Abstract
Messenger RNA degradation is an important component of overall gene expression. During the final step of eukaryotic mRNA degradation, exoribonuclease 1 (Xrn1) carries out 5′ → 3′ processive, hydrolytic degradation of RNA molecules using divalent metal ion catalysis. To initiate studies of the 5′ → 3′ RNA decay machinery in our lab, we expressed a C-terminally truncated version of Saccharomyces cerevisiae Xrn1 and explored its enzymology using a second-generation, time-resolved fluorescence RNA degradation assay. Using this system, we quantitatively explored Xrn1’s preference for 5′-monophosphorylated RNA substrates, its pH dependence, and the importance of active site mutations in the molecule’s conserved catalytic core. Furthermore, we explore Xrn1’s preference for RNAs containing a 5′ single-stranded region both in an intermolecular hairpin structure and in an RNA−DNA hybrid duplex system. These results both expand and solidify our understanding of Xrn1, a centrally important enzyme whose biochemical properties have implications in numerous RNA degradation and processing pathways.
Graphical Abstract
Processive mRNA degradation is critical to the regulation of overall gene expression. In eukaryotes, this process is carried out by an ancient and conserved set of RNA processing enzymes.1–4 In the predominant 5′ → 3′ exonucleolytic decay pathway, exoribonuclease 1 (Xrn1) serves as the major cytosolic exoribonuclease responsible for deadenylation-dependent, decapped mRNA turnover.1,5–7 Xrn1 also plays roles in mRNA processing, rRNA processing, tRNA quality control, lncRNA decay, No-Go decay, nonsense-mediated decay, miRNA-mediated decay, siRNA-mediated decay, and innate immunity.1,8–12 The ramifications of Xrn1’s varied roles are reflected by its importance in diverse biological processes, including autophagy,13 apoptosis and cancer,14,15 fertility in Drosophila,16 stress response in Arabidopsis,17–19 and antiviral defense.20,21 Additionally, a growing number of studies also point toward a central role for Xrn1 in recently proposed models of “circular” gene expression.22–26 In these models, Xrn1 and other decay factors shuttle between the nucleus and cytoplasm and buffer transcript levels by functioning both as promoter-specific transcriptional enhancers and as RNA decay factors. The observation that Xrn1 is localized in diverse locations throughout the cell, including in the nucleus,27 in the cytoplasm,27 in different types of mRNP assemblies,1 and at plasma membrane-associated eisosomes,28 portrays dynamic spatial regulation of this critical component of cellular RNA metabolism.29
Xrn1 from Saccharomyces cerevisiae was first isolated by Audrey Stevens in 197830 and subsequently further biochemically and genetically characterized by her lab and collaborators.31–34 Cumulatively, these studies revealed that Xrn1 is a processive 5′ → 3′ exoribonuclease with high specificity for 5′-monophosphorylated RNA substrates, yielding 5′-nucleotide monophosphate (5′NMP) products. Phylogenetic studies in the late 1990s revealed a conserved acidic N-terminal region, including a series of conserved Asp, Glu, His, and Lys residues that when mutated show a loss of RNA degradation activity in the cell.35
Two crystal structures of C-terminally truncated forms of Xrn1, one of Drosophila melanogaster36 and the other of Kluyveromyces lactis,37 have provided insight into the structural basis for mRNA degradation. Domain analyses and comparisons between the structures show strong conservation of the N-terminus and a high degree of conservation in the enzyme’s active site.35–38 The N-terminal region of the enzyme (amino acids 1−773 in S. cerevisiae) constitutes the conserved catalytic core of the enzyme, harboring seven acidic residues (D35, D86, E176, E178, D206, D208, and D291) that have been implicated in binding two catalytic magnesium ions.36–38 Four other basic residues (K93, Q97, R100, and R101) are proposed to interact with the negatively charged 5′-phosphate of a substrate RNA, generating electrostatic forces that help drive translocation during each catalytic cycle.36,37 Finally, two aromatic residues, H41 and W638, are oriented coaxially to the RNA nucleobases in the substrate-bound structure, suggesting an extended π-stacking network that may contribute to translocation and proper orientation of the scissile phosphate during hydrolysis.36,37
Within Xrn1’s N-terminal domain is an expansion segment only present in yeast, encompassing amino acids 352−499 in S. cerevisiae, which has recently been structurally shown to interact with the 60S subunit of the yeast ribosome during No-Go decay.38 Immediately following the internal expansion are regions with homology to PAZ and Tudor domains, which have been shown to specifically bind the 40S subunit of the eukaryotic ribosome.36,38 A winged-helix motif and SH-like motif are present from amino acids 963−1132 and 1132−1240, respectively,36 though the functions of these motifs are currently poorly characterized. The C-terminal region of Xrn1, consisting roughly of residues 1240 onward, is believed to be poorly structured and serve as a protein scaffold for association of RNA decay factors such as the decapping complex and other mRNA decay factors.37,39 The C-terminal domain (CTD) of Xrn1 is known to be less conserved and not essential for catalysis35 and in developing models serves as a species-specific scaffold for other proteins involved in mRNA decay.37,40,41 Structural studies have demonstrated that C-terminal truncations up to residue 1245 in K. lactis, homologous to residue 1242 in Drosophila and residue 1240 in S. cerevisiae, are tolerated and the enzyme retains catalytic activity.35–37 This leads to a general model in which the conserved N-terminal domain of the Xrn1 is involved in its catalytic function while the C-terminus takes part in diverse, species-specific regulatory functions.37,40,41
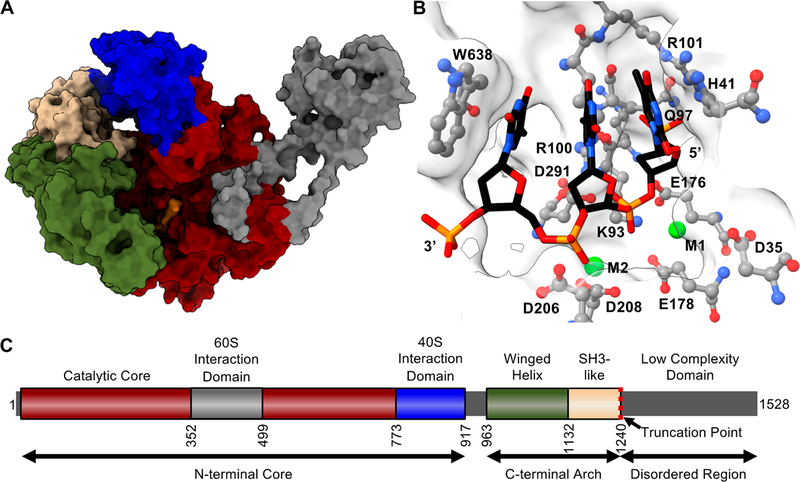
Combined, existing structural data and the high degree of conservation of Xrn1 allowed us to implement the I-TASSER42 (Iterative Threading ASSEmbly Refinement) tool to generate the structural model for the C-terminally truncated form of S. cerevisiae Xrn1, ScXrn1ΔC shown in Figure 1. Over the aligned regions of the sequence of ScXrn1ΔC, the K. lactis37 and Drosophila36 structures demonstrate 56% and 33% sequence identity, respectively, with a root-mean-square deviation (RMSD) of 3.65 Å to the more closely related K. lactis structure, which like ScXrn1ΔC includes the expansion segment of residues 352−499 that is unique to yeast. Here we have used this structural model to guide updated characterization of the enzymology of this centrally important cellular enzyme, synthesize 50 years of biochemical investigations, and address outstanding questions regarding the kinetic behavior of Xrn1, its substrate specificity, and how structured RNAs can obstruct the progress of its helicase activity.
Figure 1.
Structural model of ScXrn1ΔC. (A) Structural model of ScXrn1ΔC generated using the I-TASSER iterative threading model.42 Regions are colored to correspond to domain architecture in panel C; a DNA trinucleotide substrate, dTdTdT, from the Drosophila structure (PDB entry 2Y35)36 is colored orange. (B) Active site of the I-TASSER-generated structure of ScXrn1ΔC with the TTT trinucleotide substrate from the Drosophila structure (PDB entry 2Y35)36 and metal ions from both Drosophila and Kluveromyces (PDB entry 3PIF)37 structures modeled in. (C) Domain architecture of ScXrn1. The N-terminal core (residues 1−773) of the enzyme includes the active site. The following region of residues 774−917 forms a domain recently shown to interact with the 40S subunit of the ribosome.38 Residues 963−1132 and 1133−1240 form motifs most closely resembling winged-helix and SH3 motifs, respectively. The truncation point of ScXrn1ΔC is indicated by a red dashed line at residue 1240.
MATERIALS AND METHODS
Expression of a S. cerevisiae Xrn1 (1−1240), ScXrn1ΔC.
Residues 1−1240 of S. cerevisiae Xrn1 (UniProt ID Xrn1_Yeast, P22147) were purchased subcloned into a pET26b(+) vector (GenScript) containing an in-frame C-terminal hexahistidine affinity tag. The protein was recombinantly expressed in BL21 (DE3) Rosetta cells. Cells were grown to an OD600 of 0.6, and then induction was performed using 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 18 °C. Pelleted cells were chemically lysed using 2 mg mL−1 deoxycholic acid for 30 min on ice. The cell lysate was sonicated for 12 rounds of 15 s at 50 W on ice. The cell lysate was clarified by centrifugation at 29000g for 75 min. The soluble fraction was purified by nickel affinity chromatography in a buffer containing 300 mM NaCl, 50 mM Tris (pH 8.0), 250 mM imidazole, and 10 mM β-mercaptoethanol. Secondary chromatography was performed using a Superdex 200 gelfiltration column (GE Life Sciences) in 300 mM NaCl, 50 mM Tris (pH 7.5), and 2 mM DTT. Protein was concentrated to 2 mg mL−1 in a buffer containing 150 mM NaCl, 25 mM Tris (pH 7.5), 1 mM DTT, and 50% (v/v) glycerol and then stored at −80 °C.
Point mutants of Xrn1 were generated using the Q5 Site-Directed Mutagenesis Kit according to the manufacturer’s protocols (New England BioLabs). Constructs were sequenced to confirm proper incorporation of each mutation. Mutant proteins were purified as described above.
Expression of Bdellovibrio bacteriovorus RNA Pyro-phosphohydrolase, BdRppH.
A plasmid containing BdRppH (Uniprot ID Q6MPX4_BDEBA, Q6MPX4) in frame with a hexahistidine tag was a kind gift of J. Belasco at New York University. BdRppH was purified in a manner identical to that of Xrn1 as described above. The protein was concentrated to 12 mg mL−1 in a buffer containing 150 mM NaCl, 25 mM Tris (pH 7.5), 1 mM DTT, and 50% (v/v) glycerol and then stored at −80 °C.
In Vitro RNA Transcription.
DNA templates were ordered as gBlock DNA fragments (IDT) and subcloned into pUC19; 200 μL polymerase chain reactions (PCRs) using primers containing an upstream T7 promoter were carried out to generate dsDNA templates for transcription. Typical PCR conditions: 100 ng of plasmid DNA, 0.5 μM forward and reverse DNA primers, 500 μM dNTPs, 25 mM TAPS-HCl (pH 9.3), 50 mM KCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, and Taq DNA polymerase (New England BioLabs). dsDNA amplification was confirmed by 1.5% agarose gel electrophoresis. Transcriptions were performed in a 1 mL volume using 200 μL of PCR product. Transcription conditions: ~0.1 μM template DNA, 10 mM NTPs, 75 mM MgCl2, 30 mM Tris (pH 8.0), 10 mM DTT, 0.1% spermidine, 0.1% Triton X-100, and T7 RNA polymerase. Reaction mixtures were incubated for 10 h at 37 °C. After transcription, inorganic pyrophosphate was removed by centrifugation at 5000g for 5 min. RNA was purified by high-pressure liquid chromatography (HPLC). For HPLC purification, 1 mL transcription reaction mixtures were first diluted in 75% ethanol, brought to a final concentration of 300 mM sodium acetate, then precipitated for ≥3 h at −80 °C, and centrifuged to pellet RNA. Pelleted RNA was resuspended in 2 mL of 300 mM EDTA and 100 mM triethylammonium acetate (TEAA) (pH 7.0) immediately before HPLC purification. RNAs were purified using an Agilent 1260 Infinity II HPLC instrument using a 1 to 50% acetonitrile gradient in 100 mM TEAA (pH 7.0) with a PLRP-S Varian 8 μm, 150 mm × 7.5 mm column. RNA-containing fractions were collected and verified by 8 M urea, TBE, and 10% dPAGE and then visualized by methylene blue staining. Homogenous fractions were pooled and buffer-exchanged into nuclease-free molecular biology grade water (Applied Biosystems) and then concentrated using spin concentrators (Sartorius). RNAs were stored at −80 °C with working stocks stored at −20 °C.
In Vitro Exonuclease Activity Assay.
Assays were performed using 2 μg of RNA (ranging from ~40 to ~60 pmol depending on the construct used) in 40 μL of EC3K+ buffer containing 100 mM KCl, 50 mM Tris (pH 7.9, unless otherwise indicated), 10 mM MgCl2, and 1 mM DTT. RNAs were heated to and held at 85 °C for 2 min and then cooled to 4 °C for 2 min. Then, 2 μL of 12 mg mL−1 BdRppH was added (unless indicated for controls). Then, 2 μL of 2 mg mL−1 Xrn1 was added. Reaction mixtures were incubated at 37 °C for 2 h, and then reactions quenched with an equal volume of dPAGE loading dye containing 8 M urea, 50 mM EDTA, 0.1% (w/v) bromophenol blue, and 0.1% (w/v) xylene cyanol. Reaction mixtures were electrophoresed on an 8 M urea, TBE, 10% dPAGE gel and then visualized by methylene blue staining.
Time-Resolved Fluorescence-Based RNA Degradation (TRFRD) Assay.
Assays were performed at 37 °C for 150 min in 100 μL volumes or as indicated. Prior to each experiment, RNAs (typically 200 pmol) were folded in 50 μL of EC3K+ buffer containing 100 mM KCl, 50 mM Tris (pH 7.9, unless otherwise indicated), 10 mM MgCl2, and 1 mM DTT. RNAs were heated to and held at 85 °C for 2 min and then cooled to 4 °C for 2 min. Following reannealing, RNAs were diluted into an additional 50 μL of EC3K+ buffer supplemented with 100 μM DFHBI. The iSpinach aptamer was allowed to bind the DFHBI at 37 °C for 25 min. Fluorescence was measured by excitation at 466 nm and emission at 503 nm at 1 min intervals at 37 °C using a Tecan Infinite M1000 plate reader. To ensure that substrate RNAs became monophosphorylated, 250 pmol of BdRppH was added to the reaction mixture and then incubated for 30 min at 37 °C in a plate reader as described above. To initiate the degradation reaction, 5 pmol of Xrn1 was added and the reaction monitored. Reactions were quenched with an equal volume of dPAGE loading dye containing 8 M urea, 50 mM EDTA, 0.1% (w/v) bromophenol blue, and 0.1% (w/v) xylene cyanol. Reaction mixtures were electrophoresed on an 8 M urea, TBE, 10% dPAGE gel and then visualized by methylene blue staining.
In Vitro RNA Monophosphorylation.
HPLC-purified RNAs were treated with 1/10 of their molar concentration of BdRppH for 3 h at 37 °C in 1× EC3K+ buffer containing 100 mM KCl, 50 mM Tris (pH 7.9), 10 mM MgCl2, and 1 mM DTT. Monophosphorylated RNAs were then precipitated and repurified by HPLC as described above.
Xrn1 Kinetics.
Kinetic traces were fit to a single-exponential, three-parameter equation of the form A = A0 exp(−kt) + b in MATLAB version R2018b using a nonlinear regression model, where A0 is the initial amount of substrate and b is the undegraded population of RNA, typically <25%, yielding a typical r2 of ≥0.99. Similarly, for normalized data, the initial concentration of RNA was defined to be 1 and rates were reported as a percent of the maximum rate in an experimental group.
Protein Thermal Denaturation Assays.
The stabilities of truncated wild-type and mutated enzymes were analyzed at various pHs and temperatures to ensure mutagenesis and buffer conditions did not alter enzyme stability resulting in differential activity. Each protein was prepared as recommended using an Applied Biosystems Protein Thermal Shift Dye Kit at 0.5 mg mL−1 using an Illumina Eco Real-Time PCR System in EC3K+ buffers at the indicated pHs. The temperature was linearly increased over 15 min from 30 to 95 °C, while the fluorescence at 608 nm was recorded. Each protein denaturation experiment was performed at least three times to ensure reproducibility.
RNA Phosphate Analogue Titration.
To determine the pKa of methyl/dimethyl phosphate and β-glycerol phosphate, a 10 mM solution was prepared and degassed. These solutions were then titrated with 100 mM aqueous phosphoric acid or 100 mM sodium hydroxide. The pH was measured using a Sartorius PY-P11–2S electrode and a Sartorius pHBasic pH meter at ~293 K. The pKa was calculated as the zero point of the second derivative of the titration curve.
HPLC Detection of Xrn1 Decay Products.
All HPLC experiments were performed using an Agilent 1260 Infinity II instrument equipped with an Agilent PLRP-S column (150 mm × 7.5 mm) and a diode array UV−vis detector calibrated for detection at 254 nm. A typical reverse-phase separation and elution program involved two solvent lines, 0.1 M TEAA at pH 7.5 on line A and 100% acetonitrile on line B. The column was kept at 70 °C and equilibrated with a 98:2 A/B mixture for 15 min at a flow rate of 2 mL/min. After injection, the following program was run: 98:2 A/B for 10 min, followed by a ramp over 30 min to 75:25 A/B and then held at a constant composition. For the ion-exchange reverse-phase (IERP) method, the same column was treated with 5 mM tetrabutylammonium phosphate (TBAP) at pH 7.5 (line A) and acetonitrile (line B) at a 98:2 A:B ratio. This same composition was kept for 15 min after injection, at which point the composition was linearly changed over 25 min to 50:50 acetonitrile:water (no TBAP) and then held constant.
Quantum Mechanical Electrostatic Potential Surface Calculations.
Calculations were performed using Gaussian 0967 at the B3LYP/6–31+G(d,p) level. Generated potential files were then superimposed onto isobutyl phosphate in Chimera.
RESULTS
Expression of a S. cerevisiae Xrn1 (1−1240), ScXrn1ΔC.
To initiate biochemical studies of Xrn1, we expressed and purified ScXrn1ΔC and used the enzyme to determine the relative rates of degradation for different types of in vitro-transcribed RNA substrates. First, to express ScXrn1ΔC, we obtained a commercially generated expression vector encoding ScXrn1 residues 1−1240 in a pET26b+ backbone that supplies a C-terminal hexahistidine affinity tag. Attachment of the affinity tag in this location is important as prior structural studies36,37 have shown Xrn1’s N-terminus forms part of the molecule’s conserved active site and attachment of fusion peptides or proteins to this location likely interferes with the enzyme’s function. We expressed C-terminally His6-tagged ScXrn1ΔC in BL21 E. coli and used standard Ni-NTA affinity purification followed by size-exclusion chromatography to isolate a monomeric form of the protein (Figure S2). The enzyme was stored at 2 mg mL−1 in buffered 50% glycerol, aliquoted for use as needed, and stored at −20 to −80 °C.
Design of RNA Substrates.
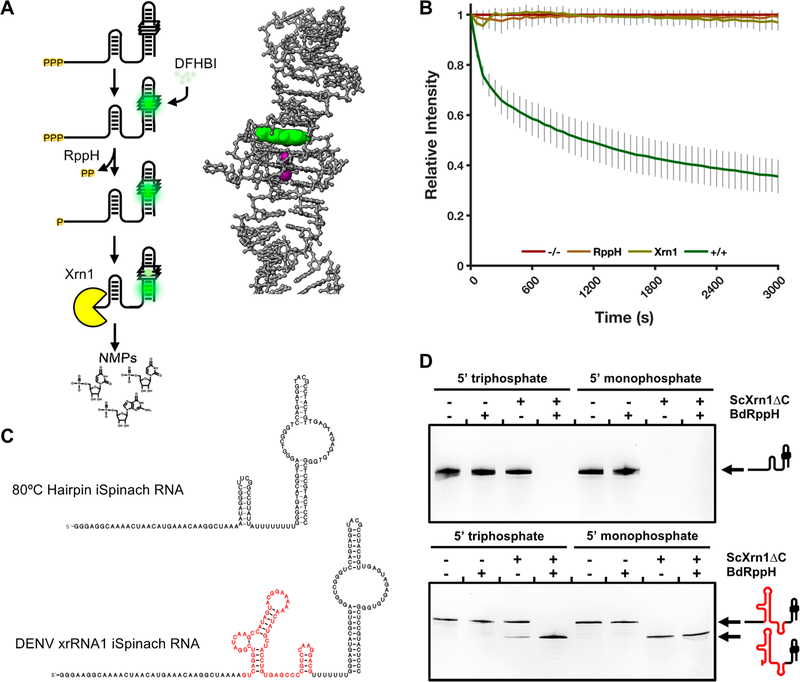
To characterize the exoribonucleolytic activity of ScXrn1ΔC, we implemented a second-generation time-resolved fluorescence-based RNA degradation (TRFRD) assay.43 In this assay, RNA constructs are in vitro transcribed with a 5′ single-stranded region, or “leader”, followed by a variable structure domain (VSD), and ending with an aptamer structure capable of binding to a small molecule fluorophore and enhancing its fluorescence (Figure 2A). Most often in this work the VSD used is a previously reported thermostable hairpin;44 in other experiments, an Xrn1-resistant RNA structure (xrRNA) structure from Dengue virus that cannot be degraded by Xrn1 and produces truncated intermediate RNAs is used (Figure 2C and Figure S3).43,45 Immediately downstream of the VSD, a poly-U spacer was included to promote independent folding of the VSD and the 3′ fluorescence aptamer, which in these studies was a recently described iSpinach aptamer.46 Throughout this work, DENVxrRNA1 and 80HP “constructs” and “substrates” refer to the entire leader-VSD-aptamer RNA construct containing the specified VSD and the 3′ iSpinach aptamer.
Figure 2.
Second-generation time-resolved, fluorescent RNA degradation assay. (A) Diagram of RNA processing in the TRFRD assays and an inset structure of the DFHBI-bound iSpinach aptamer (PDB entry 4TS2). DFHBI is colored green, and K+ ions are colored purple. (B) Normalized fluorescence trace of a TRFRD assay using the 80HP construct. The “−/−” trace shown in red is a no enzyme control. The “+/+” trace shown in green contains both RppH and ScXrn1ΔC. Conditions: 2 μM 80HP RNA, 2 μM BdRppH, and 1 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C. Error bars represent one standard deviation for nine replicates. (C) Secondary structures of the two main RNA constructs used in this study, 80HP and DENVxrRNA1. The Xrn1-resistant structure is colored red. The layout of each construct is described in the text. (D) dPAGE of RNA constructs demonstrating the phosphorylation state dependency of ScXrn1ΔC. Conditions: 0.5 μM RNA, 0.5 μM BdRppH, and 0.25 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C for 2 h.
In a typical TRFRD experiment, the 3′ iSpinach aptamer functions by binding 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), resulting in the emission of green light (Λmax = 503 nm). Degradation of the iSpinach aptamer results in a loss of fluorescence. Because Xrn1 degrades RNA processively in the 5′ → 3′ direction, the fluorescent signal of the iSpinach aptamer situated at the 3′-end of the RNA can be used to monitor total degradation of bulk RNA in real time (Figure 2A,B). We typically conduct these types of experiments in a 96-well plate in 100−200 μL volumes and monitor the reaction using a standard microplate reader. Here we have used this method to allow for collection of time-resolved kinetic data for ScXrn1ΔC and ScXrn1ΔC mutants acting under different conditions and on a diverse set of RNA substrates.
ScXrn1ΔC Prefers 5′-Monophosphorylated RNAs.
Xrn1 has been previously shown to be selective for 5′-monophosphorylated RNAs.30,36,37,43,45 To confirm the specificity of ScXrn1ΔC for 5′-monophosphorylated RNAs, we tested the activity of the enzyme versus RNA substrates with two different 5′-end chemistries, generating transcripts with both 5′-triphosphorylated and 5′-monophosphorylated ends. 5′-Triphosphate ends are obtained naturally following in vitro transcription using T7 bacteriophage RNA polymerase, while 5′-monophosphorylated RNAs can be generated using the RNA pyrophosphohydrolase from B. bacteriovorus (BdRppH).43,47 Treatment with BdRppH was performed by including it in reaction mixtures with Xrn1 as we have done previously,43,48 or in many cases throughout this work allowing an BdRppH reaction to go to completion and re-HPLC purifying exclusively 5′-monophosphorylated products. In a typical experiment in which 5′-monophosphorylated RNAs are generated in situ, the fluorescence of a solution containing RNA is monitored without enzymes, in the presence of BdRppH and ScXrn1ΔC independently, and in the presence of both enzymes together (Figure 2B). In the first three cases, the measured fluorescence remains relatively unchanged over the course of the experiment. Only when both BdRppH and ScXrn1ΔC are present together is a substantial loss of fluorescence observed corresponding to the 5′ → 3′ degradation of the iSpinach aptamer and upstream RNA.
To further confirm Xrn1’s selectivity for 5′-monophosphorylated products, single-time point experiments using both the 80HP and the DENVxrRNA1 constructs were performed using denaturing polyacrylamide gel electrophoresis (dPAGE) (Figure 2D). These experiments were performed using both the in vitro-transcribed 5′-triphosphorylated and purified 5′-monophosphorylated versions of substrate RNAs. As expected, treatment with ScXrn1ΔC alone resulted in the loss of the band corresponding to the full-length 80HP RNA only when the 5′-monophosphorylated species is used (Figure 2D). When the DENVxrRNA1 substrate was used, the resulting gel shifts indicate a similar dependence on 5′-monophosphorylation, though some degree of degradation can be seen to take place in the ScXrn1ΔC-only lane using 5′-triphosphorylated RNA (Figure 2D, lane 3). This is consistent with prior observations43,45 and reflects slow background hydrolysis of the 5′-triphosphate during the course of the reaction.49,50 Cumulatively, these results show that ScXrn1ΔC discriminates against 5′-triphosphorylated RNAs and efficiently degrades 5′-monophosphorylated RNA species.
ScXrn1ΔC Decay Products and Intermediates.
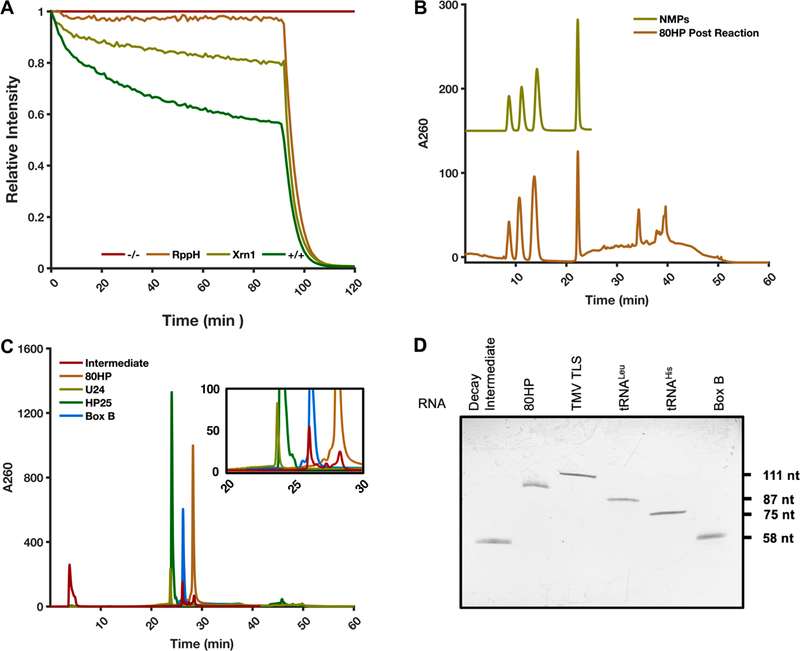
Xrn1 has been shown to produce 5′-nucleotide monophosphate (5′NMP) products and, in a growing number of examples, varying degrees of partially degraded intermediates.43,45,51–53 In the TRFRD experiments presented here and in previously examined cases,43,48 many degradation experiments reach a plateau with residual fluorescence remaining even at longer time points, typically 15−25% of the initial intensity. This feature has been observed using different fluorescent aptamers, different species of recombinant Xrn1, and different equipment,43,48 this study included. One explanation for this result could be that an undegraded population of RNA somehow evades degradation by Xrn1. To test this hypothesis and determine if the remaining fluorescence signal may be due to an undegraded population of RNA, a reaction using the 80HP construct was allowed to plateau and then was supplemented with the nonspecific endoribonuclease RNase A. Treatment with RNase A results in a total loss of fluorescence indicating complete degradation of residual 80HP RNA (Figure 3A). This result is consistent with a control experiment using RNase A alone, which also results in a total loss of fluorescence (data not shown). These results indicate that RNase A is capable of degrading the population of RNA that remains once decay by Xrn1 has finished, suggesting a population of RNA that specifically resists Xrn1-catalyzed degradation.
Figure 3.
Interrogation of ScXrn1ΔC products. (A) Typical TRFRD assay using ScXrn1ΔC with the addition of RNase A at 90 min. The “−/−” trace shown in red is a no enzyme control. The “+/+” trace shown in green contains both RppH and ScXrn1ΔC. Conditions: 2 μM 80HP RNA, 2 μM BdRppH, 0.05 μM ScXrn1ΔC, and 5 units of RNase A in 1xEC3K+ buffer at pH 7.9 and 37 °C. (B) HPLC trace of products from bulk reaction of 80HP RNA with ScXrn1ΔC (bottom trace). The top trace is the four purified 5′NMPs resolved using TBAP-modified C-18 chromatography. (C) HPLC analysis of the 80HP RNA with and without ScXrn1ΔC in the presence of DFHBI using PLRP-S chromatography overlaid with the trace of three oligonucleotide-length markers: U24 (24 nucleotides), HP25 (25 nucleotides), and Box B (58 nucleotides). The inset is a region of the trace where the DFHBI-dependent resistant RNA population appears during the reaction. We have not yet fully characterized the intermediate products, but their size roughly corresponds to the size of the iSpinach aptamer, indicating degradation up to the quadruplex DFHBI binding pocket. (D) dPAGE of the intermediate compared against the full-length 80HP construct and various RNA constructs as size markers. The intermediate appears to be approximately the same size as the 58-nucleotide Box B RNA.
To detect possible partially degraded RNA products, we analyzed several typical TRFRD reactions by HPLC. To this end, the 80HP construct was degraded in the absence of DFHBI and the resulting reaction mixture separated by HPLC using a C18 reverse-phase column treated with the cationic modifier tetrabutylammonium phosphate (TBAP). This allows resolution of the reaction’s products in a manner akin to anion-exchange chromatography. The resulting chromatogram, monitored at 260 nm, reveals all four 5′NMPs present and resolved early in the elution as well as a major peak at 35 min corresponding to the undegraded 80HP construct (Figure 3B). To better analyze longer undegraded intermediates, the 80HP construct was reacted with ScXrn1ΔC in a TRFRD assay in the presence of DFHBI and then resolved by HPLC using a higher-resolution polystyrene divinylbenzene (PLRP-S) column. Under these conditions, the 80HP starting material elutes near 28 min (Figure 3C). In the plus-DFHBI sample, a population of mixed-length products appears in the range of 25−28 min. When compared to three RNAs used as size standards, U24 (24 nucleotides), HP25 (25 nucleotides), and Box B (58 nucleotides), the major intermediate elutes at nearly the same time as the Box B RNA, suggesting a similar size. When the intermediate peaks were collected and resolved by dPAGE, the observed band again corresponds to a product of length similar to that of the 58-nucleotide Box B RNA (Figure 3D). Presuming that this fragment is produced by 5′ → 3′ degradation, this product corresponds to degradation up to the 5′-region of the iSpinach aptamer domain. There are two major conclusions from our HPLC analyses. (1) ScXrn1ΔC produces 5′NMP products, and (2) a number of peaks corresponding to partially degraded intermediates appear in our HPLC chromatograms in a DFHBI-dependent manner. When integrated, these fragments correspond to 23.1% of the starting RNA, comparable to the observed residual fluorescence observed on our TRFRD assays. Cumulatively, these results are consistent with a model in which ScXrn1ΔC loads onto seemingly all of the original substrate RNAs but sometimes stalls within the 5′-region of the iSpinach aptamer leading to the observation of a distribution of partially degraded RNA species by HPLC. These data help account for the residual fluorescence at the end point of the TRFRD decay and suggest that to an extent, Xrn1 degradation may stall at specific locations within other structured RNA substrates, though the low yield of such products and the structural complexity of the RNAs investigated here deterred us from probing this systematically in this work. Overall, these results offer a high-resolution portrayal of the dynamics of ScXrn1ΔC degradation and the products left after decay.
Kinetics of ScXrn1ΔC-Mediated RNA Degradation.
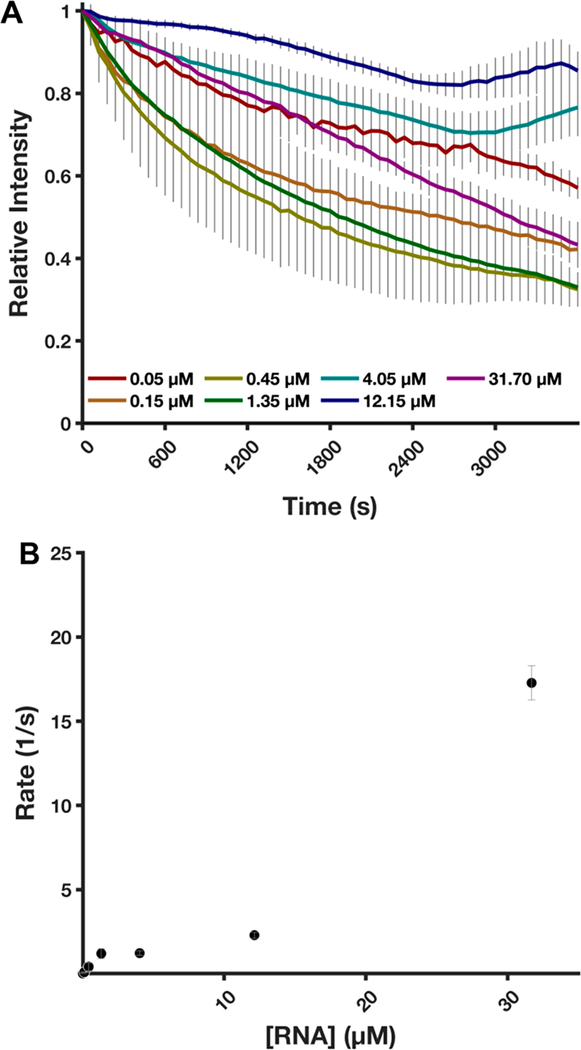
We next used the TRFRD assay to conduct experiments aimed at determining the kinetic parameters of ScXrn1ΔC. By measuring the loss of fluorescence from the RNA constructs and fitting the data to first-order exponential decay curves, we were able to estimate per phosphodiester bond hydrolysis rates for wild-type ScXrn1ΔC. In an attempt to determine the Vmax of the enzyme, 0.05 μM ScXrn1ΔC was reacted with increasing concentrations of 80HP RNA ranging from 0.05 to 31.75 μM, representing ScXrn1ΔC:RNA stoichiometries of 1:1 to 1:634. Over the range of RNA concentrations tested here, we fail to observe a plateau in rates that would indicate saturation of the enzyme and operation in a Michaelis−Menten regime. At lower concentrations of RNA, an exponential loss of fluorescence is observed, while at higher RNA concentrations, the decay curve becomes more linear (Figure 4A). This suggests that ScXrn1ΔC may be approaching Vmax, though this could not be confirmed due to inherent limitations of the assay in that higher concentrations of RNA exceed the fluorescence detection limits of the plate reader used in our experiments. As each molecule of RNA acts as a single substrate for ScXrn1ΔC, it was necessary that the number of RNA molecules be in high excess, rather than the number of phosphodiester bonds that are formally the substrate of Xrn1 (Figure 4). Using the loss of fluorescence as a surrogate for the total degradation of an RNA molecule, the rate of phosphodiester bond cleavage could be calculated as the product of the per molecule degradation rate and the 130 phosphodiester bonds per 80HP molecule. On the basis of these experiments, we are able to provide a lower boundary of 17.3 ± 0.6 s−1 for the Vmax of ScXrn1ΔC (Figure 4). This finding is consistent with a recently proposed in vivo phosphodiester hydrolysis rate54 for Xrn1 of 38−55 s−1. Additionally, due to the low-level presence of the DFHBI-dependent RNA intermediate, this further suggests the measured rate of phosphodiester bond hydrolysis is a lower estimate of the true activity of ScXrn1ΔC.
Figure 4.
ScXrn1ΔC kinetics. (A) Normalized trace of the TRFRD assay obtained during kinetic experiments varying the concentration of 5′-monophosphorylated RNA as indicated. The relative stoichiometries of ScXrn1ΔC molecules to RNA molecules are as follows: 1:1, 1:3, 1:9, 1:27, 1:81, 1:243, and 1:634. Conditions: 0.05 μM ScXrn1ΔC, 5′-monophosphorylated 80HP RNA concentrations as shown, 1xEC3K+ buffer, pH 7.9, 37 °C. (B) Quantification of rates of per phosphodiester bond hydrolysis. An enzyme-limited regime, or saturation, was not achieved using RNA concentrations of ≤31.7 μM. From the data, a lower bound for ScXrn1ΔC activity can be estimated to be 17.3 ± 0.6 s−1. Error reported as one standard deviation for nine replicates excluding 31.7 μM where n = 3.
pH Dependence of ScXrn1ΔC.
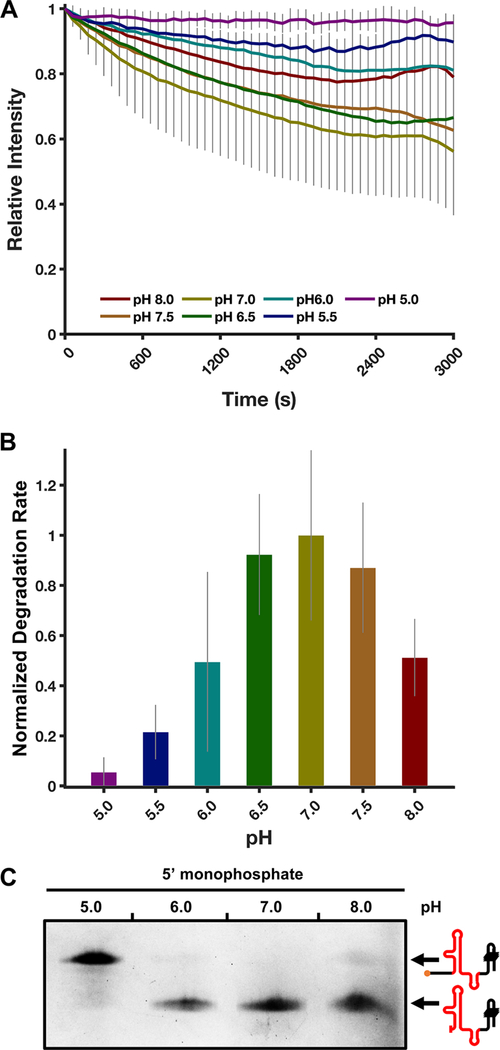
To examine the pH dependence of ScXrn1ΔC, we performed standard TRFRD assays using the 80HP RNA construct at varying pHs ranging from 5.0 to 8.0 in 0.5 pH unit steps. In these experiments, we observe a fairly broad pH tolerance with the activity of ScXrn1ΔC peaking at pH 7.0 (Figure 5A,B). Reactions carried out at pH 6.5 and 7.5 yielded rates of 92 ± 24% and 87 ± 25%, respectively, relative to that at pH 7.0. Further changing the pH to 6.0 or 8.0 resulted in measured rates of 50 ± 36% or 51 ± 15%, respectively, and as the pH was further decreased to 5.5, the corresponding rate was measured to be 22 ± 11% of that at pH 7.0. Finally, at pH 5.0, the measured rate of RNA degradation was not significantly different from zero. Again, the results of our single-time point dPAGE experiment are in agreement with these results; at pH 7.0, the full-length DENVxrRNA1 construct is degraded completely up to the xrRNA structure, and in experiments at pH 6.0 and 8.0, some full-length RNA remains (Figure 5C). At pH 5.0, very little RNA degradation is apparent.
Figure 5.
ScXrn1ΔC exhibits pH-dependent activity. (A) Normalized trace of the TRFRD assay conducted using 80HP RNA and ScXrn1ΔC at different pHs. Conditions: 2 μM purified, 5′-monophosphorylated 80HP RNAs, and 0.05 μM ScXrn1ΔC in 1xEC3K+ buffer at the indicated pH and 37 °C. (B) Relative rates of ScXrn1ΔC-catalyzed degradation of 80HP RNA at the indicated pHs. Error reported as one standard deviation for nine replicates. (C) dPAGE of the DENVxrRNA1 RNA constructs showing the pH dependency of ScXrn1ΔC. Conditions: 0.5 μM RNA, 0.5 μM BdRppH, and 0.25 μM ScXrn1ΔC in 1xEC3K+ buffer at the indicated pH and 37 °C for 2 h.
As a means to test whether the enzyme may be undergoing pH-dependent denaturation, thermal denaturation experiments were performed under the varying pH conditions using a commercial protein stability assay in which binding of a hydrophobic dye is used as a readout for denaturation. The data obtained from these experiments do not depict pH-dependent sensitivity in the thermal denaturation of ScXrn1ΔC, suggesting its stability at all pHs tested (Figure S4B).
We hypothesized that pH-dependent differences in observed rates may indicate titration of a functional group important to Xrn1 catalysis. Candidate functional groups include amino acids within the active site with pKas between pH 5 and 7, namely histidine; however, the divalent metal ion mechanism by which Xrn1 is proposed to operate does not include a catalytic histidine.55,56 Our findings therefore suggested that some other functional group may be a factor in determining ScXrn1ΔC degradation rates. Correspondingly, we hypothesized that the observed pH dependence in ScXrn1ΔC activity may be due to differences in the protonation state of an RNA substrate’s 5′-monophosphate. The pKa values of 5′-monophosphate analogues have been reported to lie between pH 6.0 and 7.0,57 suggesting a transition from a single negatively charged terminal phosphate to a double negatively charged dianionic species could fall within the tested pH range.
Titration of 5′-Monophosphate Analogues.
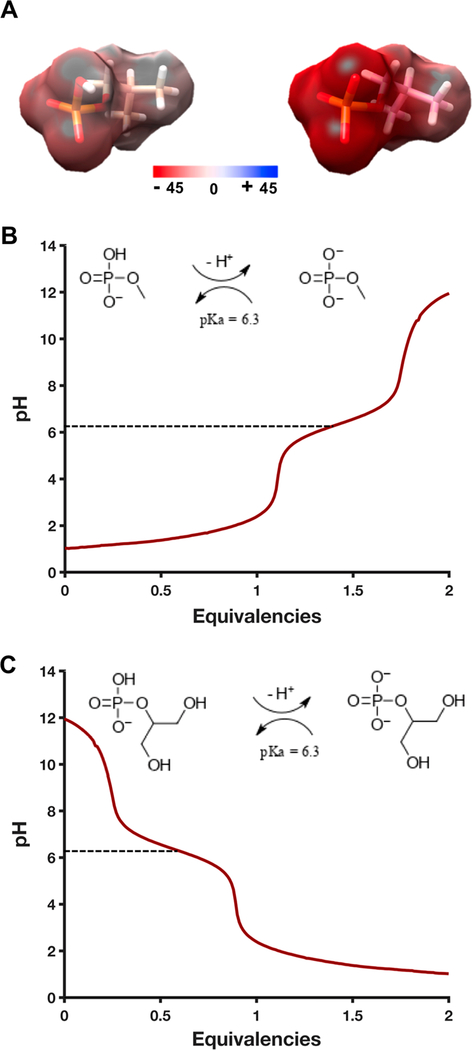
To gain experimental insight into the protonation dynamics of several model 5′-phosphate analogues, pH titrations were performed on two compounds: methyl phosphate and β-glycerol phosphate. The pKa2 of these two chemicals, corresponding to the transition from a charged state of −1 to −2 on each phosphate, was determined by acid or base titration to occur at pH 6.3 (Figure 6B,C). This result is consistent with the decrease in SCXrn1ΔC activity observed below pH 6.5 (Figure 5B). The pH of the cytosol is variable though frequently approximated to be ~7.2,58,59 suggesting that dianionic phosphate species likely predominate in the cell.
Figure 6.
pH titration of 5′-phosphate analogues, methyl phosphate and β-glycerol phosphate. (A) Quantum mechanical calculation of the surface potential of isobutyl phosphate showing the potential difference of the monoanionic (left) and dianionic (right) phosphate groups, calculated using Gaussian 0967 at the B3LYP/6–31+G(d,p) level. (B) pH titration of a 10 mM methyl/dimethyl phosphate solution with 100 mM sodium hydroxide. pKa2 is indicated by a dashed line at pH 6.3. The inset scheme shows the relevant protonation/deprotonation reaction being monitored. (C) pH titration of a 10 mM β-glycerol phosphate solution with 100 mM phosphoric acid. pKa2 is indicated by a dashed line at pH 6.3. Again, the inset scheme depicts the relevant equilibrium.
To depict the effect of pH change distributed throughout the resonant 5′-phosphate group, quantum mechanical calculations were performed on a third chemical analogue of the 5′-phosphate group, isobutyl phosphate, in the presence and absence of the pKa2 proton. Intuitively, the presence of this proton significantly decreases the presented negative change density of the phosphate from −45 to −10 hartree (Figure 6A). Combined, these results suggest the charged state of the 5′-monophosphate may play an important role in determining how ScXrn1ΔC recognizes and distinguishes a dianionic 5′-monophosphate from the single negatively charged polyanionic backbone of RNA.
ScXrn1ΔC Activity Is Dependent on a Set of Conserved Active Site Residues.
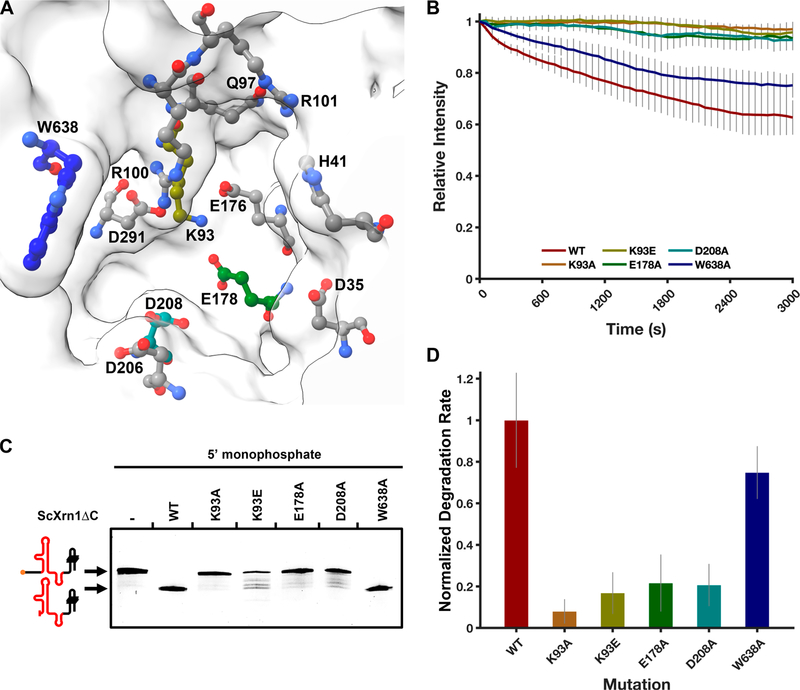
To confirm the veracity of the I-TASSER-generated structure of ScXrn1ΔC, we tested the importance of several conserved active site residues: K93, E178, D208, and W638. Either directly or on the basis of primary sequence alignment and structural homology to Drosophila or Kluyveromyces, Asp35, His41, Lys93, Gln97, Arg100, Arg101, Glu176, Glu178, Asp206, Asp208, Asp291, and Trp638 mutations have all been previously tested35–37 (Figures 1B and 7A). Disruption of several of these residues has been reported to abolish the activity of Xrn1 in exonuclease assays36,37 and in some cases result in decreased viability in yeast.35 We sought to determine the importance of several of these residues on the rate of ScXrn1ΔC. To test homology-predicted structure−function relationships, we performed site-directed mutagenesis on ScXrn1ΔC to generate the constructs listed above and submitted the 80HP construct to degradation by the resulting mutants using TRFRD assays. These experiments revealed that mutating the proposed basic patch Lys93 to either alanine or glutamate disrupted catalysis with a rate of 8 ± 6% or 17 ± 10% of that of the wild-type enzyme, respectively (Figure 7A,B). Similarly, mutation of acidic residues implicated in metal ion coordination, Glu178 or Asp208 to alanine, reduced the activity of the enzyme to 22 ± 14% or 21 ± 10% of that of the wild-type enzyme, respectively. Finally, mutation of the “gatekeeping” Trp638 to alanine reduced the activity of the enzyme only modestly to 75 ± 13% of that of the wild-type enzyme.
Figure 7.
ScXrn1ΔC activity is dependent on a set of conserved active site residues. (A) Model of the ScXrn1ΔC active site generated using the ITASSER iterative threading model. Previously mutagenized residues implicated in the mechanism of Xrn1 are shown: Asp35, His41, Lys93, Gln97, Arg100, Arg101, Glu176, Glu178, Asp206, Asp208, Asp291, and Trp638.35–37 Carbon atoms of the corresponding residues tested in this study are colored to correspond to the plots and bar graphs in this figure. (B) Normalized TRFRD assay traces of both WT and mutant ScXrn1ΔC constructs using the 80HP RNA. Conditions: 2 μM purified, 5′-monophosphorylated 80HP RNA, and 0.05 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C. (C) dPAGE of both the wild type and mutants of ScXrn1ΔC using the DENVxrRNA1 construct. Conditions: 0.5 μM DENVxrRNA1 RNA and 0.25 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C for 2 h. (D) Relative rates of RNA degradation carried out by the WT and mutant ScXrn1ΔC’s K93A, K93E, E178A, D208A, and W638A. Error reported as one standard deviation for nine replicates.
When the DENVxrRNA1 RNA was submitted for degradation by these mutated enzymes, similar trends can also be observed in a single-time point experiment analyzed by dPAGE (Figure 7C). While the wild-type enzyme totally degrades the substrate RNA up to, but not beyond, the xrRNA structure, K93A, K93E, E178A, and D208A mutants all show a loss of activity. These results are in agreement with those of our previous TRFRD assays. Specifically, K93A appears to be catalytically inactive, while K93E, E178A, and D208A all appear to “stutter” and produce intermediates in the gel, suggestive of defects in the processivity of the enzyme. Interestingly, the lane containing the W638A mutant shows a total loss of the full-length band, similar to the wild-type enzyme, having no obvious effect on the activity of the enzyme in this experiment. The TRFRD assay portrays decreased the activity of the W638A mutant, highlighting the utility of the time-resolved assay in these types of experiments.
To be rigorous, we sought to confirm that the loss of activity due to mutations observed in TRFRD assays was not due to disruption of the folded structures of the mutated ScXrn1ΔCs. This was interrogated by thermal denaturation experiments using the wild-type enzyme and those five point mutants using the same commercial protein stability assay mentioned above. Using this method, the Tm of wild-type ScXrn1ΔC was observed to be 42 °C (Figure S4A). The Tms of each of the mutants were measured at 44 °C, corresponding to a slight increase in observed thermostability. Accordingly, we hypothesize that the changes in activity associated with active site mutagenesis are not a result of decreased structural stability, but rather these mutations have deleterious effects most directly related to the chemical mechanism of ScXrn1ΔC.
Xrn1 More Effectively Acts on Substrate RNA with Longer Single-Stranded Leaders.
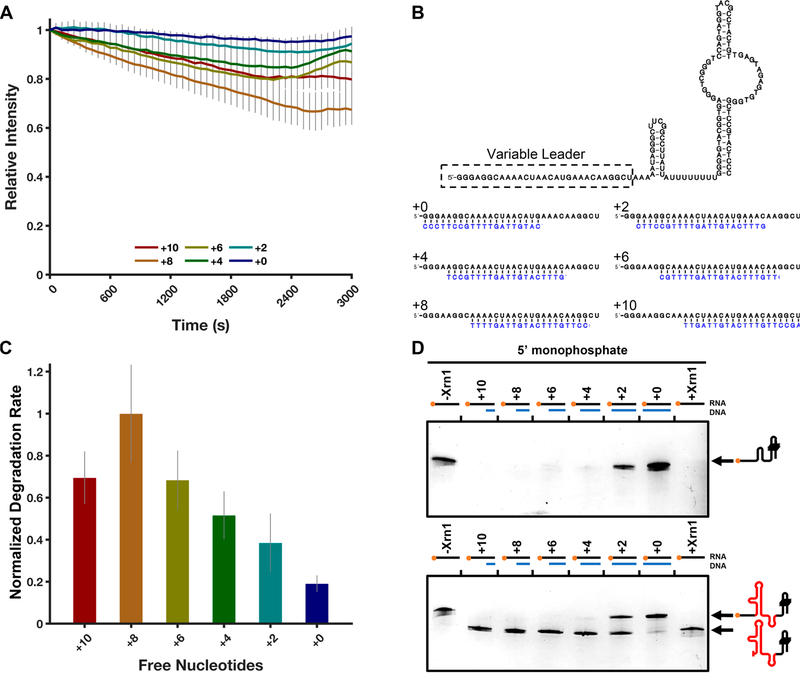
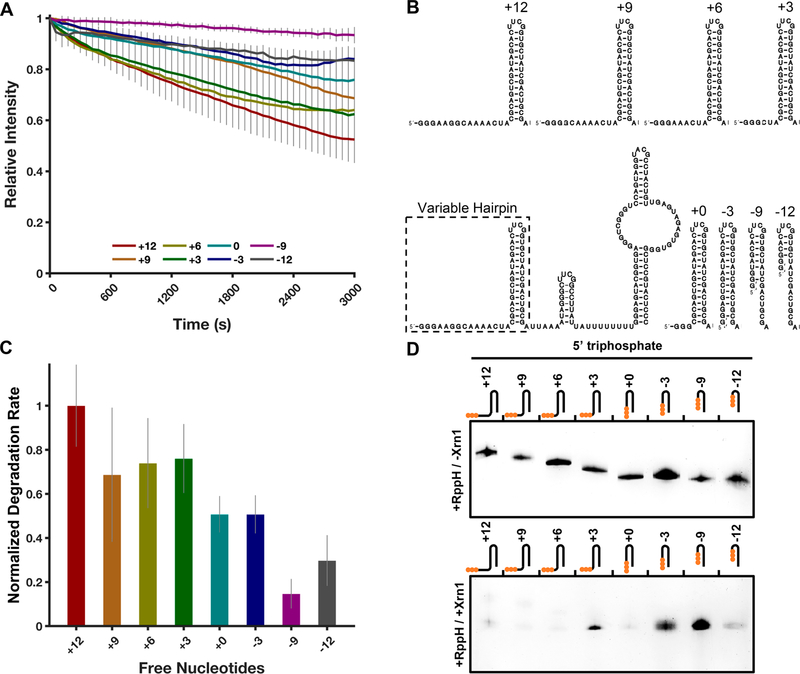
Xrn1 has been proposed to favor RNA substrates with single-stranded 5′-ends, and prior experiments have shown Xrn1 to require a minimum of four single-stranded nucleotides at the 5′-end of RNA for it to be properly recognized and loaded for decay.36 Given previous studies implicating sequestration of the 5′ single-stranded region of RNA in decreased processivity, we chose to probe this requirement using our TRFRD assay. To test this scenario in an intermolecular system, a set of primers were designed to anneal to the 5′ single-stranded region of the 80HP construct and were systematically walked back by two-nucleotide increments, creating RNA−DNA hybrid duplexes with 10 free nucleotides down to zero free nucleotides (Figure 8B). RNAs with decreasingly exposed 5′-ends were then subjected to TRFRD analysis. The results of this set of experiments show a trend where the more exposed nucleotides are present, the greater the rate of degradation by ScXrn1ΔC (Figure 8A,C). Specifically, the determined relative rates in decreasing leader length are as follows: 70 ± 13% (+10 nucleotides), 100 ± 23% (+8 nucleotides), 68 ± 14% (+6 nucleotides), 52 ± 11% (+4 nucleotides), 38 ± 14% (+2 nucleotides), and 19 ± 4% (+0 nucleotides) relative to the highest rate of degradation observed in the +8 RNA. The corresponding experiments, when captured at an intermediate time point and resolved by dPAGE, reveal similar results for both the 80HP construct and the DENVxrRNA1 construct (Figure 8D) where RNAs with longer free single-stranded regions are more rapidly degraded than RNAs with shorter single-stranded regions.
Figure 8.
ScXrn1ΔC prefers RNAs with single-stranded ends. (A) Normalized TRFRD traces for degradation of DNA primer-annealed 80HP constructs by ScXrn1ΔC. Conditions: 2 μM purified, 5′-monophosphorylated 80HP RNA annealed to 2 μM indicated primer, and 0.05 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C. (B) Secondary structure of the 80HP construct used in these experiments showing the indicated primers (blue) used to duplex the 5′ single-stranded region. (C) Relative rates of degradation for constructs with the indicated number of free 5′-nucleotides. Error reported as one standard deviation for nine replicates. (D) dPAGE of the 80HP and DENVxrRNA1 RNA constructs in panel B showing the single-stranded leader length dependency of ScXrn1ΔC. Conditions: 0.5 μM RNA, 0.5 μM BdRppH, and 0.25 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C for 30 min.
As the experiment described above depends on intermolecular interactions, we also designed a system that would probe the same phenomenon using strictly intramolecular interactions, which may be more likely to be encountered in vivo. We created a set of modified 80HP constructs, in which an additional hairpin was installed upstream of the 80HP structure. In this construct, the 5′ single-stranded region was systematically truncated in steps of three nucleotides from +12 bases upstream of the hairpin structure to −12 nucleotides into it in an attempt to sequester the 5′ single-stranded region into a structure (Figure 9B). The −6 nucleotide construct was omitted due to difficulties transcribing the RNA. All buried hairpin constructs retain a 5′-GGG sequence as an artifact of our transcript design and thus vary slightly in the number of predicted single-stranded G’s at their 5′-ends. The general trends observed in the TRFRD assays using these constructs were similar to those observed in the previous experiment; as single-stranded nucleotides are removed, the observed rates of degradation decrease. The determined relative rates of the most extended construct to the shortest are as follows: 100 ± 19% (+12 nucleotides), 69 ± 30% (+9 nucleotides), 74 ± 20% (+6 nucleotides), 76 ± 16% (+3 nucleotides), 51 ± 8% (+0 nucleotides), 51 ± 9% (−3 nucleotides), 15 ± 7% (−9 nucleotides), and 30 ± 12% (−12 nucleotides). For the construct containing a totally buried 5′-region (+0 nucleotides), the relative rate of degradation measured was 51 ± 8% of that of the fully extended +12 species, a less pronounced effect than observed in the corresponding intermolecular experiment (in Figures 8C and 9C, compare +0 free nucleotides). As the single-stranded leader becomes more sequestered into the stem structure, at −9 and −12 nucleotides, the measured rate of degradation continues to decrease to <30% of that measured for the +12 species. Similar trends were observed in a single-time point dPAGE experiment in which the shorter leader RNAs showed a decreased extent of degradation compared to more extended constructs (Figure 9D). Together, these results demonstrate that RNAs presenting non-base-paired single-stranded 5′-ends are more quickly degraded by ScXrn1ΔC in vitro.
Figure 9.
ScXrn1ΔC prefers RNAs with exposed 5′-ends. (A) Normalized trace of the TRFRD assay using a buried hairpin RNA and ScXrn1ΔC. Conditions: 2 μM purified, 5′-triphosphorylated buried hairpin RNA as indicated, and 0.05 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C. (B) Secondary structure of the buried hairpin constructs used in these experiments showing the indicated truncations made to the 5′ single-stranded region. (C) Relative rates of degradation for the indicated constructs. Error reported as one standard deviation for nine replicates. (D) Single-time point dPAGE analysis of the RNA constructs in panel B. Conditions: 0.5 μM RNA, 0.5 μM BdRppH, and 0.25 μM ScXrn1ΔC in 1xEC3K+ buffer at pH 7.9 and 37 °C for 30 min. Top, no ScXrn1ΔC added (controls). Bottom, reactions with both BdRppH and ScXrn1ΔC included.
DISCUSSION
Xrn1 is a central component of regulated gene expression.1–4 To improve our understanding of Xrn1 and begin dissection of the yeast 5′ → 3′ mRNA decay machinery in our lab, we expressed a C-terminally truncated form of S. cerevisiae Xrn1, ScXrn1ΔC. We used ScXrn1ΔC in conjunction with in vitro-transcribed RNAs to probe the biochemical properties of the enzyme using an updated TRFRD assay. Overall, our data reflect known features of Xrn1 enzymology through a trend toward a broader substrate and conditional tolerance than is commonly cited.36 The sum of our studies serves as a platform for future investigations regarding the biophysical characteristics of Xrn1 and its applications in biotechnology.
In the first experiments described here, TRFRD assays and dPAGE analyses were used to demonstrate ScXrn1ΔC’s previously established preference for 5′-monophosphorylated RNAs. Such substrates can be generated in situ or purified and used directly. The inability of ScXrn1ΔC to load and degrade 5′-triphosphorylated and other RNAs with bulky 5′-ends, such as capped mRNAs in vivo, can be rationalized on the basis of the size constraints of the enzyme’s active site. This feature is critical to Xrn1’s regulation, allowing for mRNA decapping and other RNA processing events that generate 5′-monophosphorylated RNA species to operate in rate-limiting biological steps.60
Implementation of an updated TRFRD assay in this work allowed us to characterize ScXrn1ΔC and the decay rates of model substrate RNAs. In this study, we employed highly structured RNAs containing both a previously characterized thermostable hairpin44 and a fluorescent iSpinach aptamer. To better understand the origin of the remaining fluorescent signal left following a plateau in ScXrn1ΔC-mediated degradation, we conducted experiments demonstrating that the signal is due to an undegraded population of RNAs that includes the majority of the iSpinach aptamer domain. The observation that the formation of these products depends on the inclusion of DFHBI in these reactions suggests that binding of the ligand to the aptamer may rigidify its structure and promote resistance to degradation. An analogous effect has been observed for riboswitches in which ligand binding has been shown to enhance their thermostability.61
Though this was not further examined in this work, there is a growing list of RNA structures and RNA−protein interactions that have been reported to resist degradation by Xrn1. In some cases, these structures appear to have evolved specifically to resist decay by forming a complex slip-knot-like structure that Xrn1 cannot unwind, therefore acting as a structural block,43,45,51,52 but in others stalling of Xrn1, while still structure-based, may be more artifactual. The precision of the point in an RNA at which Xrn1 stops appears to depend on the inherent structure of the RNA structure being tested, with some RNAs like the Flaviviridae and Dianthoviridae xrRNAs producing single, discrete stops, while other structures such as IRESs62 and poly(G) tracks63,64 producing multiple products are indicative of a different type of impediment. On the basis of these observations and the data presented here, we propose that Xrn1’s progress through an RNA is attenuated by local RNA structure, though higher-resolution experiments are needed to fully probe this effect.
Using the second-generation TRFRD assay, we extrapolated a lower limit for the Vmax of phosphodiester bond cleavage by ScXrn1ΔC of 17.3 ± 0.6 s−1. In vivo, Xrn1 participates in several different versions of co-translational mRNA decay, including No-Go decay and nonsense-mediated decay. A structure of the yeast ribosome in complex with ScXrn1 was recently determined by cryo-EM.38 In this structure, ScXrn1 is positioned at the end of the mRNA exit tunnel, poised to degrade a mRNA immediately following translation. The lower limit for Vmax of ScXrn1ΔC measured here, 17.3 ± 0.6 nucleotides s−1, suggests that degradation is capable of proceeding at least as fast as translation, a result supported by the observation of three-nucloetide periodicity in mRNAs subject to co-translational decay.65 It is interesting to note that like the decoding center, the active site of Xrn1 also specifically accommodates three nucleotides of RNA.
We next conducted experiments probing the pH dependence of ScXrn1ΔC, revealing a broad pH tolerance that roughly corresponds to the pKa of a 5′-monophosphate as informed by chemically similar analogues. How Xrn1 specifically selects for the 5′-end of an incoming RNA substrate is not yet fully understood though likely involves a conformational search driven by the attraction between a negatively charged RNA species and Xrn1’s electropositive, Mg2+-containing active site. In this model, the RNA phosphodiester backbone is uniformly negatively charged, raising the question of how Xrn1 can distinguish the 5′-end of an incoming RNA. A simple explanation, and one that fits well with our data, is that a dianionic 5′-monophosphate may provide a discernible chemical beacon and help facilitate the search process inside the cell.
We then used TRFRD assays to probe the effect of mutations within the conserved active site of ScXrn1ΔC. Phosphodiester hydrolysis by Xrn1 is proposed to result from a classic two-metal ion mechanism55 involving active site chelation of two Mg2+ ions that together activate water and stabilize the phosphorane intermediate involved in hydrolysis. The loss of catalytic activity observed using the E178A and D208A mutants is consistent with the requirement for these residues in orchestrating this type of chemistry. In addition to being an RNA hydrolase, Xrn1 is also an RNA helicase that simultaneously translocates along an RNA that is being degraded. This processivity of Xrn1 has been proposed to be the result of a “Brownian ratchet”-type mechanism that is driven by electrostatic attraction between a positively charged, basic patch located at the base/end of the enzyme’s active site and the 5′-monophosphate of a substrate RNA. In addition, a π-stacking network may be formed among H41, three RNA nucleobases that can be accommodated in the enzyme’s active site, and a conserved “gatekeeping” tryptophan (W638 in ScXrn1ΔC).36 The observation that both K93A and K93E mutations compromise enzyme activity is consistent with the hypothesis that electrostatics factor heavily in driving Xrn1 processivity, though this contrasts with previous findings demonstrating tolerance for an analogous mutation in the Drosophila enzyme.36 The observation that mutation of W638 only slightly impairs ScXrn1ΔC activity argues against dependence on the formation of a π-stacking network for Xrn1 processivity.
Finally, we tested ScXrn1ΔC’s ability to degrade RNAs with different lengths of exposed 5′-ends. The trends observed in our experiments show that ScXrn1ΔC prefers substrates with longer 5′-ends but can access and degrade most 5′-monophosphorylated substrates regardless of the structural context of the 5′-end. The ends of even fully base-paired nucleic acids are known to “fray” or “breathe”,66 and perhaps in vitro, this accounts for ScXrn1ΔC’s ability to access the RNA’s 5′-ends. However, here it may be important to recognize the simplicity of our experiments compared to the complex, crowded environment of the cell where biophysical mechanisms governing substrate selection may vary significantly from observations made using our purified system.
CONCLUSION
Xrn1 is a key player in cellular RNA metabolism. Here we have documented many aspects of ScXrn1ΔC enzymology in an effort to provide a foundation for further investigation into biophysical mechanisms and future technical applications involving Xrn1. Xrn1 is an interesting enzyme given its important biological roles and dual function as both an RNA hydrolase and an RNA helicase. Further in vitro characterization in our lab will focus on the thermodynamic basis for the degradation of structured RNAs and continue to probe the robust helicase activity of Xrn1, while high-throughput sequencing results from throughout the community continue to reveal novel aspects of Xrn1’s function and regulation in the cell.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the current and former members of the Chapman lab for thoughtful discussions and technical assistance and David Costantino for his careful reading of the manuscript. The expression vector for BdRppH was a kind gift from Dr. Joel Belasco at New York University.
Funding
This work was supported by National Institutes of Health Grant R00GM115757 to E.G.C.
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.9b01035.
ScXrn1ΔC rate data, iSpinach−DFHBI binding curve, SDS gels demonstrating ScXrn1ΔC and BdRpph purification, secondary structures of RNA constructs, and ScXrn1ΔC thermal denaturation experiments (PDF)
Accession Codes
ScXrn1ΔC XRN1_YEAST, P22147; BdRppH Q6MPX4_BDEBA, Q6MPX4.
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.9b01035
Contributor Information
Conner J. Langeberg, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States.
William R. W. Welch, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States
John V. McGuire, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States
Alison Ashby, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States.
Alexander D. Jackson, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States
Erich G. Chapman, Department of Chemistry and Biochemistry, University of Denver, Denver, Colorado 80208, United States.
REFERENCES
- (1).Parker R, and Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11, 121–127. [DOI] [PubMed] [Google Scholar]
- (2).Garneau NL, Wilusz J, and Wilusz CJ (2007) The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 8, 113–126. [DOI] [PubMed] [Google Scholar]
- (3).Lebreton A, Tomecki R, Dziembowski A, and Séraphin B (2008) Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996. [DOI] [PubMed] [Google Scholar]
- (4).Fabian MR, Sonenberg N, and Filipowicz W (2010) Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 79, 351–379. [DOI] [PubMed] [Google Scholar]
- (5).Larimer FW, Hsu CL, Maupin MK, and Stevens A (1992) Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120, 51–57. [DOI] [PubMed] [Google Scholar]
- (6).Stevens A (2001) 5′-exoribonuclease 1: Xrn 1. Methods Enzymol. 342, 251–259. [DOI] [PubMed] [Google Scholar]
- (7).Houseley J, and Tollervey D (2009) The Many Pathways of RNA Degradation. Cell 136, 763–776. [DOI] [PubMed] [Google Scholar]
- (8).Bousquet-Antonelli C, Presutti C, and Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102, 765–775. [DOI] [PubMed] [Google Scholar]
- (9).Coller J, and Parker R (2004) Eukaryotic mRNA Decapping. Annu. Rev. Biochem. 73, 861–890. [DOI] [PubMed] [Google Scholar]
- (10).Gatfield D, and Izaurralde E (2004) Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429, 575–578. [DOI] [PubMed] [Google Scholar]
- (11).Doma MK, and Parker R (2006) Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Huntzinger E, Kashima I, Fauser M, Sauliere J, and̀ Izaurralde E (2008) SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14, 2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Delorme-Axford E, and Klionsky DJ (2019) On the edge of degradation: Autophagy regulation by RNA decay. Wiley Interdiscip. Rev.: RNA 10, e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Waldron JA, Jones CI, Towler BP, Pashler AL, Grima DP, Hebbes S, Crossman SH, Zabolotskaya MV, and Newbury SF (2015) Xrn1/Pacman affects apoptosis and regulates expression of hid and reaper. Biol. Open 4, 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Pashler AL, Towler BP, Jones CI, and Newbury SF (2016) The roles of the exoribonucleases DIS3L2 and XRN1 in human disease. Biochem. Soc. Trans. 44, 1377–1384. [DOI] [PubMed] [Google Scholar]
- (16).Zabolotskaya MV, Grima DP, Lin M-D, Chou T-B, and Newbury SF (2008) The 5′−3′ exoribonuclease Pacman is required for normal male fertility and is dynamically localized in cytoplasmic particles in Drosophila testis cells. Biochem. J. 416, 327–335. [DOI] [PubMed] [Google Scholar]
- (17).Nagarajan VK, Jones CI, Newbury SF, and Green PJ (2013) XRN 5′→3′ exoribonucleases: Structure, mechanisms and functions. Biochim. Biophys. Acta, Gene Regul. Mech. 1829, 590–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Merret R, Descombin J, Juan Y. t., Favory JJ, Carpentier MC, Chaparro C, Charng Y. y., Deragon JM, and Bousquet-Antonelli C (2013) XRN4 and LARP1 are required for a heat-triggered mRNA decay pathway involved in plant acclimation and survival during thermal stress. Cell Rep. 5, 1279–1293. [DOI] [PubMed] [Google Scholar]
- (19).Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen D, and Seki M (2015) Loss of Arabidopsis 5′−3′ Exoribonuclease AtXRN4 Function Enhances Heat Stress Tolerance of Plants Subjected to Severe Heat Stress. Plant Cell Physiol. 56, 1762–1772. [DOI] [PubMed] [Google Scholar]
- (20).Molleston JM, and Cherry S (2017) Attacked from all sides: RNA decay in antiviral defense. Viruses 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Rowley PA, Ho B, Bushong S, Johnson A, and Sawyer SL (2016) XRN1 Is a Species-Specific Virus Restriction Factor in Yeasts. PLoS Pathog. 12, e1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, and Glaunsinger BA (2011) Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the Mammalian Exonuclease Xrn1. PLoS Pathog. 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, and Cramer P (2013) Global analysis of Eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol. Cell 52, 52–62. [DOI] [PubMed] [Google Scholar]
- (24).Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, and Choder M (2013) Gene expression is circular: Factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000. [DOI] [PubMed] [Google Scholar]
- (25).Braun KA, and Young ET (2014) Coupling mRNA Synthesis and Decay. Mol. Cell. Biol. 34, 4078–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Abernathy E, Gilbertson S, Alla R, and Glaunsinger B (2015) Viral nucleases induce an mRNA degradation-transcription feedback loop in mammalian cells. Cell Host Microbe 18, 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Heyer WD, Johnson AW, Reinhart U, and Kolodner RD (1995) Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol. Cell. Biol. 15, 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Grousl T, Opekarová M, Stradalova V, Hasek J, and́ Malinsky J (2015) Evolutionarily conserved 5′−3′ exoribonuclease Xrn1 accumulates at plasma membrane-associated eisosomes in postdiauxic yeast. PLoS One 10, e0122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sheth U, and Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (Washington, DC, U. S.) 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Stevens A (1978) An exoribonuclease from saccharomyces cerevisiae: Effect of modifications of 5′ end groups on the hydrolysis of substrates to 5′ mononucleotides. Biochem. Biophys. Res. Commun. 81, 656–661. [DOI] [PubMed] [Google Scholar]
- (31).Stevens A (1980) Purification and characterization of a Saccharomyces cerevisiae exoribonucleasewhich yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J. Biol. Chem. 255, 3080–3085. [PubMed] [Google Scholar]
- (32).Lasater LS, and Eichler DC (1984) Isolation and properties of a single-strand 5′→3′exoribonuclease from Ehrlich ascites tumor cell nucleoli. Biochemistry 23, 4367–4373. [DOI] [PubMed] [Google Scholar]
- (33).Stevens A, and Maupin MK (1987) A 5′ → 3′ exoribonuclease of Saccharomyces cerevisiae: Size and novel substrate specificity. Arch. Biochem. Biophys. 252, 339–347. [DOI] [PubMed] [Google Scholar]
- (34).Poole TL, and Stevens A (1997) Structural modifications of RNA influence the 5′ exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 235, 799–805. [DOI] [PubMed] [Google Scholar]
- (35).Page AM, Davis K, Molineux C, Kolodner RD, and Johnson AW (1998) Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 26, 3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jinek M, Coyle SM, and Doudna JA (2011) Coupled 5′ nucleotide recognition and processivity in Xrn1- mediated mRNA decay. Mol. Cell 41, 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chang JH, Xiang S, Xiang K, Manley JL, and Tong L (2011) Structural and biochemical studies of the 5′→3′ exoribonuclease Xrn1. Nat. Struct. Mol. Biol. 18, 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Tesina P, Heckel E, Cheng J, Fromont-Racine M, Buschauer R, Kater L, Beatrix B, Berninghausen O, Jacquier A, Becker T, and Beckmann R (2019) Structure of the 80S ribosomeXrn1 nuclease complex. Nat. Struct. Mol. Biol. 26, 275–280. [DOI] [PubMed] [Google Scholar]
- (39).Braun JE, Truffault V, Boland A, Huntzinger E, Chang C. Te, Haas G, Weichenrieder O, Coles M, and Izaurralde E (2012) A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat. Struct. Mol. Biol. 19, 1324–1331. [DOI] [PubMed] [Google Scholar]
- (40).Mittag T, and Parker R (2018) Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly. J. Mol. Biol. 430, 4636–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Jonas S, and Izaurralde E (2013) The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27, 2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015) The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Chapman EG, Moon SL, Wilusz J, and Kieft JS (2014) RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. eLife 3, e01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Tuerk C, Gauss P, Thermes C, Groebe DR, Gayle M, Guild N, Stormo G, d’Aubenton-Carafa Y, Uhlenbeck OC, and Tinoco I (1988) CUUCGG hairpins: extraordinarily stable RNA secondarystructures associated with various biochemical processes. Proc. Natl. Acad. Sci. U. S. A. 85, 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, and Kieft JS (2014) The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science (Washington, DC, U. S.) 344, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Autour A, Westhof E, and Ryckelynck M (2016) ISpinach: A fluorogenic RNA aptamer optimized for in vitro applications. Nucleic Acids Res. 44, 2491–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Messing SAJ, Gabelli SB, Liu Q, Celesnik H, Belasco JG, Piñeiro SA, and Amzel LM (2009) Structure and Biological Function of the RNA Pyrophosphohydrolase BdRppH from Bdellovibrio bacteriovorus. Structure 17, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Kieft JS, Rabe JL, and Chapman EG (2015) New hypotheses derived from the structure of a flaviviral Xrn1-resistant RNA: Conservation, folding, and host adaptation. RNA Biol. 12, 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Hulett HR (1970) Non-enzymatic hydrolysis of adenosine phosphates. Nature 225, 1248–1249. [DOI] [PubMed] [Google Scholar]
- (50).Miller DL, and Westheimer FH (1966) The Hydrolysis of γ-Phenylpropyl Di-and Triphosphates1. J. Am. Chem. Soc. 88, 1507–1511. [DOI] [PubMed] [Google Scholar]
- (51).Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi PY, Nix JC, Beckham JD, and Kieft JS (2016) Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science (Washington, DC, U. S.) 354, 1148–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Steckelberg AL, Akiyama BM, Costantino DA, Sit TL, Nix JC, and Kieft JS (2018) A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure. Proc. Natl. Acad. Sci. U. S. A. 115, 6404–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Dilweg IW, Gultyaev AP, and Olsthoorn RC (2019) Structural features of an Xrn1-resistant plant virus RNA. RNA Biol. 16, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Hoek TA, Khuperkar D, Lindeboom RGH, Sonneveld S, Verhagen BMP, Boersma S, Vermeulen M, and Tanenbaum ME (2019) Single-Molecule Imaging Uncovers Rules Governing Nonsense-Mediated mRNA Decay. Mol. Cell 75, 324–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Steitz TA, and Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U. S. A. 90, 6498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Palermo G, Cavalli A, Klein ML, Alfonso-Prieto M, Dal Peraro M, and De Vivo M (2015) Catalytic metal ions and enzymatic processing of DNA and RNA. Acc. Chem. Res. 48, 220–228. [DOI] [PubMed] [Google Scholar]
- (57).Thaplyal P, and Bevilacqua PC (2014) Experimental Approaches for Measuring pKa ‘s in RNA and DNA. Methods Enzymol. 549, 189–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Casey JR, Grinstein S, and Orlowski J (2010) Sensors and regulators of intracellular pH. Nat. Rev. Mol. Cell Biol. 11, 50–61. [DOI] [PubMed] [Google Scholar]
- (59).Madshus IH (1988) Regulation of intracellular pH in eukaryotic cells. Biochem. J. 250, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Zhang S, Williams CJ, Wormington M, Stevens A, and Peltz SW (1999) Monitoring mRNA decapping activity. Methods 17, 46–51. [DOI] [PubMed] [Google Scholar]
- (61).Nozinovic S, Reining A, Kim YB, Noeske J, Schlepckow K, Woḧnert J, and Schwalbe H (2014) The importance of helix p1 stability for structural pre-organization and ligand binding affinity of the adenine riboswitch aptamer domain. RNA Biol. 11, 655–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJT, Keene JD, Wilusz CJ, Bradrick SS, and Wilusz J (2015) XRN1 Stalling in the 5′ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability. PLoS Pathog. 11, e1004708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Charley PA, Wilusz CJ, and Wilusz J (2018) Identification of phlebovirus and arenavirus RNA sequences that stall and repress the exoribonuclease XRN1. J. Biol. Chem. 293, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Muhlrad D, Decker CJ, and Parker R (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Pelechano V, Wei W, and Steinmetz LM (2015) Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161, 1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Von Hippel PH, Johnson NP, and Marcus AH (2013) Fifty years of DNA “breathing”: Reflections on old and new approaches. Biopolymers 99, 923–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, and Had M (2009) Gaussian 09, Gaussian, Inc., Wallingford, CT. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.