Abstract
MhsT of Bacillus halodurans is a transporter of hydrophobic amino acids and a homologue of the eukaryotic SLC6 family of Na+‐dependent symporters for amino acids, neurotransmitters, osmolytes, or creatine. The broad range of transported amino acids by MhsT prompted the investigation of the substrate recognition mechanism. Here, we report six new substrate‐bound structures of MhsT, which, in conjunction with functional studies, reveal how the flexibility of a Gly‐Met‐Gly (GMG) motif in the unwound region of transmembrane segment 6 (TM6) is central for the recognition of substrates of different size by tailoring the binding site shape and volume. MhsT mutants, harboring substitutions within the unwound GMG loop and substrate binding pocket that mimick the binding sites of eukaryotic SLC6A18/B0AT3 and SLC6A19/B0AT1 transporters of neutral amino acids, exhibited impaired transport of aromatic amino acids that require a large binding site volume. Conservation of a general (G/A/C)ΦG motif among eukaryotic members of SLC6 family suggests a role for this loop in a common mechanism for substrate recognition and translocation by SLC6 transporters of broad substrate specificity.
Keywords: amino acids uptake, MhsT, neurotransmitter, sodium symporters, substrate recognition, X‐ray crystallography
Subject Categories: Membrane & Intracellular Transport; Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Structural and functional analyses of the Bacillus halodurans MhsT provide insight into sequence motifs underlying substrate recognition and their conservation in eukaryotic SLC6‐family Na+‐dependent symporters.
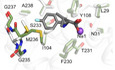
Introduction
The solute carrier 6 (SLC6) subfamily is part of a larger amino acid–polyamine–organocation (APC) transporter superfamily (Broer et al, 2006; He et al, 2009). Twenty different human genes encode SLC6 transporters that are responsible for the active transport of a variety of solutes including neurotransmitters such as serotonin, dopamine, and norepinephrine, as well as creatine, taurine, choline, betaine, and amino acids (Broer et al, 2006). Amino acid transporters of the SLC6 family, such as the γ‐aminobutyric acid transporter (GABA transporter, GAT), glycine transporters (GlyT), and the neutral amino acid transporters SLC6A18 and SLC6A19, participate in the active reuptake of amino acids in kidneys, small intestine (Romeo et al, 2006), and brain tissue (Broer, 2008; Andersen et al, 2011). Malfunctions of SLC6 transporters are associated with a number of neurological and metabolic diseases, such as schizophrenia (Mateos et al, 2005), epilepsy (Meldrum, 1995a; Meldrum, 1995b), depression (Laasonen‐Balk et al, 1999; Gether et al, 2006; Kristensen et al, 2011), and aminoacidurias (Broer et al, 2004; Broer et al, 2008). Loss‐of‐function mutations of the neutral amino acid transporter SLC6A19 (B0AT1), the major amino acid uptake system in the gut, are causative of Hartnup disorder, where amino acid uptake is insufficient and in particular affecting tryptophan levels and therefore biosynthesis of niacin, melatonin, and serotonin (Broer, 2009).
Structural characterization of eukaryotic SLC6 proteins has been based on crystal structures of the dopamine transporter from Drosophila melanogaster (dDAT) (Penmatsa et al, 2013) and the human serotonin transporter (hSERT) (Coleman et al, 2016). They both belong to the neurotransmitter:sodium symporter (NSS) family and feature the so‐called LeuT fold (Yamashita et al, 2005) that was first identified in the crystal structure of the NSS homologue LeuT from Aquifex aeolicus (Abramson & Wright, 2009).
The LeuT fold displays an inverted pseudo‐twofold symmetry between two transmembrane helix (TM) bundles that is conserved in many other families of symporters and exchangers (Faham et al, 2008; Weyand et al, 2008; Ressl et al, 2009; Shaffer et al, 2009; Gao et al, 2010; Tang et al, 2010). In the LeuT structure, the primary substrate (S1) binding site and the two sodium ion binding sites (Na1 and Na2) are located between the so‐called bundle and scaffold domains (Fig 1A and B). Proposed first for LeuT (Shi et al, 2008), a second, allosteric substrate site S2 has been associated with an extracellular vestibule of the transporter (Quick et al, 2018; Fitzgerald et al, 2019). It has not been possible to capture crystal structures of MhsT or LeuT in S2‐bound states, which from single‐molecule studies appear dynamic in nature (Fitzgerald et al, 2019), but inhibitor binding at an overlapping site has been observed (Singh et al, 2007; Zhou et al, 2007; Quick et al, 2009; Penmatsa et al, 2013; Penmatsa et al, 2015; Coleman et al, 2016).
Figure 1. Overview of MhsT structure and function.
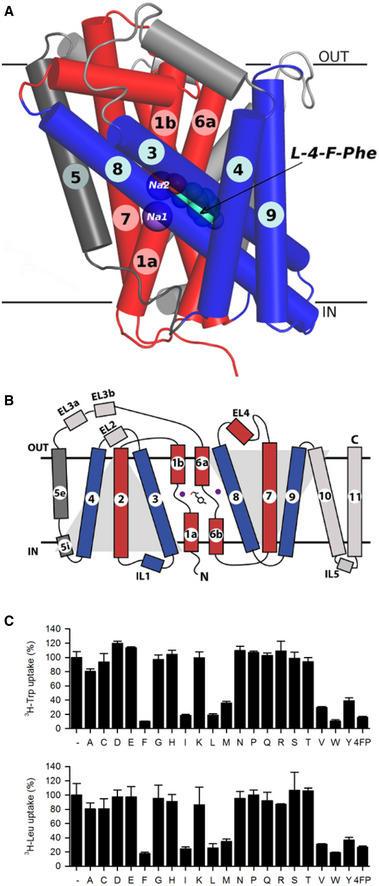
- MhsT is shown in a ribbon representation (scaffold helices in blue, bundle helices in red, and TM5 in dark gray, sodium ions shown as purple spheres, L‐4‐Fluoro‐phenylalanine, bound between unwound region of TM1 and TM6, shown as green spheres.
- Schematic representation of the MhsT structure (LeuT fold). L‐4‐Fluoro‐phenylalanine shown as gray sticks. Remaining color coding same as in panel A.
- Results of a competitive uptake assay showing inhibition of 0.2 µM L‐[3H]Trp or L‐[3H]Leu uptake in MhsT‐WT‐expressing MQ614 cells, measured for 30 s in the absence (−) or presence of 12 µM of the natural amino acids (the one‐letter code is used) or 4‐fluoro‐phenylalanine (4FP). Data are the mean ± SEM of three independent experiments each performed as technical triplicates.
We have reported structures of another SLC6 orthologue, MhsT (multiple hydrophobic amino acid substrate transporter) from Bacillus halodurans, revealing an occluded, inward‐oriented state with bound Na+ and L‐tryptophan in the Na1 and Na2 sites and the substrate site S1, respectively (Malinauskaite et al, 2014). It was previously reported that MhsT displays a broad specificity for hydrophobic amino acids (Quick & Javitch, 2007; Malinauskaite et al, 2014) (Fig 1C). Interestingly, based on sequence conservation, MhsT is an orthologue of the human SLC6A18 and SLC6A19 (also known as B0AT3 and B0AT1 with sequence identities of 36 and 30%, respectively). These are major amino acid transporting systems for non‐polar amino acids in the brush border membrane of epithelial cells (Broer, 2008), in kidney proximal tubules (Nash et al, 1998; Romeo et al, 2006) and intestine (jejunum) (Romeo et al, 2006). SLC6A19 was reported to facilitate Na+‐dependent transport of its substrates (L‐Leu, L‐Ile, L‐Val, L‐Met, L‐Phe, L‐Trp, L‐Thr, and L‐His) with millimolar apparent affinities (Broer, 2009). SLC6A18 exhibits 50% sequence identity to SLC6A19 and, to some extent, an overlapping substrate specificity. It has been shown to transport aliphatic amino acids (L‐Ala, L‐Met, L‐Val, L‐Ile, L‐Gly, L‐Ser, L‐Leu) also with millimolar apparent affinities (Singer et al, 2009).
Here, we report crystal structures of MhsT complexed with six different substrates, exploiting further the inward‐oriented, occluded state, in an effort to identify the structural elements of the S1 binding site underlying substrate recognition, promiscuity, and transport. We find that the architecture of the S1 site is broadly defined by two possible states that are dependent on the nature of the hydrophobic amino acid substrate: smaller aliphatic amino acids (L‐Val, L‐Leu, and L‐Ile) and larger aromatic amino acids (L‐Trp, L‐Phe, L‐Tyr, and the L‐Tyr analog L‐4‐F‐Phe). The shape and volume regulation of the S1 site centers on a key role of the unwound segment of TM6 and in both cases is compatible with a flexible transport mechanism. Additionally, we used site‐directed mutagenesis to investigate the role of various residues involved in substrate binding and their influence on substrate specificity and transport.
Results
MhsT crystallization, processing, and refinement
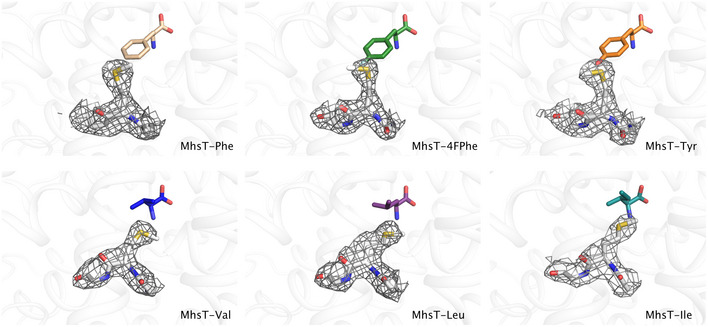
Previously, crystals of MhsT bound to L‐Trp and Na+ were obtained using high concentrations of lipid and detergent (HiLiDe) (Gourdon et al, 2011) and the lipid cubic phase (LCP) (Cherezov, 2011) methods. Using the HiLiDe crystallization conditions, we have obtained six structures of MhsT in complex with six different ligands in the S1 site (Fig EV1 and Appendix Fig S1): the tyrosine analog 4‐fluoro‐L‐phenylalanine (4F‐Phe), L‐Tyr, L‐Phe, L‐Ile, L‐Leu, and L‐Val. Protein‐substrate crystals (Appendix Fig S2) were obtained in similar conditions to those previously reported (Malinauskaite et al, 2014).
Figure EV1. F o−F c simulated annealing omit maps of the MhsT substrates.
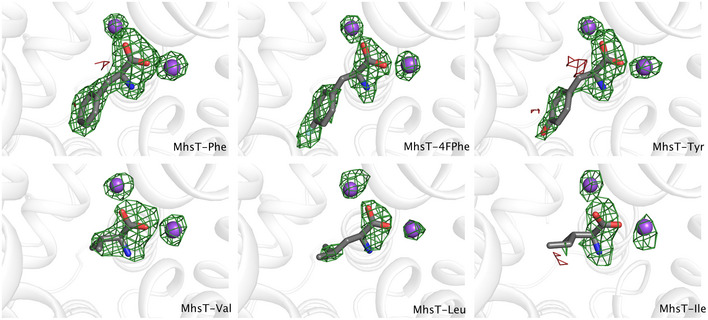
F o−F c simulated annealing omit maps for MhsT in complex with L‐Phe, L‐4‐F‐Phe, L‐Tyr, and L‐Val contoured at ± 3.0 r.m.s.d. Omit maps for MhsT in complex with L‐Leu and L‐Ile contoured at ± 2.5 r.m.s.d.
Structures of MhsT in complex with 4F‐Phe, Tyr and Phe were determined at about 2.3 Å resolution (Table 1) in the P2 space group with one molecule in the asymmetric unit similar to the Trp‐bound complex (Malinauskaite et al, 2014). The complexes with Leu and Val substrates crystallized in a different P21 crystal form with two molecules in the asymmetric unit related by twofold NCS, and their structures were determined at 2.35 and 2.60 Å resolution, respectively (Table 1). The dataset for MhsT‐Leu showed pseudomerohedral twinning with a twin fraction close to 0.5 and therefore was refined using the appropriate twin law (h, ‐k, ‐l) resulting in a large drop of R‐factors. The dataset for MhsT‐Val exhibited no notable twinning. The MhsT‐Ile complex crystallized in a distinct P21 crystal form with translational non‐crystallographic symmetry (tNCS) that exhibited strong radiation sensitivity and crystal‐to‐crystal non‐isomorphism, but a dataset with completeness of 80% at 3.1 Å resolution could be obtained by merging of data collected from two crystals (Table 1). The presence of translational NCS in this crystal form combined with the limited completeness of the crystallographic data impaired refinement, but the structure determination was sufficiently clear to discern important features (see below).
Table 1.
Data reduction and refinement statistics.
| Processing | MhsT‐Tyr | MhsT‐4FPhe | MhsT‐Phe | MhsT‐Ile c | MhsT‐Leu b | MhsT‐Val b |
|---|---|---|---|---|---|---|
| Beamline | DLS‐I24 | DLS‐I04 | DLS‐I24 | SLS‐ PXI | DLS‐I04 | DLS‐I24 |
| Space group | P2 | P2 | P2 | P21 | P21 | P21 |
| Unit cell dimensions | ||||||
| a, b, c (Å) | 44.1, 49.9, 110.3 | 44.2, 49.9, 109.7 | 44.4, 49.9, 110.1 | 44.0, 97.3, 110.9 | 44.2, 215.6, 50.2 | 44.1, 216.1, 50.4 |
| α, β, γ (°) | 90.0, 96.8, 90.0 | 90.0, 96.1, 90.0 | 90.0, 96.8, 90.0 | 90.0, 96.1, 90.0 | 90.0, 90.05, 90.0 | 90.0, 90.02, 90.0 |
| Wavelength (Å) | 0.96864 | 0.96858 | 0.96860 | 1.00001 | 0.97949 | 0.97950 |
| No. reflections a | 69,099 (6,707) | 40,499 (4,060) | 41,999 (4,369) | 37,578 (6,463) | 114,897 (5,823) | 97,704 (9,611) |
| Resolution a | 29.8–2.30 (2.38–2.30) | 43.9–2.26 (2.33–2.26) | 45.5–2.25 (2.32–2.25) | 43.7–3.10 (3.31–3.10) | 45.5–2.35 (2.43–2.35) | 29.1–2.60 (2.72–2.60) |
| R merge a | 0.119 (0.734) | 0.118 (0.677) | 0.120 (0.820) | 0.195 (0.815) | 0.096 (0.679) | 0.117 (0.950) |
| R pim a | 0.077 (0.595) | 0.091 (0.532) | 0.083 (0.563) | 0.124 (0.532) | 0.066 (0.502) | 0.075 (0.607) |
| I/σI a | 7.8 (1.6) | 5.3 (1.3) | 7.3 (2.1) | 3.6 (1.2) | 8.1 (1.4) | 8.0 (1.2) |
| Completeness (%) a | 97.4 (95.2) | 92.9 (93.9) | 98.2 (99.3) | 81.6 (83.8) | 98.5 (98.5) | 99.5 (99.7) |
| Redundancy a | 3.3 (3.34) | 2.3 (1.3) | 2.9 (3.0) | 2.7 (2.5) | 3.0 (2.6) | 3.4 (3.4) |
| CC1/2 a | 0.991 (0.472) | 0.991 (0.472) | 0.989 (0.488) | 0.989 (0.636) | 0.996 (0.511) | 0.994 (0.489) |
| Wilson B‐factor | 39.2 | 34.3 | 38.2 | 63.0 | 37.4 | 49.3 |
| Twin fraction | N/A | N/A | N/A | N/A | 0.443 | (0.065) |
| Refinement | ||||||
| Resolution (Å) a | 29.8–2.30 (2.38–2.30) | 39.3–2.26 (2.34–2.26) | 45.4–2.25 (2.33–2.25) | 43.7–3.10 (3.31–3.10) | 45.5–2.35 (2.43–2.35) | 49.1–2.60 (2.69–2.60) |
| R work/R free | 0.223/0.263 (0.254/0.305) | 0.223/0.261 (0.291/0.299) | 0.224/0.255 (0.305/0.356) | 0.277/0.305 (0.340/0.355) | 0.185/0.222 (0.260/0.304) | 0.207/0.237 (0.306/0.355) |
| No. atoms | ||||||
| Protein | 3,324 | 3,324 | 3,315 | 6,630 | 6,678 | 6,674 |
| Ligand | 13 | 13 | 12 | 18 | 18 | 16 |
| Sodium | 2 | 2 | 2 | 4 | 4 | 4 |
| Detergent | 103 | 195 | 107 | 109 | 176 | 357 |
| Water | 63 | 28 | 59 | 17 | 46 | 73 |
| B‐ factors (Å2) | ||||||
| Protein | 36.2 | 38.2 | 45.0 | 56.7 | 41.4 | 56.9 |
| Ligand | 26.1 | 28.7 | 31.8 | 52.6 | 34.1 | 48.8 |
| Sodium | 28.8 | 26.7 | 34.4 | 52.8 | 33.3 | 49.6 |
| Detergent | 47.9 | 55.8 | 64.1 | 58.2 | 51.7 | 68,4 |
| Water | 38.5 | 43.3 | 47.9 | 52.2 | 37.9 | 55.4 |
| R.m.d. deviations | ||||||
| Bond length (Å) | 0.002 | 0.003 | 0.002 | 0.002 | 0.002 | 0.003 |
| Bond angles (o) | 0.542 | 0.601 | 0.583 | 0.578 | 0.505 | 0.595 |
| Ramachandran | ||||||
| Favoured (%) | 97.7 | 98.82 | 98.2 | 95.7 | 96.9 | 96.4 |
| Outliers (%) | 0 | 0 | 0 | 0.23 | 0 | 0 |
Values in parentheses are for highest resolution shell.
MhsT‐Leu was refined against the h,‐k,‐l twin law with 44.3% twinning, whereas MhsT‐Val was refined without use of twin law.
The refinement of the MhsT‐Ile complex was hindered by low completeness and the presence of translational NCS. Initial difference maps were consistent with the binding mode observed for the other aliphatic substrates.
Substrate‐bound MhsT
All crystal structures of MhsT captured the protein in an inward‐facing occluded state, with a closed extracellular vestibule, an ordered N‐terminal tail associated with the intracellular surface, and an unwound TM5 (Fig 1A) within a conserved ProX9Gly motif as previously described (Malinauskaite et al, 2014). This state allows initial solvation of the Na+ ion at the Na2 site from the cytoplasmic environment. The various substrate‐bound structures are overall similar and superimpose with a low Cα r.m.s.d. (Appendix Table S1). However, local differences are observed that allow the S1 site to accommodate substrates of different size/volume (Fig 2).
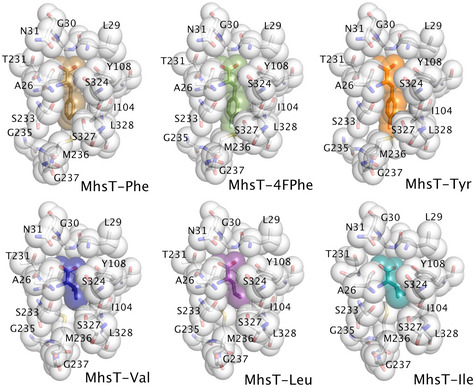
Figure 2. Volumes of the binding pockets visualized by the van der Waals spheres of the ligand and coordinating residues of MhsT in complex with L‐Phe, L‐4‐F‐Phe, L‐Tyr, L‐Val, L‐Leu, and L‐Ile.
The substrates are bound with the amino acid moiety within the S1 site similar to the MhsT‐Trp structure (Malinauskaite et al, 2014) (Fig EV1, Appendix Figs S1 and S3). The substrate amino group forms hydrogen bonds with backbone amide oxygen atoms of Ala26 in TM1, Phe230, Thr231, and the side chain of Ser233 in TM6, while the carboxyl group interacts via hydrogen bonds to the side chain of Tyr108, the backbone amide nitrogen of Gly30, and Na+ in the Na1 site. Additionally, the positive dipole on TM1b interacts with the negatively charged carboxyl group of the substrate, whereas the negative dipoles of TM1a and TM6a point toward the positively charged amino group.
The side chains of hydrophobic substrates are located to a separate pocket defined by the side chains of Ile104 in TM3, Phe229, Phe230, Ser233, Met236, Ala238 in TM6, Val331, Ser324, Ser327, and Leu328 in TM8, Leu393 in TM10, and the backbone atoms of Ala26‐Leu29, Phe229, Thr231 and Leu324. The hydrophobic nature of the side chain binding pocket explains the clear preference of MhsT for hydrophobic substrates over polar/charged amino acids (Appendix Fig S1).
Discrimination of aromatic and aliphatic amino acid binding modes
The different substrate complexes of MhsT illustrate the changes required for the substrate binding pocket to accommodate the disparately sized substrates. Although the seven substrates (including L‐Trp‐bound structure; PDB ID: 4US3) bind the S1 pocket in a similar mode concerning the amino acid group, significant structural changes within the hydrophobic cavity are observed that serve to modulate the S1 site on the basis of the nature and size of the substrate’s side chain, i.e., aliphatic or aromatic.
Binding of aromatic amino acids was characterized using MhsT structures with bound 4F‐Phe, Tyr, Phe (this study), and Trp (Malinauskaite et al, 2014). A comparison of these four structures shows a small conformational change of the rotamer of Met236 in MhsT‐4F‐Phe and MhsT‐Tyr compared to the two other structures (Fig 3A). For the 4F‐Phe and Tyr complexes, the sulfur atom of Met236 is pointing in the direction of the fluorine atom of 4‐F‐Phe making a polar interaction, and the hydroxyl group of Tyr making a favorable hydrogen bond, which is not possible for Phe and Trp substrates, where instead the hydrophobic methyl group of Met236 points in the direction of the ligands for hydrophobic interactions.
Figure 3. Binding of aliphatic and aromatic substrates to MhsT.
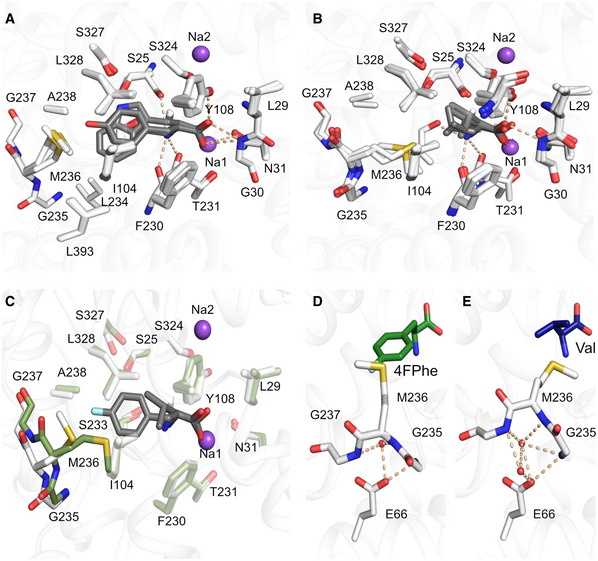
-
A, BGrouping of binding sites: (A) aromatic ligands consisting of L‐Trp, L‐Phe, L‐4‐F‐Phe, and L‐Tyr, (B) aliphatic ligands consisting of L‐Val, L‐Leu, and L‐Ile.
-
COverlay of MhsT‐Val (in green) and MhsT‐4FPhe (in white) structures to visualize changes upon binding of different sized ligands. The unwound region of TM6 is non‐transparent.
-
D, EConformation of the unwound part of TM6 in case of an aromatic substrate (L‐4‐F‐Phe as an example) and (E) aliphatic substrate (L‐Val as an example). Glu66 is coordinating the GMG loop through water molecules—in the case of aromatic substrates only one water molecule is found, whereas with aliphatic substrates two water molecules are present.
Data information: Protein is shown as white ribbon with the binding site defined with white sticks. Substrates are visualized as gray sticks and the sodium ions as purple spheres.
Binding of the aliphatic amino acids was characterized using structures obtained for the MhsT‐Val, MhsT‐Leu and MhsT‐Ile complexes (Fig 3B). Comparison of aliphatic and aromatic substrate complexes highlighted that the non‐helical fragment of TM6, formed by residues Leu234‐Gly235‐Met236‐Gly237‐Ala238, changes its relative position within the binding site depending on the size of the ligand (Figs 3C and EV2). This loop will be referred to as the GMG motif, as the shift of these three residues is most significant. The movement is most pronounced when the MhsT‐Val complex is compared to the MhsT‐Trp structure. The presence of a small substrate in the binding site prompts the inward movement of the GMG loop thus compensating for the smaller substrate side chains. The unwound region of TM6 is displaced by approx. 2 Å toward the bound substrate when comparing the position of Met236 Cα. The inward movement of the GMG loop reduces the volume of the binding pocket significantly, with volumes approaching 230 Å3 for the aromatic substrates and diminishing to 144 Å3 in the case of valine (Appendix Table S1, Fig 2, Appendix Figs S3 and S4). Taken together the seven substrate‐bound structures describe a bi‐modal binding site that distinguishes between apolar aliphatic and aromatic amino acids by a main chain movement of the GMG motif, and by finetuning of the position of individual side chains through rotamer changes of the M236 side chain.
Figure EV2. 2F o−F c electron density maps contoured at 1.0 r.m.s.d. for the unwound region of TM6 of MhsT in complex with L‐Phe, L‐4‐Phe, L‐Tyr, L‐Val, L‐Leu, and L‐Ile.
Overall changes in the volume of the MhsT hydrophobic cavity upon binding of different substrates could follow an “induced‐fit” mechanism (Klingenberg, 2005; Nyola et al, 2010) or conformational selection (Hammes et al, 2009; LeVine & Weinstein, 2014). Specifically, the hydrophobic nature of the substrate and its binding pocket disfavors the possibility of increased solvation compensating for the deficit in substrate volume, thus promoting an intrinsic structural fit of the substrate binding site—a mechanism, which is also reminiscent of the movement of the unwound segment of TM1 to compensate the empty hydrophobic‐lined substrate binding site in the occluded return state of LeuT and other SLC6 family transporters (Malinauskaite et al, 2016).
The conserved Glu66
Proximal to the substrate binding site, however, a glutamate residue (Glu66) is buried in the MhsT structure and interacts with the backbone of the GMG motif of TM6. Interestingly, Glu66MhsT is conserved throughout the SLC6 family (Appendix Fig S5A). Mutagenesis of the equivalent glutamate residue in SERT (Korkhov et al, 2006), DAT (Chen et al, 2001; Sen et al, 2005), NET (Sucic et al, 2002), and GAT1 (Keshet et al, 1995) diminishes transport activity, supporting its important role in substrate translocation. In SERT, interaction between this glutamate (Glu136SERT) and TM6 has been proposed to be crucial for conformational transitions of the protein (Korkhov et al, 2006), allowing for changes between the outward and inward‐facing states associated with the transport cycle. MhsT offers the opportunity to analyze interactions of this residue in the occluded inward‐oriented state.
Even though the unwound region of TM6 adopts different conformations in the aliphatic and aromatic substrate‐bound MhsT complexes, interactions with Glu66 are maintained. These interactions proceed through both direct and more flexible water‐mediated hydrogen bonds (Figs 3D and E, and EV3). With smaller aliphatic substrates bound to MhsT, the space formed through the displacement of unwound TM6 (including GMG) is filled with an additional ordered water molecule interacting with both Glu66 and unwound TM6. The water molecules could have two functions (i) to stabilize the different conformations of unwound TM6 (B‐factor analysis of all complexes revealed that the conformation of unwound TM6 is largely rigid for all bound substrates, Fig 4A and B) and (ii) to preserve the interaction with Glu66 and its critical role in transport.
Figure EV3. F o−F c simulated annealing omit maps of the water molecules near the GMG loop contoured at ± 3.0 r.m.s.d. of MhsT in complex with L‐Phe, L‐4‐F‐Phe, L‐Tyr, L‐Val, L‐Leu, and L‐Ile.
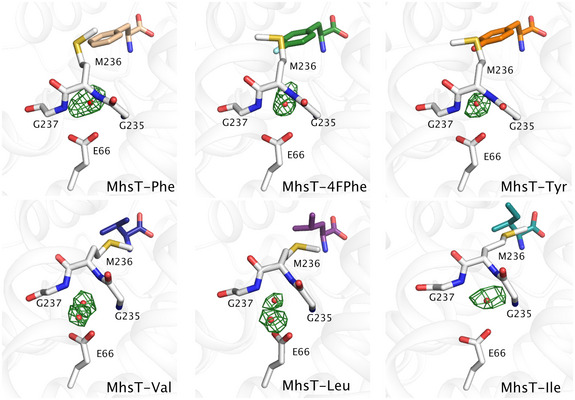
Figure 4. Structural flexibility analysis of the MhsT‐substrate complexes.
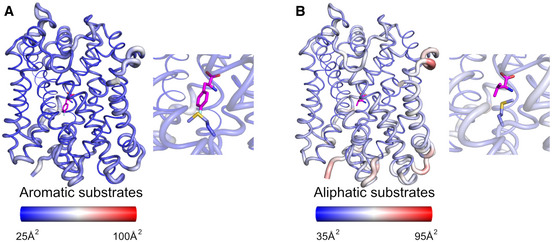
-
A, BStructural flexibility analysis of MhsT in complex with (A) L‐4‐F‐Phe and (B) L‐Val as representatives of the aromatic and aliphatic substrates, respectively. Ligands are shown as magenta sticks. The analysis was visualized by atomic putty and thermal gradient representation of B‐factor in Pymol. Red color indicates a high degree of disorder, while blue color indicates a low degree of disorder. In the inset, the substrates as well as the GMG loop are highlighted.
Indeed, a comparison of the various known LeuT and MhsT structures in different states highlights a consistent interaction of Glu66 with the unwound TM6 in spite of the significant rearrangements of TM2, TM6a and TM6b during the transport cycle (Appendix Fig S6). The interaction is also maintained when different sized substrates are bound, underlying the importance of this interaction. The Glu66 residue could be viewed as a fulcrum at the protein core about which changes related to the transport mechanism happen.
Conservation of the GMG loop
Unlike the considerable conservation of the unwound region of TM1 in SLC6 transporters, which has been reported as important also for the substrate‐free return transition (Malinauskaite et al, 2016), the unwound region of TM6 is far more diverse in sequence. The central residue, Met236 in MhsT, is typically a non‐polar residue ranging in size from leucine (e.g., GABA transporter) to tryptophan (e.g., glycine transporter) with a majority of SLC6 transporters, like LeuT, having a phenylalanine (e.g., serotonin and dopamine transporters). A C‐terminally flanking glycine, Gly237 in MhsT, is fully conserved in the SLC6 family (Appendix Fig S5B), while the N‐terminally flanking residue, Gly235 in MhsT, is typically a cysteine, glycine, or alanine. The flanking residues may determine the degree of flexibility of the loop, which, combined with the diversity of the unwound TM6 sequence, is likely related to the substrate specificity of the transporter, eventually with promiscuity to a broader range of substrates as for the GMG motif of MhsT.
Like MhsT, the closely related amino acid transporters SLC6A18 (B0AT3) and SLC6A19 (B0AT1) are neutral amino acid transporters, although with a specific preference for aliphatic amino acids (Camargo et al, 2005; Broer et al, 2006; Singer et al, 2009). Sequence alignment of MhsT, SLC6A18 and SLC6A19 identified similarities and differences between their binding pockets (defined as residues within 4 Å from bound substrate in MhsT, Appendix Table S2). The substrate binding site of MhsT exhibits ~ 50% conservation compared to SLC6A18 and SLC6A19. In both transporters, the unwound TM6 motif is AFG, with the reduced flexibility and bulkier central Phe residue putatively defining the substrate specificity toward a preference for aliphatic amino acids similar to LeuT. Interestingly, in both SLC6A18 and SLC6A19 the AFG motif is followed by an additional glycine that may provide additional, spatial flexibility (Appendix Fig S5).
To probe the role of the unwound region of TM6 and the composition of the substrate binding site on substrate specificity of the SLC6 transporters, we generated several mutant forms of MhsT in which the binding site mimicks the binding sites of SLC6A18 and SLC6A19. Comparable levels of expression of all of the constructs were confirmed suggesting that alterations of transport rates were caused by lowered activity of the protein. These MhsT mutants were subsequently used for transport studies (Fig EV4B–J).
Figure EV4. Competitive uptake assay of L‐[3H]Leu by WT or mutated variants of MhsT in the presence or absence of selected amino acids.
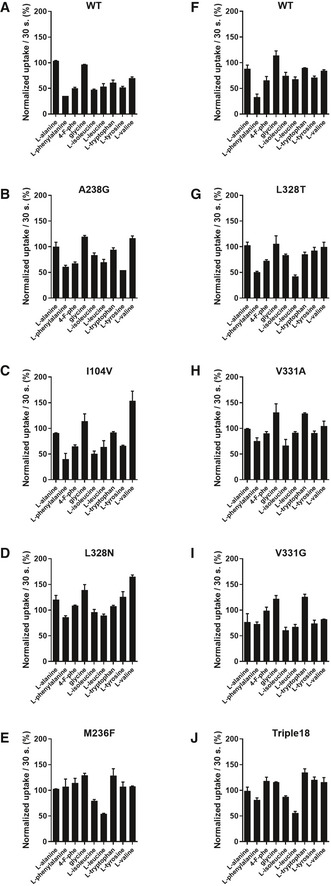
-
A–JMhsT‐WT uptake of L‐leucine is inhibited to various degree by all of the hydrophobic amino acids. The M236F mutant shows complete inability to compete l‐leucine with hydrophobic amino acids, indicating impaired binding or transport. The triple18 variant harbors the following substitutions: L328T, M236F, and V331A. Uptake measurements were performed in Escherichia coli strain YG228 expressing the indicated variants. Data are the mean ± SEM of two independent experiments each performed as technical duplicates.
In vivo uptake experiments
A competitive uptake assay of L‐[3H]Leu and non‐radiolabeled amino acids allowed for an investigation of the substrate specificity of the generated MhsT variants. From the group of single mutants, the most striking effect was observed for the M236F variant, which showed complete insensitivity toward the aromatic amino acids (Figs 5A and EV4), while the interaction of hydrophobic but non‐aromatic substrates seems to be unaffected in this variant. Measurements of initial rates of transport in MQ614 cells expressing MhsT‐WT or ‐M236F confirmed comparable kinetics of the two proteins for Leu (Fig 5). The V max of Leu transport was calculated to be 3.23 ± 0.14 nmol/min × mg of cell protein−1 for MhsT‐WT and 3.33 ± 0.13 nmol/min × mg of cell protein−1 for MhsT‐M236F. The Km and V max of Trp uptake by MhsT‐WT were 1.07 ± 0.19 μM and 2.59 ± 0.12 nmol/min × mg of cell protein−1, respectively, but could not be determined for MhsT‐M236F, suggesting complete abolishment of L‐Trp uptake by M236F variant (Fig 5C).
Figure 5. Specificity of MhsT‐M236F.
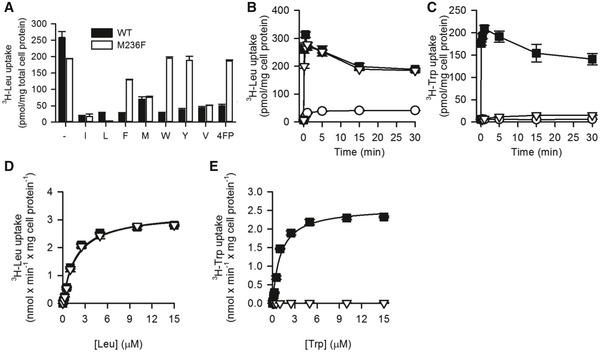
-
AUptake of 0.1 μM L‐[3H]Leu was measured for 10‐s periods in the presence or absence of the indicated natural amino acids and 4‐fluorophenylalanine (4FP, all compounds tested at 100 μM final concentration) in MQ614 expressing MhsT‐WT or –M236F.
-
B, CTime course of 0.1 µM L‐[3H]Leu (B) or 0.1 µM L‐[3H]Trp (C) by MQ614 expressing MhsT‐WT (■) or –M236F (∇), or MQ614 transformed with the control plasmid, pQE60 (○).
-
D, EKinetics of L‐[3H]Leu (D) or L‐[3H]Trp (E) uptake.
Data information: Data are the mean ± SEM of triplicate determinations of a representative experiment.
Discussion
The total of seven substrate‐bound crystal structures of MhsT, six of them determined here, highlight the structural reorganization, centered on the unwound region of TM6, that is required to accommodate the transport of aliphatic and aromatic residues. This movement tailors the S1 binding site volume and provides a structural basis for the promiscuity of the transporter. A single point mutation of Met236 to Phe, found in ~ 50% of human SLC6 and also in, e.g. LeuT, abolishes the transport of aromatic amino acids by MhsT, but maintains transport of aliphatic side chains.
Comparison of the various substrate‐bound MhsT structures illustrates that the GMG motif present in the unwound region of TM6, and conserved to some extent among other bacterial homologues and eukaryotic members of NSS family (Appendix Fig S5B), serves as a regulator of the binding site pocket volume and modulates the chemical environment. Involvement of the non‐helical fragment of TM6 in substrate binding was also observed in crystal structures of outward‐oriented LeuT with different amino acids [L‐Leu (Yamashita et al, 2005), Gly, L‐Ala, L‐Met] and their derivatives [L‐4‐F‐Phe (Singh, 2008)].
However, it seems that the two transporters use different mechanisms to accommodate for different substrates sizes, and while both MhsT and LeuT transport a broad range of hydrophobic amino acids, their specificities seem to be shifted toward opposing ends of the spectrum. LeuT is more suitable to the transport of smaller, aliphatic substrates (Gly, L‐Ala, L‐Leu) with a maximal catalytic efficiency for L‐Ala, whereas MhsT has higher apparent affinity for bulky, aromatic ligands (L‐Tyr, L‐Phe and L‐Trp). Formation of the LeuT complex with L‐Ala and Gly is possible due to torsions of two spatially neighboring residues, Phe259 and Ile359, by about 30° and 15°, respectively, when compared to the L‐Leu complex. These two residues were characterized as the volumetric sensors in LeuT and mediators of the allosteric communication between the substrates in the S1 site and the intracellular gate (LeVine & Weinstein, 2014; LeVine et al, 2019). Indeed, the Phe259 residue in LeuT corresponds to Met236 in MhsT, with both of them flanked by Gly residues (Appendix Fig S5B). While binding of the various aliphatic substrates by LeuT is associated with the same binding site architecture as for the LeuT‐Leu complex, the much bulkier 4F‐Phe requires the largest rearrangements of LeuT; Ile359 undergoes a ~ 180° rotation of its side chain, while the backbone of the unwound region of TM6 is slightly shifted outward in order to accommodate the substrate within the protein cavity (Singh et al, 2008). This is similar, but significantly less pronounced, than the outward movement of unwound TM6 region in MhsT with aromatic substrates. Singh et al (2008) speculated that the displacement of the unwound TM6 region in the LeuT‐L‐4F‐Phe structure marked a strained occluded state, with a reduced rate of transition to the inward‐open state, consistent with the lower tyrosine turnover rate. Furthermore, the larger L‐Trp acts as a competitive inhibitor for LeuT and traps the transporter in an outward‐open conformation. In light of our findings for MhsT, this movement of unwound TM6 in LeuT could also be viewed as an extension of the binding pocket in order to accommodate larger substrate side chains.
The movement of the unwound region of TM6 was also observed in crystal structures of the D. melanogaster dopamine transporter (dDAT) in complex with substrates like dopamine or amphetamines (Wang et al, 2015) and inhibitors like antidepressants (Penmatsa et al, 2013, 2015) or tropane inhibitors. In the case of dopamine, methamphetamine, or D‐amphetamine, the non‐helical fragment of TM6 is moved into the binding pocket in a similar way as for MhsT in complex with aliphatic substrates. The binding of substrates is accompanied by specific conformations of Phe319dDAT (Phe230MhsT) and Phe325dDAT (Met236MhsT) that additionally reduce the size of the binding pocket. Inhibitors lock dDAT in an outward‐open conformation, either by blocking the binding site with multiple aromatic rings, like in the case of antidepressants, where the unwound part of TM6 is moved out and the two abovementioned Phe turn away from the pocket, or by hindering the movement of the extracellular gate as in the case of tropane inhibitors. Here, the unwound part of TM6 and the two Phe residues are in similar conformations as the in the case of the substrates (Penmatsa et al, 2013).
Since the GMG motif in MhsT and its corresponding loop in LeuT and dDAT can compensate for different substrate sizes, it is tempting to speculate if the mechanism applies also for other transporters. Probably, the functional properties of the (G/A/C)ΦG motif (GMG for MhsT, GFG for LeuT, and AFG, AWG, CLG, CQG, GFG, and GLG for human SLC6 members) would be relevant in other transporters with a broad range of substrates, especially if they are of different sizes. Functional studies in Escherichia coli cells expressing WT MhsT or mutant variants mimicking the neutral amino acid transporters SLC6A18 and SLC6A19 revealed loss‐of‐function to transport aromatic amino acids (Fig EV4) with M236F (of the GMG motif) being the most significant in its effect on function of MhsT (Figs 5B–E and EV4E). Other single mutations had moderate to no effect (Fig EV4B–J). Comparable levels of expression of all of the constructs were confirmed suggesting that any alterations of the transport rates were caused by lowered activity of the protein. The observed phenotype can be easily explained by an alteration of the binding site as well as a change within the electrostatic and steric environment of the hydrophobic pocket.
Based on the available crystal structures and competitive uptake assays, we can assume the Val side chain represents the smallest ligand suitable for transport by MhsT‐WT (Appendix Fig S1 and Fig 5A). The size of Gly or Ala may be too small to be compensated by an inward tilt of the GMG loop, as Met236 presumably cannot stabilize the requirement of a smaller binding site. The M236F substitution did not result in a gain of glycine or alanine transport activity showing that also other features in the S1 site play a role in defining the substrate specificity. Possibly, mutagenesis of several residues, e.g. placed at the interface of more distant TM helices, would be required to alter the overall shape and dynamics of the binding site and thereby its binding specificity along with transport activity.
In a computational study on LeuT, the binding site residues placed in TM6 (Phe259 of the GFG motif) and TM8 (Ile359) were found to have the highest contributions in communication between the substrate binding site and the intracellular gate of LeuT, thus serving as important residues in allosteric signaling during the substrate transport (LeVine & Weinstein, 2014; LeVine et al, 2019). Also, among the residues present within the LeuT S1 site, Phe259 was reported to have the highest contribution to coordination of the intracellular gate. In general, the findings highlight TM6 as a major mediator in coordination of the intracellular gate by residues from S1 and S7 sites of LeuT with F259 (MhsT M236) being proposed to serve as a regulatory residue which, e.g. in the case of L‐Trp binding to LeuT, leads to inhibition (LeVine & Weinstein, 2014).
As observed also in the MhsT‐substrate complexes, the unwound region of TM6 interacts with a highly conserved Glu66 from TM2. The residue, when mutated in SERT, was shown to reduce or abolish the transport, while not affecting the substrate binding suggesting it is crucial for conformational transitions of the protein (Korkhov et al, 2006). The negative influence of the conserved Glu substitutions was also reported for DAT (Chen et al, 2001), (Sen et al, 2005) and NET (Sucic et al, 2002). Our MhsT structures reveal that depending on the size of the bound substrate water molecules and H‐binding networks involved in the coordination of the unwound region of TM6 to Glu66 are slightly changed, but maintained (Fig EV3). These interactions and the presence of substrate and Na+ inside the binding pockets support transition from the substrate‐bound, outward facing state toward the inward‐facing conformation and substrate release. The water molecules mediating contacts to TM6 are also found in the LeuT structures (Dang et al, 2010), and a comparison of the various LeuT and MhsT structures in different conformational states highlights that the interaction of the buried glutamate on TM2 with the unwound TM6 is maintained through the transport cycle (Appendix Fig S6). Given the importance of this interaction to transport, it is remarkable that in MhsT the significant movements of unwound TM6 are accommodated and play a crucial role in substrate specificity. The corresponding residue in other SLC6 transporters (Appendix Fig S5A) is Glu84DAT, Glu136SERT, and Glu62LeuT. In most cases, it interacts with another glutamate residue on TM10 (Glu490DAT, Glu508SERT, Glu419LeuT) and the interaction has been noted as important for protein stability and conformational changes (Koldso et al, 2013). The mutation of the interacting glutamate residue on TM10 to a lysine is one of the disease mutations causing Hartnup disease in SLC6A19 [E501K (Seow et al, 2004)].
Why is Glu66 at all conserved as a glutamic acid and cannot be replaced by, e.g., a glutamine residue? Presumably, Glu66 (and its equivalent residue in other SLC6 transporters) is even protonated and neutral in the buried environment [PROPKA estimates a pKa of 7.6 (molecule A) and 8.1 (molecule B) for the L‐Val complex and 6.9 (partially protonated) for the Trp complex (Olsson et al, 2011)], but the hydrogen position of such a functionality is dislocated and can shift to support a rapid and dynamic change in hydrogen bonding capacities that promotes transport, unlike for a glutamine side chain that would have to rotate fixed hydrogen bonding geometries to align with dynamic transitions associated with transport.
Many transporters belonging to other SLC families share the LeuT fold despite of low sequence similarity, implying that structural as well as mechanistic similarities are present, e.g., the arginine/agmatine antiporter, AdiC (Gao et al, 2010; Kowalczyk et al, 2011), and the L‐5‐benzyl‐hydantoin bound sodium‐benzylhydantoin transporter, Mhp1 (Weyand et al, 2008). In both cases, the substrates are bound to the transporters in a similar manner as LeuT and MhsT, with an additional π‐cation interaction in the case of AdiC and a π‐stacking interaction in the case of Mhp1. Similarly, looking at the apo state of the organocation transporter, ApcT (Shaffer et al, 2009), a water filled cavity large enough to accommodate an L‐Phe substrate molecule is found at the same location. However, the betaine transporter, BetP (Ressl et al, 2009), the galactose transporter, vSGLT (Faham et al, 2008), and l‐carnitine transporter, CaiT (Tang et al, 2010) transporters have binding pockets shifted toward TM2 and TM7, but with the TM1 and TM6 still participating in substrate binding. Notably, however, the conserved (G/A/C)ΦG sequence in TM6 of SLC6 transporters is not conserved within the unwound region of TM6 of transporters that do not belong to the SLC6 family, despite that fact that they share the overall LeuT fold. This is pointing to a specific role of an unwound region in TM6 in the case of SLC6 transporters including the NSS and AAT subfamilies. Furthermore, the conserved Glu residue in TM2 (Glu66 in MhsT) is only conserved among the SLC6 members, further corrobating the notion of distinct transport mechanisms along the lines of the existing gene families. The common LeuT fold therefore appears to represent an archetype of transporters that, by rather small structural modifications, has evolutionarily developed into distinct transporter families with varying substrate translocation mechanisms for different physiological conditions and substrates.
Materials and Methods
Reagents
All chemicals were obtained from Sigma, unless stated otherwise. Amino acids stocks were prepared at 10 mM concentration in H2O and were stored at 4°C.
Protein expression and purification for structural studies
The mhsT gene was cloned into the pNZ8048 vector containing a chloramphenicol resistance gene, an N‐terminal His‐tag, and a TEV protease cleavage site. The protein was expressed in Lactococcus lactis NZ9000 strain and purified as described previously (Fig EV5A–C) (Malinauskaite et al, 2014).
Figure EV5. Purification of the MhsT‐substrate complexes and expression of the MhsT mutants.
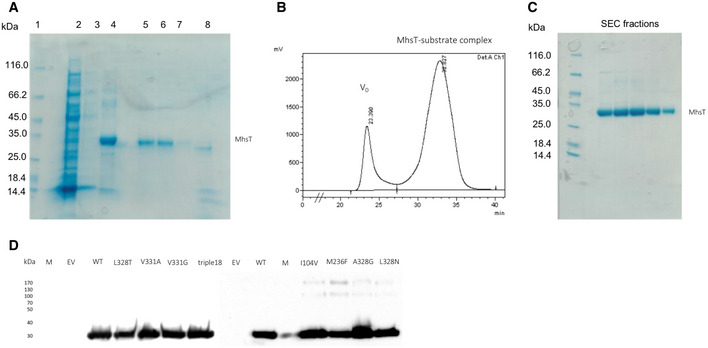
- SDS–PAGE of the Ni2+‐affinity chromatography and reverse Ni2+‐affinity chromatography of the MhsT‐substrate complexes. Samples order: 1—Protein marker; 2, 3, 4—Fractions from Ni2+‐ affinity chromatography (respectively: flow‐through, wash, and elution in 250 mM imidazole buffer); 5, 6, 7, 8—Fractions from reverse Ni2+‐affinity chromatography (respectively: flow‐through, wash (two lanes), and elution of the uncleaved protein and TEV protease in 250 mM imidazole buffer.
- Size exclusion chromatogram (TSKgel G3000SW gel filtration column, flow rate = 0.5 ml/min).
- SDS–PAGE of the SEC peak fractions.
- An example of Western blot analysis of MhsT expression in cells. The immunological detection of the relative amount of MhsT‐WT and selected MhsT mutants confirms comparable protein expression levels.
Source data are available online for this figure.
Protein relipidation and crystallization
Purified MhsT, concentrated to approx. 9 mg/ml, was relipidated overnight in 1,2‐dioleoyl‐sn‐glycero‐3‐phosphocholine (DOPC) at w/w ratio 3:04 and 3:08 protein:lipids, according to the method described previously (Malinauskaite et al, 2014). Prior to crystallization experiment, protein sample was spun down at 290,000 × g for 20 min and mixed with either n‐octyl‐β‐D‐glucopyranoside (OG; Anatrace) or n‐nonyl‐β‐D‐glucoside (NG; Anatrace) detergent to final detergent concentration of 4 CMC.
Crystallization was achieved at 19°C by vapor diffusion in hanging drop using immersion oil as a sealing agent. The reservoir buffer composition covered 14–24% PEG400, 0.3–0.5 M NaCl, 100 mM Tris–HCl, or HEPES‐NaOH pH 7.0, 5%, or 10% glycerol, 5% Trimethylamine N‐oxide (TMANO). Obtained crystals were harvested at 4°C and tested for diffraction at the Diamond synchrotron facility using beamline I24 and I04, Swiss Light Source using beamline PXI.
Data processing and structure refinement
Obtained datasets were processed in P2 or P21 space groups using XDS package (Kabsch, 2010) and CCP4 program suite (Collaborative Computational Project N, 1994). The initial phases for the structures were obtained using Phaser (McCoy, 2007) with MhsT‐Trp structure (PDB ID: 4US3) (Malinauskaite et al, 2014) as a search model excluding TM5 from the model. Initial models were rebuilt manually in COOT (Emsley et al, 2010), and atomic models were refined using phenix.refine (Adams et al, 2010). The quality of the datasets and presence of twinning were checked in phenix.xtriage. The dataset for MhsT‐Leu was refined using the (h –k –l) twin law. The final data and refinement statistics are presented in Table 1. Attached figures were prepared in PYMOL (Schrodinger, 2015), the multiple sequence alignment was performed using MUSCLE (Edgar, 2004), and the LOGO representation was obtained in CLC Main Workbench (CLC Bio, Qiagen).
Protein expression for functional studies
Desired mutations were introduced into mhsT wild‐ type gene via site‐directed mutagenesis (Mutagenesis QuickChange Lightning Kit, Agilent Technologies, Inc) using designed primers. Final constructs were verified via DNA sequencing. Subsequently, the mhsT variants were cloned into pQO6TEV vector, which is a modified version of pQE30, and were expressed in E. coli strain MQ614 [aroP mtr tnaB271∷Tn5 tyrP1 pheP::cat] as described previously (Malinauskaite et al, 2014). Briefly, an overnight preculture of E. coli cells was diluted in LB medium supplemented with 100 µg/ml ampicillin to an OD420 of 0.1. The culture was incubated at 37°C with shaking, until the OD reached 1 when the expression was induced by addition of 0.3 mM IPTG. Induced cells were incubated at 37°C with shaking for two additional hours. Afterward, the cells were harvested by centrifugation for 10 min at 13,200 × g, at 4°C. The pellet was washed twice in 100 mM Tris/MES, pH 7.5 and stored on ice until the uptake experiment.
Transport measurements in intact Escherichia coli cells
Uptake of L‐[3H]Trp (18 Ci/mmol) or L‐[3H]Leu (120 Ci/mmol; both American Radiolabeled Chemicals, Inc.) was measured in intact E. coli MQ614 (Quick et al, 2006) or E. coli YG228 [Koyanagi et al, 2004] transformed with pQE60 or its derivatives harboring indicated MhsT variants. Cells were prepared for uptake studies as described (Quick et al, 2006), and uptake was performed in 10 mM Tris/Mes, pH 8.5, 150 mM NaCl at a final total cellular protein concentration of 0.035 mg/ml in the presence or absence of substrates or inhibitors as indicated. 100 μl samples were assayed for the indicated time periods, and the uptake reactions were quenched by the addition of 100 mM KPi, pH 6.0, and 100 mM LiCl. Cells were collected on Advantce MFS GF75 glass fiber filters. The accumulated radioactivity was determined (as counts per minute, cpm) in a Hidex 300 SL scintillation counter. Known amounts of radioactivity were used to determine the cpm‐to‐pmol conversion.
Immunological protein detection
Relative amounts of the respective MhsT variants in the membrane of MQ614 were detected by Western blotting using a monoclonal antibody against the N‐terminal His tag present in all MhsT constructs (Fig EV5D). 10 μg of total membrane protein in membrane vesicles of MQ614 harboring the indicated MhsT variant (or the control plasmid) was subjected to 11% SDS–PAGE followed by incubation of the membrane with the His probe antibody (Santa Cruz Biotechnology, Inc.) and horseradish peroxidase‐based chemiluminescence detection (SuperSignal® West Pico kit, Thermo Scientific).
Data analysis
All uptake measurements were performed in duplicate or triplicate and repeated at least five times. Data (shown as mean ± SEM of triplicate determination) are from representative experiments in which all constructs were assayed in parallel. The Michaelis–Menten constant (K m) and maximum velocity (V max) of transport were determined by fitting the data of 10‐s uptake measurements plotted as function of the concentration of the respective substrate to the Michaelis–Menten equation in GraphPad Prism 7.0. They are shown as mean ± SEM of the fit.
Author contributions
DF and CN involved in equal contributions on production of MhsT‐substrate complexes and crystallization. DF and JL involved in data collection, processing, and model refinement of MhsT‐Tyr, MhsT‐4FPhe, and MhsT‐Leu. CN involved in data collection, twinning analysis, processing, and model refinement of MhsT‐Tyr, MhsT‐Phe, MhsT‐Leu, MhsT‐Val, and data processing, twinning analysis, and model refinement of MhsT‐Ile with inputs from JL and PN. AEB involved in crystallization of MhsT‐Ile, RB: expression, purification, and initial crystallization of MhsT‐Val. DF, IOS, MQ involved in E. coli uptake experiments with inputs from JAJ. LM involved in initial experiments on crystallization. PN: involved in project design and supervision. DF and CN drafted the manuscript with input from JL, MQ, and PN.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Acknowledgements
The authors are grateful to technical assistance by Tetyana Klymchuk, Lotte T. Pedersen, Anna Marie Nielsen, and Audrey Warren and support for computing by Jesper L. Karlsen. We thank Steffen Sinning and Birgit Schiøtt for fruitful discussions. Work on the project was supported by a Short Term EMBO Fellowship [ASTF 80‐2016] and Boehringer Ingelheim Fonds travel grant (2015) to DF, a PhD fellowship from the Lundbeck Foundation to CN (2015‐3225), a PhD fellowship from the Boehringer Ingelheim Fonds to LM (2010), and a post‐doctoral fellowship from the Lundbeck Foundation to JAL (2015‐2704). The research was supported by NIH grants U54GM087519 and R01DA04510 to JAJ, R01GM119396 to MQ, and by the Lundbeck Foundation grants 2011‐3868 and 2016‐2518 to PN.
The EMBO Journal (2021) 40: e105164.
Contributor Information
Matthias Quick, Email: mq2102@cumc.columbia.edu.
Poul Nissen, Email: pn@mbg.au.dk.
Data availability
The atomic coordinates and structure factors of the six MhsT‐substrate complexes produced in this study have been deposited to the Protein Data Bank https://www.rcsb.org/ under accession codes: 6YU2 (MhsT‐Ile), 6YU3 (MhsT‐Phe), 6YU4 (MhsT‐4FPhe), 6YU5 (MhsT‐Val), 6YU6 (MhsT‐Leu), and 6YU7 (MhsT‐Tyr).
References
- Abramson J, Wright EM (2009) Structure and function of Na(+)‐symporters with inverted repeats. Curr Opin Struct Biol 19: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J, Stuhr‐Hansen N, Zachariassen L, Toubro S, Hansen SM, Eildal JN, Bond AD, Bogeso KP, Bang‐Andersen B, Kristensen AS et al (2011) Molecular determinants for selective recognition of antidepressants in the human serotonin and norepinephrine transporters. Proc Natl Acad Sci USA 108: 12137–12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer A, Klingel K, Kowalczuk S, Rasko JE, Cavanaugh J, Broer S (2004) Molecular cloning of mouse amino acid transport system B0, a neutral amino acid transporter related to Hartnup disorder. J Biol Chem 279: 24467–24476 [DOI] [PubMed] [Google Scholar]
- Broer A, Cavanaugh JA, Rasko JE, Broer S (2006) The molecular basis of neutral aminoacidurias. Pflugers Arch 451: 511–517 [DOI] [PubMed] [Google Scholar]
- Broer S (2008) Apical transporters for neutral amino acids: physiology and pathophysiology. Physiology 23: 95–103 [DOI] [PubMed] [Google Scholar]
- Broer S, Bailey CG, Kowalczuk S, Ng C, Vanslambrouck JM, Rodgers H, Auray‐Blais C, Cavanaugh JA, Broer A, Rasko JE (2008) Iminoglycinuria and hyperglycinuria are discrete human phenotypes resulting from complex mutations in proline and glycine transporters. J Clin Investig 118: 3881–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer S (2009) The role of the neutral amino acid transporter B0AT1 (SLC6A19) in Hartnup disorder and protein nutrition. IUBMB Life 61: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo SM, Makrides V, Virkki LV, Forster IC, Verrey F (2005) Steady‐state kinetic characterization of the mouse B(0)AT1 sodium‐dependent neutral amino acid transporter. Pflugers Arch 451: 338–348 [DOI] [PubMed] [Google Scholar]
- Chen N, Vaughan RA, Reith ME (2001) The role of conserved tryptophan and acidic residues in the human dopamine transporter as characterized by site‐directed mutagenesis. J Neurochem 77: 1116–1127 [DOI] [PubMed] [Google Scholar]
- Cherezov V (2011) Lipidic cubic phase technologies for membrane protein structural studies. Curr Opin Struct Biol 21: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Green EM, Gouaux E (2016) X‐ray structures and mechanism of the human serotonin transporter. Nature 532: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N (2010) Structure of a fucose transporter in an outward‐open conformation. Nature 467: 734–738 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321: 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald GA, Terry DS, Warren AL, Quick M, Javitch JA, Blanchard SC (2019) Quantifying secondary transport at single‐molecule resolution. Nature 575: 528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhou L, Jiao X, Lu F, Yan C, Zeng X, Wang J, Shi Y (2010) Mechanism of substrate recognition and transport by an amino acid antiporter. Nature 463: 828–832 [DOI] [PubMed] [Google Scholar]
- Gether U, Andersen PH, Larsson OM, Schousboe A (2006) Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol Sci 27: 375–383 [DOI] [PubMed] [Google Scholar]
- Gourdon PA, Lauwring J, Langmach KL, Bublitz M, Pedersen BP, Xiang‐Yu L, Yatime L, Nybolm M, Nielsen TT, Olesen C et al (2011) HiLiDe—systematic approach to membrane protein crystallization in lipid and detergent. Cryst Growth Des 11: 2098–2106 [Google Scholar]
- Hammes GG, Chang YC, Oas TG (2009) Conformational selection or induced fit: a flux description of reaction mechanism. Proc Natl Acad Sci USA 106: 13737–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Vasiliou K, Nebert DW (2009) Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genom 3: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010) Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet GI, Bendahan A, Su H, Mager S, Lester HA, Kanner BI (1995) Glutamate‐101 is critical for the function of the sodium and chloride‐coupled GABA transporter GAT‐1. FEBS Lett 371: 39–42 [DOI] [PubMed] [Google Scholar]
- Klingenberg M (2005) Ligand‐protein interaction in biomembrane carriers. The induced transition fit of transport catalysis. Biochemistry 44: 8563–8570 [DOI] [PubMed] [Google Scholar]
- Koldso H, Christiansen AB, Sinning S, Schiott B (2013) Comparative modeling of the human monoamine transporters: similarities in substrate binding. ACS Chem Neurosci 4: 295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov VM, Holy M, Freissmuth M, Sitte HH (2006) The conserved glutamate (Glu136) in transmembrane domain 2 of the serotonin transporter is required for the conformational switch in the transport cycle. J Biol Chem 281: 13439–13448 [DOI] [PubMed] [Google Scholar]
- Kowalczyk L, Ratera M, Paladino A, Bartoccioni P, Errasti‐Murugarren A, Valencia E, Portella G, Bial S, Zorzano A, Fita I et al (2011) Molecular basis of substrate‐induced permeation by an amino acid antiporter. Proc Natl Acad Sci USA 108: 3935–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi T, Katayama T, Suzuki H, Kumagai H (2004) Identification of the LIV‐I/LS system as the third phenylalanine transporter in Escherichia coli K‐12. J Bacteriol 186: 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jorgensen TN, Sorensen L, Eriksen J, Loland CJ, Stromgaard K, Gether U (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63: 585–640 [DOI] [PubMed] [Google Scholar]
- LeVine MV, Weinstein H (2014) NbIT–a new information theory‐based analysis of allosteric mechanisms reveals residues that underlie function in the leucine transporter LeuT. PLoS Comput Biol 10: e1003603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeVine MV, Terry DS, Khelashvili G, Siegel ZS, Quick M, Javitch JA, Blanchard SC, Weinstein H (2019) The allosteric mechanism of substrate‐specific transport in SLC6 is mediated by a volumetric sensor. Proc Natl Acad Sci USA 116: 15947–15956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laasonen‐Balk T, Kuikka J, Viinamaki H, Husso‐Saastamoinen M, Lehtonen J, Tiihonen J (1999) Striatal dopamine transporter density in major depression. Psychopharmacology 144: 282–285 [DOI] [PubMed] [Google Scholar]
- Malinauskaite L, Quick M, Reinhard L, Lyons JA, Yano H, Javitch JA, Nissen P (2014) A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat Struct Mol Biol 21: 1006–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinauskaite L, Said S, Sahin C, Grouleff J, Shahsavar A, Bjerregaard H, Noer P, Severinsen K, Boesen T, Schiott B et al (2016) A conserved leucine occupies the empty substrate site of LeuT in the Na(+)‐free return state. Nat Commun 7: 11673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos JJ, Lomena F, Parellada E, Font M, Fernandez E, Pavia J, Prats A, Pons F, Bernardo M (2005) Decreased striatal dopamine transporter binding assessed with [123I] FP‐CIT in first‐episode schizophrenic patients with and without short‐term antipsychotic‐induced parkinsonism. Psychopharmacology 181: 401–406 [DOI] [PubMed] [Google Scholar]
- McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum B (1995a) Epilepsy. Taking up GABA again. Nature 376: 122–123 [DOI] [PubMed] [Google Scholar]
- Meldrum BS (1995b) Excitatory amino acid receptors and their role in epilepsy and cerebral ischemia. Ann N Y Acad Sci 757: 492–505 [DOI] [PubMed] [Google Scholar]
- Nash SR, Giros B, Kingsmore SF, Kim KM, el‐Mestikawy S, Dong Q, Fumagalli F, Seldin MF, Caron MG (1998) Cloning, gene structure and genomic localization of an orphan transporter from mouse kidney with six alternatively‐spliced isoforms. Recept Channels 6: 113–128 [PubMed] [Google Scholar]
- Nyola A, Karpowich NK, Zhen J, Marden J, Reith ME, Wang DN (2010) Substrate and drug binding sites in LeuT. Curr Opin Struct Biol 20: 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH (2011) PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 7: 525–537 [DOI] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E (2013) X‐ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503: 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E (2015) X‐ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat Struct Mol Biol 22: 506–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Yano H, Goldberg NR, Duan L, Beuming T, Shi L, Weinstein H, Javitch JA (2006) State‐dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter:sodium symporter from Fusobacterium nucleatum . J Biol Chem 281: 26444–26454 [DOI] [PubMed] [Google Scholar]
- Quick M, Javitch JA (2007) Monitoring the function of membrane transport proteins in detergent‐solubilized form. Proc Natl Acad Sci USA 104: 3603–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Winther AM, Shi L, Nissen P, Weinstein H, Javitch JA (2009) Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor‐bound conformation. Proc Natl Acad Sci USA 106: 5563–5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick M, Abramyan AM, Wiriyasermkul P, Weinstein H, Shi L, Javitch JA (2018) The LeuT‐fold neurotransmitter:sodium symporter MhsT has two substrate sites. Proc Natl Acad Sci USA 115: E7924–E7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C (2009) Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature 458: 47–52 [DOI] [PubMed] [Google Scholar]
- Romeo E, Dave MH, Bacic D, Ristic Z, Camargo SM, Loffing J, Wagner CA, Verrey F (2006) Luminal kidney and intestine SLC6 amino acid transporters of B0AT‐cluster and their tissue distribution in Mus musculus. Am J Physiol Renal Physiol 290: F376–F383 [DOI] [PubMed] [Google Scholar]
- Schrodinger, LLC (2015) The PyMOL molecular graphics system, Version 1.8
- Sen N, Shi L, Beuming T, Weinstein H, Javitch JA (2005) A pincer‐like configuration of TM2 in the human dopamine transporter is responsible for indirect effects on cocaine binding. Neuropharmacology 49: 780–790 [DOI] [PubMed] [Google Scholar]
- Seow HF, Broer S, Broer A, Bailey CG, Potter SJ, Cavanaugh JA, Rasko JE (2004) Hartnup disorder is caused by mutations in the gene encoding the neutral amino acid transporter SLC6A19. Nat Genet 36: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E (2009) Structure and mechanism of a Na+‐independent amino acid transporter. Science 325: 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA (2008) The mechanism of a neurotransmitter:sodium symporter–inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell 30: 667–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer D, Camargo SM, Huggel K, Romeo E, Danilczyk U, Kuba K, Chesnov S, Caron MG, Penninger JM, Verrey F (2009) Orphan transporter SLC6A18 is renal neutral amino acid transporter B0AT3. J Biol Chem 284: 19953–19960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E (2007) Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448: 952–956 [DOI] [PubMed] [Google Scholar]
- Singh SK (2008) LeuT: a prokaryotic stepping stone on the way to a eukaryotic neurotransmitter transporter structure. Channels 2: 380–389 [DOI] [PubMed] [Google Scholar]
- Singh SK, Piscitelli CL, Yamashita A, Gouaux E (2008) A competitive inhibitor traps LeuT in an open‐to‐out conformation. Science 322: 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, Paczkowski FA, Runkel F, Bonisch H, Bryan‐Lluka LJ (2002) Functional significance of a highly conserved glutamate residue of the human noradrenaline transporter. J Neurochem 81: 344–354 [DOI] [PubMed] [Google Scholar]
- Tang L, Bai L, Wang WH, Jiang T (2010) Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol 17: 492–496 [DOI] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E (2015) Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 521: 322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J et al (2008) Structure and molecular mechanism of a nucleobase‐cation‐symport‐1 family transporter. Science 322: 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E (2005) Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature 437: 215–223 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Zhen J, Karpowich NK, Goetz RM, Law CJ, Reith ME, Wang DN (2007) LeuT‐desipramine structure reveals how antidepressants block neurotransmitter reuptake. Science 317: 1390–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Data Availability Statement
The atomic coordinates and structure factors of the six MhsT‐substrate complexes produced in this study have been deposited to the Protein Data Bank https://www.rcsb.org/ under accession codes: 6YU2 (MhsT‐Ile), 6YU3 (MhsT‐Phe), 6YU4 (MhsT‐4FPhe), 6YU5 (MhsT‐Val), 6YU6 (MhsT‐Leu), and 6YU7 (MhsT‐Tyr).


