasion and migration in OS cells. Furthermore, we also evaluated whether the miR‐338‐3p/MTHFD1L axis is implicated in the regulatory action of Arbutin in the progression of OS.
Materials and methods
Arbutin and cell culture
Arbutin (cat. no HY‐N0192; purity > 98%) was obtained from MedChemExpress LLC (Monmouth Junction, NJ, USA). It was dissolved in DMSO solution (Sigma, St Louis, MO, USA) and then stored at 4 °C. Two OS cell lines (MG‐63 and SW1353) were obtained from American Type Culture Collection (Manassas, VA, USA). The culture medium was Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA, USA) containing 1% antibiotics (penicillin and streptomycin) and 10% FBS (Invitrogen). The culture condition was 37 °C in carbon dioxide incubator with 5% CO2.
Transfection
Small interfering (si) ‐MTHFD1L to knock down MTHFD1L expression, miR‐338‐3p inhibitor to reduce miR‐338‐3p expression and their corresponding controls were all synthesized by GenePharma company (Shanghai, China). Cells (5 × 105) were inoculated in six‐well plates to achieve more than 80% confluency proceeding to transfection using Lipofectamine 2000 (Life Technologies, Grand Island, NE, USA). After 24 h of transfection, quantitative real‐time PCR was conducted to quantify the MTHFD1L or miR‐338‐3p expression.
Cell viability analysis
Cell Counting Kit‐8 (CCK‐8) (Beyotime, Beijing, China) was used to estimate the proliferative ability of cells. Cells (1000 cells for each well) treated by Arbutin were seeded in 96‐well plates in 37 °C with 5% CO2, followed by the treatment of CCK‐8 solution (10 μL per well) at indicated time points and 1.5‐h additional culture. The absorbance (A) value was recorded by a Tecan Group microplate reader (Shanghai, China) with the wavelength of 450 nm.
Cell migration and invasion assay
Transwell chamber (8‐μm pore size; BD Biosciences, Franklin Lakes, NJ, USA) was applied to detect the migratory and invasive capabilities of OS cells. In brief, Matrigel was bio‐coated on the top surface of transwell chambers for cell invasion analysis. The treated cells were subsequently cultivated at 37 °C with 5% CO2 for 48 h. These cells were resuspended into Dulbecco's modified Eagle's medium without serum, 100 μL (1 × 105) of cell suspension was placed into the top surface, while 500 μL of conventional culture medium with 10% FBS was put on the lower chamber. Overnight, invaded cells passed through Matrigel or migrated cells passed through polyethylene membrane were subsequently fixed by 4% paraformaldehyde. The crystal violet staining was used to show the invasive and migratory cells using a light microscope (Motic, Xiamen, China).
Luciferase activity assay
Dual‐luciferase Reporter Assay Kit was used to determine the relevance between miR‐338‐3p and MTHFD1L in OS cells. First, the sequences of MTHFD1L carrying the binding sites to miR‐338‐3p or not were amplified into luciferase reporter vector pmirGLO, which was named wild‐type (wt)‐MTHFD1L and mutant (mut)‐MTHFD1L. After 24 h, cells transfected with miR‐338‐3p inhibitor, the specific negative control (NC) and wt‐MTHFD1L or mut‐MTHFD1L were collected to measure the luciferase activity.
Quantitative real‐time PCR
The expression of MTHFD1L of transfected OS cells was examined with quantitative real‐time PCR test. Total RNA of cells with various treatments was isolated by TRIzol reagent and then reverse transcribed into cDNA by TaqMan Reverse Transcription Kit (Thermo Fisher Scientific, San Jose, CA, USA). SYBR Premix Ex Taq II (Takara, Dalian, China) was used to conduct quantitative real‐time PCR on the basis of protocols provided by the manufacturer on the 7900HT real‐time PCR system. Glyceraldehyde‐3 phosphate dehydrogenase was used as a housekeeping gene, and the relative expression of MTHFD1L was normalized to glyceraldehyde‐3 phosphate dehydrogenase, calculated by the method.
Western blot analysis
Cell lysates were isolated with radioimmunoprecipitation assay buffer supplemented with protease inhibitor. The bicinchoninic acid method was used to detect the concentration of proteins. Afterward, proteins (20 μg) were loaded in 12% SDS/PAGE by electrophoresis and transferred onto poly(vinylidene difluoride) membranes. Followed by blocking using 5% fat‐free milk, these poly(vinylidene difluoride) membranes were first hybridized with indicated primary antibodies against MTHFD1L, CCND1, AKT/phosphorylated (p)‐AKT and mTOR/p‐mTOR overnight at 4 °C and probed with secondary antibody for 1 h at 37 °C. The protein bands were visualized with ECL Plus (Thermo Scientific, Rockford, IL, USA) and scanned by imagej software (National Institutes of Health, Bethesda, MD, USA). Glyceraldehyde‐3 phosphate dehydrogenase was selected as the loading control.
Bioinformatics and statistics
OS clinical data from the Gene Expression Omnibus (GEO) database were downloaded for computing the differentially expressed miRNAs. TargetScan, PICTAR5, miRanda and miRWalk were used to predict the possible upstream regulators of MTHFD1L. The potential drugs that could inhibit MTHFD1L expression were achieved via accessing the CTDbase. All data were statistically analyzed by spss version 22.0 (SPSS, Chicago, IL, USA) and exhibited as mean ± standard deviation (SD). Student’s t‐test or ANOVA with Dunn’s test was used for comparisons in two or more groups. A P value <0.05 was regarded as statistically significant.
Results
Arbutin inhibits cell proliferation in OS cells
MG‐63 and SW1353 OS cell lines were selected to evaluate whether Arbutin affects the proliferative ability of OS cells in this study. MG‐63 and SW1353 cells were first treated by specific concentrations (10, 20, 50, 100, 200 and 500 μm) of Arbutin, and cell proliferation was examined using CCK‐8. Results manifested that the proliferative capability of MG‐63 and SW1353 cells was gradually reduced with the increasing concentration of Arbutin (Fig. 1A). The concentration of 200 μm was finally selected for further experiments. CCK‐8 assay at a time‐dependent mode showed that Arbutin (200 μm) significantly inhibited cell viability of OS cells in both MG‐63 and SW1353 (Fig. 1B). Therefore, these findings demonstrated that in vitro treatment of OS with Arbutin elicited significant inhibition against cell viability.
Fig 1.
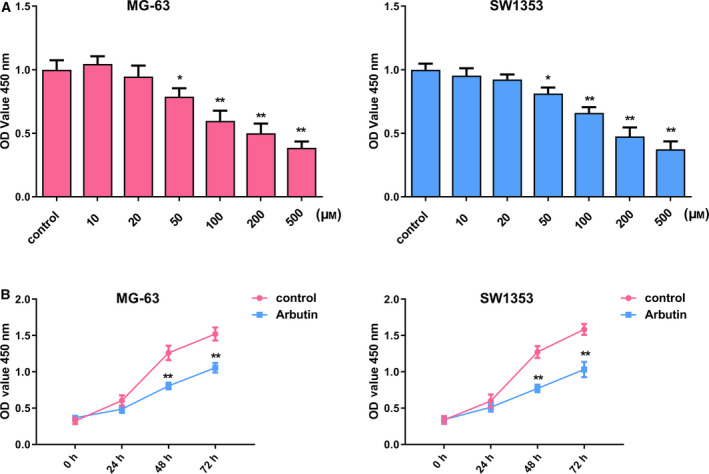
Arbutin suppresses OS cell viability. (A) The proliferative ability was explored using CCK‐8 kit in MG‐63 and SW1353 cells treated with 10, 20, 50, 100, 200 and 500 μm Arbutin. (B) The proliferation of OS cells induced by 200 μm Arbutin was measured in a time‐dependent manner by CCK‐8 assay. Data are shown as the means ± SD of three independent experiments. The data were statistically analyzed by Student’s t‐test. *P < 0.05, **P < 0.01 versus control group.
Arbutin treatment attenuates MTHFD1L expression and might inactivate the AKT/mTOR pathway
Analysis of the CTDbase revealed that Arbutin was a potential agent inhibiting the expression of MTHFD1L. Thus, to verify the correlation, we implemented western blot to evaluate the MTHFD1L expression in OS cells treated by Arbutin. Results exhibited that the treatment of Arbutin significantly declined the MTHFD1L protein expression (Fig. 2). The expression of CCND1 was also measured by western blot experiment to estimate the impact of Arbutin on cell cycle in OS cells. Arbutin exposure induced a decrease in level of CCND1 in MG‐63 and SW1353 cells (Fig. 2). In addition, as we previously described [12], silencing MTHFD1L can suppress the activity of the AKT/mTOR pathway in OS cells. Thereby, we evaluated the AKT/mTOR pathway‐related protein levels to validate whether Arbutin treatment affects the activation of AKT/mTOR signaling. As illuminated in Fig. 2, the levels of p‐AKT and p‐mTOR were all reduced after Arbutin treatment, and AKT and mTOR showed no differences compared with the NC group. Taken together, these findings validated that the treatment of Arbutin attenuated MTHFD1L and CCND1 expression and restrained the AKT/mTOR signaling in OS.
Fig 2.
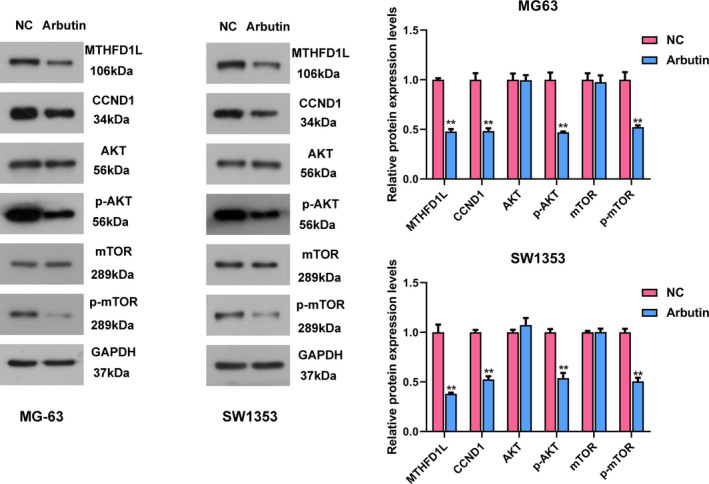
Arbutin attenuates MTHFD1L and CCND1 expression and might inactivate the AKT/mTOR pathway. The expressions of MTHFD1L, CCND1, AKT/p‐AKT and mTOR/p‐mTOR were investigated by using western blot in Arbutin‐treated OS cells. The intensity of protein bands was quantified. Data are shown as the means ± SD of three independent experiments. The data were statistically analyzed by Student’s t‐test. **P < 0.01 versus NC group.
MTHFD1L is a direct target gene of miR‐338‐3p in OS cells
miRNAs primarily perform their biological functions via regulating the target genes [19]. As searched via TargetScan, PICTAR5, miRanda and miRWalk, we obtained 16 miRNAs targeting MTHFD1L in overlapping part (Fig. 3A). Then, the two datasets GEO: GSE28423 and GSE65071 were used to identify the down‐regulated miRNAs in OS. After intersecting with down‐regulated miRNAs and potential miRNAs that target MTHFD1L, only one common miRNA, miR‐338‐3p, was left (Fig. 3B). miR‐338‐3p was significantly reduced in OS tissues relative to normal controls (Fig. 3C,D). Fig. 3E exhibited the binding sites of miR‐338‐3p and MTHFD1L. Luciferase activity analysis was subsequently conducted to verify the prediction, and results revealed that the luciferase activity was obviously increased in cells transfected with wt‐MTHFD1L and miR‐338‐3p inhibitor. There were no changes in the mut‐MTHFD1L group after the transfection of miR‐338‐3p inhibitor. Furthermore, miR‐338‐3p inhibitor strikingly increased the expression of MTHFD1L at both mRNA and protein levels (Fig. 3F,G). The cotransfection of si‐MTHFD1L and miR‐338‐3p inhibitor restored the MTHFD1L expression that was decreased or increased by si‐MTHFD1L or miR‐338‐3p inhibitor. Herein, these observations supported that MTHFD1L is a direct target of miR‐338‐3p in OS.
Fig 3.
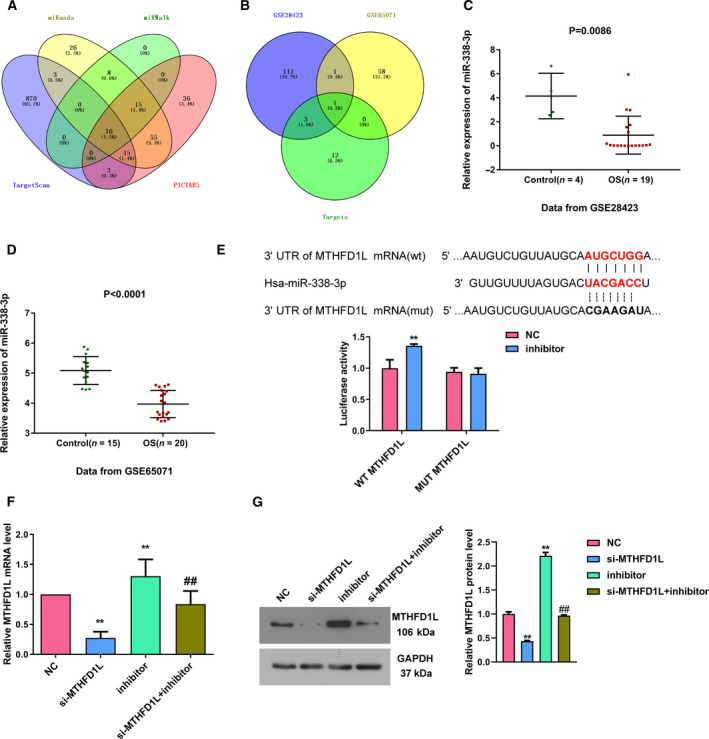
MTHFD1L is a direct target of miR‐338‐3p in OS cells. (A) The upstream miRNAs of MTHFD1L predicted by TargetScan, PICTAR5, miRanda and miRWalk. (B) Intersection among predicted upstream miRNAs of MTHFD1L, down‐regulated miRNAs identified by GEO: GSE28423 and GSE65071. (C, D) Expression of miR‐338‐3p was markedly decreased in OS tissues compared with controls. (E) Sequences of the binding sites between miR‐338‐3p and MTHFD1L are shown, and luciferase reporter gene analysis was conducted to determine the relationship between miR‐338‐3p and MTHFD1L. (F, G) After MG‐63 and SW1353 cells were transfected with si‐MTHFD1L, miR‐338‐3p inhibitor and si‐MTHFD1L + miR‐338‐3p inhibitor, the mRNA and protein levels of MTHFD1L were measured by quantitative real‐time PCR and western blotting, respectively. Data are shown as the means ± SD of three independent experiments. The data were statistically analyzed by Student’s t‐test or Dunn’s test. **P < 0.01 versus NC group; ## P < 0.01 versus si‐MTHFD1L or miR‐338‐3p inhibitor group.
Arbutin mediates the regulatory action of the miR‐338‐3p/MTHFD1L axis in OS cells
After determining the targeted correlation between miR‐338‐3p and MTHFD1L, we next examined whether miR‐338‐3p is implicated in the regulation of MTHFD1L on Arbutin‐stimulated OS cell behaviors. First, we detected the expression of miR‐338‐3p in Arbutin‐treated MG‐63 and SW1353 cells. Results manifested that miR‐338‐3p was significantly promoted in OS cells as a result of the treatment of Arbutin (Fig. 4A). Compared with the NC group, Arbutin treatment remarkably inhibited OS cells’ invasive and migratory motilities (Fig. 4B). The MTHFD1L silencing further repressed the invasion and migration of Arbutin‐treated OS cells, and this effect was restored by the addition of miR‐338‐3p inhibitor. As expected, the transfection of miR‐338‐3p inhibitor significantly elevated the invasion and migration capabilities of OS cells after Arbutin stimulation, which was reversed by MTHFD1L knockdown (Fig. 4B). Similarly, the dramatic repression of Arbutin to cell viability was also observed in OS cells (Fig. 4C). More than that, the repressive effect of Arbutin on cell proliferation was enhanced because of MTHFD1L deficiency but abrogated by the transfection of miR‐338‐3p inhibitor (Fig. 4C). Collectively, results demonstrated that Arbutin modulates the regulatory action of the miR‐338‐3p/MTHFD1L pair in OS cells.
Fig 4.
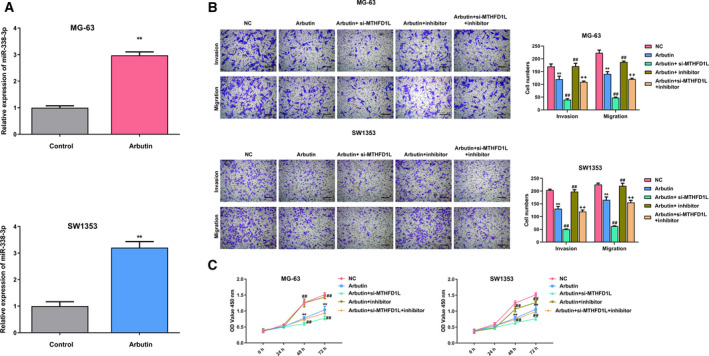
Arbutin mediates the regulatory action of miR‐338‐3p/MTHFD1L in OS cells. (A) The expression of miR‐338‐3p in Arbutin‐treated OS cells. (B) Transwell migration and invasion experiments were implemented in Arbutin‐treated MG‐63 and SW1353 cells after the transfection of si‐MTHFD1L, miR‐338‐3p inhibitor and si‐MTHFD1L + miR‐338‐3p. (C) CCK‐8 assay was conducted in Arbutin‐stimulated MG‐63 and SW1353 cells transfected with si‐MTHFD1L or miR‐338‐3p inhibitor. Scale bars, 200 μm. Data are shown as the means ± SD of three independent experiments. The data were statistically analyzed by Student’s t‐test or Dunn’s test. **P < 0.01 versus control or NC group; ## P < 0.01 versus Arbutin group; ++ P < 0.01 versus si‐MTHFD1L or miR‐338‐3p inhibitor group.
Arbutin is involved in the AKT/mTOR pathway modulated by the miR‐338‐3p/MTHFD1L axis
Given the suppressive effect of Arbutin treatment on MTHFD1L expression, we then conducted western blot to estimate whether Arbutin affects the management of the miR‐338‐3p/MTHFD1L axis in the AKT/mTOR pathway. In Arbutin‐treated MG‐63 cells, the transfection of miR‐338‐3p inhibitor significantly increased MTHFD1L expression, and these conditions were all reversed by the cotransfection of si‐MTHFD1L and miR‐338‐3p inhibitor (Fig. 5A). Down‐regulation of miR‐338‐3p also elevated the expression of CCND1, and this tendency was overturned by the involvement of si‐MTHFD1L (Fig. 5A). Moreover, the repression of Arbutin treatment on p‐AKT/p‐mTOR was enhanced by silencing MTHFD1L but overturned by miR‐338‐3p inhibitor. The individual effects of si‐MTHFD1L and miR‐338‐3p inhibitor on AKT/mTOR pathway‐related proteins expressions were abolished by the cotransfection of si‐MTHFD1L and miR‐338‐3p inhibitor (Fig. 5A). As expected, the similar phenomena were also observed in Arbutin‐stimulated SW1353 cells (Fig. 5B). All these data suggested that Arbutin might participate in the AKT/mTOR pathway modulated by the miR‐338‐3p/MTHFD1L axis.
Fig 5.
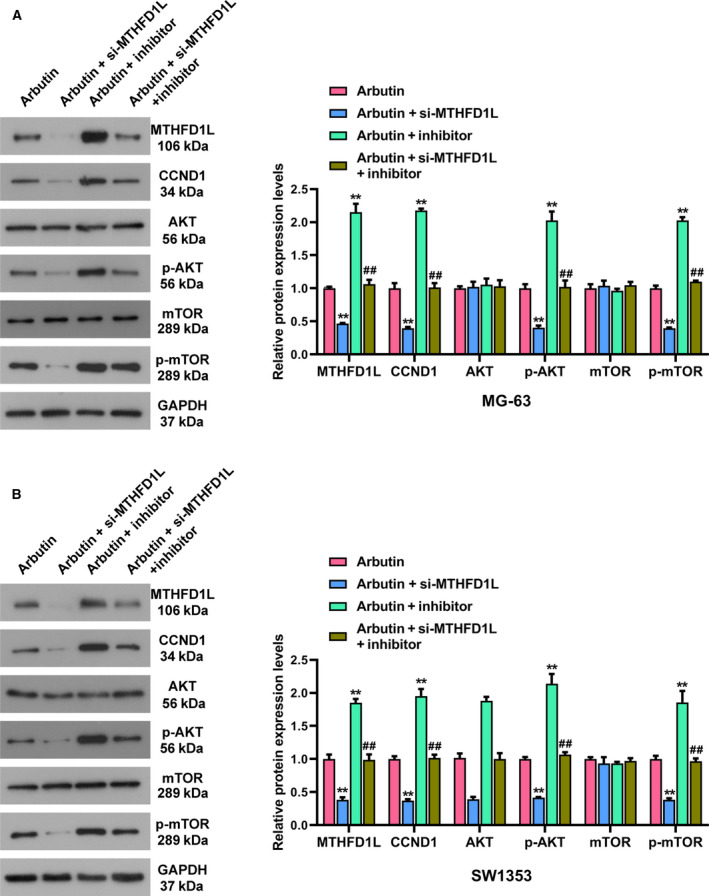
Arbutin participates in the miR‐338‐3p/MTHFD1L axis mediating the AKT/mTOR pathway. (A, B) Expressions of MTHFD1L, CCND1, AKT/p‐AKT and mTOR/p‐mTOR were measured using western blot in MG‐63 and SW1353 cells after Arbutin treatment and different transfections. Data are shown as the means ± SD of three independent experiments. The data were statistically analyzed by Dunn’s test. **P < 0.01 versus Arbutin group; ## P < 0.01 versus si‐MTHFD1L or miR‐338‐3p inhibitor group.
Discussion
In this investigation, we harvested evidences supporting the correlation between Arbutin and miR‐338‐3p/MTHFD1L in OS. In OS cells, Arbutin treatment significantly inhibited cell viability. Moreover, the expressions of MTHFD1L and AKT/mTOR signaling‐related markers were also decreased by Arbutin treatment. MTHFD1L was a potential target gene of miR‐338‐3p. Effects of miR‐338‐3p and MTHFD1L in OS cells were affected by the stimulation of Arbutin. In summary, these findings determined our hypothesis that Arbutin hindered the development of OS by modulating the effects of miR‐338‐3p/MTHFD1L to OS and inactivating AKT/mTOR signaling.
Proliferation and metastasis have been identified as main cosmetic concerns in cancer progression and therapeutics [20]. Arbutin is a naturally occurring hydroquinone, which has been proved to be a cytoprotective agent with insignificant cytotoxicity at high doses [21]. Arbutin plays an inhibitory effect on tumor cell viability, including HCT‐15 human TCC SUP cells and human A375 melanoma cells [10, 22]. Consistent with these prior studies, our data displayed that Arbutin remarkably suppressed cell proliferation of OS cells. Invasion and migration are two major forms for cancer metastasis. Our transwell assay results showed that Arbutin blocked OS cell invasion and migration. Moreover, Arbutin was selected as an inhibitor for MTHFD1L expression. Western blot concluded that the treatment of Arbutin could significantly decrease the MTHFD1L protein expression. Several studies have indicated that Arbutin exerts important roles by targeting miRNAs. Through targeting miR‐27a, Arbutin could relieve apoptosis and autophagy of protected high‐glucose‐stimulated HK‐2 cells [23]. The oxidative injury caused by hydrogen peroxide was relieved by Arbutin via regulating miR‐29a in retinal ganglion cells [24]. The key mechanism that allows miRNAs to be involved in tumorigenesis is to modulate the expression of targeted genes. Therefore, in the future, we should take measures to focus on the underlying mechanism addressed by Arbutin/miRNA/mRNA in the development of OS.
Bioinformatics analysis in this study manifested that miR‐338‐3p was obviously reduced in OS and can directly target MTHFD1L, indicating that miR‐338‐3p may be an inhibitory regulator for OS development. Existing evidences demonstrated that miR‐338‐3p retards ovarian cancer cells proliferation and metastasis via mediating the Wnt/β signaling pathway [25]. miR‐338‐3p carries 5‐fluorouracil resistance in p53 mutant colon cancer cells through regulating the mTOR [26]. miR‐338‐3p increased by baicalin exerts an anticancer role in breast cancer [20]. In addition to that, in OS, miR‐338‐3p plays the role of a tumor suppressor for OS cell malignant behaviors through inactivating the mitogen‐activated protein kinase pathway [18]. A similar role of miR‐338‐3p was found in OS cells by Cao et al. [17], who uncovered that miR‐338‐3p overexpression markedly repressed OS cells viability, migration and invasion. However, the possible mechanism of miR‐338‐3p and putative natural agents have not been clearly explored in OS. Functional in vitro experiments displayed that down‐regulation of miR‐338‐3p reversed the anticancer role of Arbutin in cell proliferation, migration and invasion. MTHFD1L expression was negatively regulated by miR‐338‐3p in OS cells. MTHFD1L was a direct target of miR‐338‐3p. To further assess the underlying mechanism addressed by Arbutin/miR‐338‐3p/MTHFD1L, we conducted in vitro assays to measure the impacts of Arbutin/miR‐338‐3p/MTHFD1L in cell viability, invasion and migration in OS cells. All data illuminated that miR‐338‐3p inhibited cell malignant behaviors in Arbutin‐treated OS cells through directly targeting MTHFD1L.
Abnormal activity of AKT/mTOR signaling is frequent in solid tumors [27]. Its activation significantly facilitates the development of various cancers, including OS [28]. Notably, the prooncogenic role of the AKT/mTOR signaling pathway in carcinogenesis has become a hotspot for the development of drug [29, 30]. Moreover, because our previous study has demonstrated that MTHFD1L expression was enriched in the AKT/mTOR pathway, and we found that silencing MTHFD1L could hinder OS cell aggressiveness by inactivating the AKT/mTOR pathway [12], thereby we implemented a western blot test to expound the correlation between Arbutin/miR‐338‐3p/MTHFD1L and AKT/mTOR signaling. AKT/p‐AKT and mTOR/p‐mTOR were crucial markers in the AKT/mTOR signaling pathway [31]. Data observed by us showed that Arbutin induced a decreased level of p‐AKT and p‐mTOR, and knockdown of MTHFD1L strengthened the inhibitory effect of Arbutin on p‐AKT and p‐mTOR expression, whereas the miR‐338‐3p inhibitor reversed it. CCND1 functions as a crucial regulator in G1‐to‐S phase and is increased in most human tumors [32, 33]. Its overexpression was related with unfavorable prognosis and tumor metastasis, and it also was involved in the regulatory action of MTHFD1L to OS [34]. The expressional pattern of CCND1 was consistent with p‐AKT and p‐mTOR in Arbutin‐induced OS cells. These observations demonstrated that Arbutin inhibited OS cell aggressiveness via miR‐338‐3p/MTHFD1L signaling.
Taken together, to the best of our knowledge, our work was the first study to unearth the involvement of Arbutin on the effects of the miR‐338‐3p/MTHFD1L axis to OS development. We concluded that Arbutin exerted antiproliferative and anti‐invasive properties in OS cells through modulating the miR‐338‐3p/MTHFD1L axis to inactivate the AKT/mTOR pathway. Nevertheless, how Arbutin modulates the miR‐338‐3p/MTHFD1L axis in OS cells has yet to be explored. Thus, our future work would emphasize this mechanism.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
C‐QW and LW initiated and designed the study. C‐QW, X‐MW and B‐LL prepared experiments and collected data. C‐QW and Y‐MZ analyzed data. All authors helped to prepare and revise the manuscript.
Data Accessibility
The datasets supporting the conclusions of this work are included within the article.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Yu D, Kahen E, Cubitt CL, McGuire J, Kreahling J, Lee J, Altiok S, Lynch CC, Sullivan DM and Reed DR (2015) Identification of synergistic, clinically achievable, combination therapies for osteosarcoma. Sci Rep 5, 16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sampo M, Koivikko M, Taskinen M, Kallio P, Kivioja A, Tarkkanen M and Böhling T (2011) Incidence, epidemiology and treatment results of osteosarcoma in Finland ‐ a nationwide population‐based study. Acta Oncol 50, 1206–1214. [DOI] [PubMed] [Google Scholar]
- 4. Isakoff MS, Bielack SS, Meltzer P and Gorlick R (2015) Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol 33, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li Y, Kong D, Wang Z and Sarkar FH (2010) Regulation of microRNAs by natural agents: an emerging field in chemoprevention and chemotherapy research. Pharm Res 27, 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Man X, Yang L, Liu S, Yang L, Li M and Fu Q (2019) Arbutin promotes MC3T3‐E1 mouse osteoblast precursor cell proliferation and differentiation via the Wnt/β‐catenin signaling pathway. Mol Med Rep 19, 4637–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim YJ, Lee EH, Kang TH, Ha SK, Oh MS, Kim SM, Yoon TJ, Kang C, Park JH and Kim SY (2009) Inhibitory effects of arbutin on melanin biosynthesis of alpha‐melanocyte stimulating hormone‐induced hyperpigmentation in cultured brownish guinea pig skin tissues. Arch Pharm Res 32, 367–373. [DOI] [PubMed] [Google Scholar]
- 8. Shahaboddin ME, Pouramir M, Moghadamnia AA, Parsian H, Lakzaei M and Mir H (2011) Pyrus biossieriana Buhse leaf extract: an antioxidant, antihyperglycaemic and antihyperlipidemic agent. Food Chem 126, 1730–1733. [DOI] [PubMed] [Google Scholar]
- 9. Yousefi F, Mahjoub S, Pouramir M and Khadir F (2013) Hypoglycemic activity of Pyrus biossieriana Buhse leaf extract and arbutin: inhibitory effects on alpha amylase and alpha glucosidase. Caspian J Intern Med 4, 763–767. [PMC free article] [PubMed] [Google Scholar]
- 10. Li H, Jeong YM, Kim SY, Kim MK and Kim DS (2011) Arbutin inhibits TCCSUP human bladder cancer cell proliferation via up‐regulation of p21. Pharmazie 66, 306–309. [PubMed] [Google Scholar]
- 11. Nawarak J, Huang‐Liu R, Kao SH, Liao HH, Sinchaikul S, Chen ST and Cheng SL (2009) Proteomics analysis of A375 human malignant melanoma cells in response to arbutin treatment. Biochim Biophys Acta 1794, 159–167. [DOI] [PubMed] [Google Scholar]
- 12. Li Y, Zhou J, Wang J, Chen X, Zhu Y and Chen Y (2020) Mir‐30b‐3p affects the migration and invasion function of ovarian cancer cells by targeting the CTHRC1 gene. Biol Res 53, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L and Ding F (2019) Hsa_circ_0008945 promoted breast cancer progression by targeting miR‐338‐3p. Onco Targets Ther 12, 6577–6589. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14. Li M, Bian Z, Jin G, Zhang J, Yao S, Feng Y, Wang X, Yin Y, Fei B, You Q et al (2019) LncRNA‐SNHG15 enhances cell proliferation in colorectal cancer by inhibiting miR‐338‐3p. Cancer Med 8, 2404–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang P, Shao G, Lin X, Liu Y and Yang Z (2017) MiR‐338‐3p inhibits the growth and invasion of non‐small cell lung cancer cells by targeting IRS2. Am J Cancer Res 7, 53–63. [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y and Qin H (2018) miR‐338‐3p targets RAB23 and suppresses tumorigenicity of prostate cancer cells. Am J Cancer Res 8, 2564–2574. [PMC free article] [PubMed] [Google Scholar]
- 17. Cao R, Shao J, Hu Y, Wang L, Li Z, Sun G and Gao X (2018) microRNA‐338‐3p inhibits proliferation, migration, invasion, and EMT in osteosarcoma cells by targeting activator of 90 kDa heat shock protein ATPase homolog 1. Cancer Cell Int 18, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jia F, Zhang Z and Zhang X (2019) MicroRNA‐338‐3p inhibits tumor growth and metastasis in osteosarcoma cells by targeting RUNX2/CDK4 and inhibition of MAPK pathway. J Cell Biochem 120, 6420–6430. [DOI] [PubMed] [Google Scholar]
- 19. Fujiwara T and Yada T (2013) miRNA‐target prediction based on transcriptional regulation. BMC Genom 14 (Suppl 2), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duan X, Guo G, Pei X, Wang X, Li L, Xiong Y and Qiu X (2019) Baicalin inhibits cell viability, migration and invasion in breast cancer by regulating miR‐338‐3p and MORC4. Onco Targets Ther 12, 11183–11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jurica K and Brčić Karačonji I (2018) In vitro safety assessment of the strawberry tree (Arbutus unedo L.) water leaf extract and arbutin in human peripheral blood lymphocytes. Cytotechnology 70, 1261–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng SL, Liu RH, Sheu JN, Chen ST, Sinchaikul S and Tsay GJ (2007) Toxicogenomics of A375 human malignant melanoma cells treated with arbutin. J Biomed Sci 14, 87–105. [DOI] [PubMed] [Google Scholar]
- 23. Lv L, Zhang J, Tian F, Li X, Li D and Yu X (2019) Arbutin protects HK‐2 cells against high glucose‐induced apoptosis and autophagy by up‐regulating microRNA‐27a. Artif Cells Nanomed Biotechnol 47, 2940–2947. [DOI] [PubMed] [Google Scholar]
- 24. Zhao W, Wang S, Qin T and Wang W (2019) Arbutin attenuates hydrogen peroxide‐induced oxidative injury through regulation of microRNA‐29a in retinal ganglion cells. Biomed Pharmacother 112, 108729. [DOI] [PubMed] [Google Scholar]
- 25. Zhang R, Shi H, Ren F, Feng W, Cao Y, Li G, Liu Z, Ji P and Zhang M (2019) MicroRNA‐338‐3p suppresses ovarian cancer cells growth and metastasis: implication of Wnt/catenin beta and MEK/ERK signaling pathways. J Exp Clin Cancer Res 38, 494. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Han J, Li J, Tang K, Zhang H, Guo B, Hou N and Huang C (2017) miR‐338‐3p confers 5‐fluorouracil resistance in p53 mutant colon cancer cells by targeting the mammalian target of rapamycin. Exp Cell Res 360, 328–336. [DOI] [PubMed] [Google Scholar]
- 27. Ocana A, Vera‐Badillo F, Al‐Mubarak M, Templeton AJ, Corrales‐Sanchez V, Diez‐Gonzalez L, Cuenca‐Lopez MD, Seruga B, Pandiella A and Amir E (2014) Activation of the PI3K/mTOR/AKT pathway and survival in solid tumors: systematic review and meta‐analysis. PLoS One 9, e95219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu K, Dai HB and Qiu ZL (2016) mTOR signaling in osteosarcoma: oncogenesis and therapeutic aspects (Review). Oncol Rep 36, 1219–1225. [DOI] [PubMed] [Google Scholar]
- 29. Ocaña A, Amir E, Seruga B, Martin M and Pandiella A (2013) The evolving landscape of protein kinases in breast cancer: clinical implications. Cancer Treat Rev 39, 68–76. [DOI] [PubMed] [Google Scholar]
- 30. Garcia‐Echeverria C and Sellers WR (2008) Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene 27, 5511–5526. [DOI] [PubMed] [Google Scholar]
- 31. Sun X, Wei B, Peng ZH, Fu QL, Wang CJ, Zheng JC and Sun JC (2019) Knockdown of lncRNA XIST suppresses osteosarcoma progression by inactivating AKT/mTOR signaling pathway by sponging miR‐375‐3p. Int J Clin Exp Pathol 12, 1507–1517. [PMC free article] [PubMed] [Google Scholar]
- 32. Santarius T, Shipley J, Brewer D, Stratton MR and Cooper CS (2010) A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 10, 59–64. [DOI] [PubMed] [Google Scholar]
- 33. Matsushime H, Quelle DE, Shurtleff SA, Shibuya M, Sherr CJ and Kato JY (1994) D‐type cyclin‐dependent kinase activity in mammalian cells. Mol Cell Biol 14, 2066–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. John RR, Malathi N, Ravindran C and Anandan S (2017) Mini review: multifaceted role played by cyclin D1 in tumor behavior. Indian J Dent Res 28, 187–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this work are included within the article.


