Abstract
Hyaluronan (HA) is a linear glycosaminoglycan (GAG) of extracellular matrix (ECM) synthesized by three hyaluronan synthases (HASes) at the plasma membrane using uridine diphosphate (UDP)-glucuronic acid (UDP-GlcUA) and UDP-N-acetylglucosamine (UDP-GlcNAc) as substrates. The production of HA is mainly regulated by hyaluronan synthase 2 (HAS2), that can be controlled at different levels, from epigenetics to transcriptional and post-translational modifications. HA biosynthesis is an energy-consuming process and, along with HA catabolism, is strongly connected to the maintenance of metabolic homeostasis. The cytoplasmic pool of UDP-sugars is critical for HA synthesis. UDP-GlcNAc is an important nutrient sensor and serves as donor substrate for the O-GlcNAcylation of many cytosolic proteins, including HAS2. This post-translational modification stabilizes HAS2 in the membrane and increases HA production. Conversely, HAS2 can be phosphorylated by AMP activated protein kinase (AMPK), a master metabolic regulator activated by low ATP/AMP ratios, which inhibits HA secretion. Similarly, HAS2 expression and the deposition of HA within the pericellular coat are inhibited by sirtuin 1 (SIRT1), another important energetic sensor, confirming the tight connection between nutrients availability and HA metabolism.
Keywords: hyaluronidase, beta glycosidases, hexosamine biosynthetic pathway, UGDH, autophagy, ubiquitin, cancer, cardiovascular diseases, metabolic reprogramming, HAS2-AS1, extracellular matrix, proteoglycan, glycosaminoglycan
Introduction
For many years, the extracellular matrix (ECM) was considered an inert scaffold made of a complex network of macromolecules where cells adhere and grow. Nowadays, ECM has acquired a central role in cell biology as it is able to regulate critical functions such as motility, survival, proliferation, and differentiation.1 Alterations of the composition of ECM are commonly found in several pathologies and the study of factors able to drive ECM remodeling can be useful to find new pharmacological targets to treat widespread diseases including cancer.2,3 ECM is also the battlefield of the immune system, and it is well known that ECM fragments (i.e., matrikines) generated by ECM remodeling are potent signaling modulators able to trigger many cellular responses via cell surface receptors.4,5 Moreover, inflammation is a common denominator of several acute as well as chronic pathologies and ECM plays an active role to modulate this pivotal process.6-9 Among the different ECM components that are known to interact with inflammatory cells, this review will focus on hyaluronan (HA).
HA is one of the most abundant and ubiquitous component of ECM and belongs to the family of glycosaminoglycans (GAGs).10 HA is made by linear repetitions of N-acetyl-D-glucosamine (GlcNAc) and d-glucuronic acid (GlcUA) joined together via beta (1,4) and beta (1,3) glycosidic bonds that, differently from other GAGs, are not further chemically modified by sulfation, or epimerization. This alternating structure produces a negatively charged polysaccharide with a molecular mass ranging from 1000 to 8000 KDa.11 Importantly, in tissues, the chain length of HA is not uniform (i.e., polydispersed) and physiologically, the most common sizes are of about 1000 to 5000 MDa corresponding to high molecular weight HA (HMW-HA).8,12 Many of the human-approved applications of HA (i.e., dermal filler or intra-articular injection) use HMW-HA (or chemically modified HMW-HA) due to its viscoelastic properties and anti-inflammatory effects.13,14
HA of less than 0.5 MDa is known as low molecular weight HA (LMW-HA) which is usually formed during HA turnover but can accumulate also in inflammation sites. Generally, LMW-HA and HA oligos activate Toll-like receptor 4 (TLR4) and 2 (TLR2) signaling, although the physical interaction between HA and these receptors is still debated. Besides enzymatic degradation, LMW-HA can be also generated by oxidative stress or ultraviolet (UV) light.10,15 It is worth mentioning that not always LMW-HA is associated with inflammation and pathological states;16 for example, during lactation the ~0.35 MDa LMW-HA contained in milk has the specific function to induce antibacterial proteins in infant gut contributing to microbiota colonization.17
Differently from other GAGs, HA does not form a proteoglycan (PG); however, several proteins interact with HA (hyaladherins) via a specific “Link domain” or module (a sequence of ~100 amino acids composed of two alpha-helices, two triple-stranded anti-parallel beta-sheets, and two disulfide bonds) or a BX7B motif (where the ‘B’s are arginine [R] or lysine (K) residues and the “X” is a sequence of seven non-acidic amino acid).18,19 Among hyaladherins, CD44 and RHAMM are considered the most important HA receptors, mediating cell proliferation and migration, respectively.20 On the other hand, HA is able to interact with other ECM components as the PGs aggrecan, versican, and neurocan, altering ECM architecture and mechanical properties (e.g., stiffness).21,22
Interestingly, HA can covalently bind with heavy chains (HC) of inter-alpha-trypsin inhibitor (IaI), making complexes that are crucial in several physiological and pathological processes including oocyte maturation, dendritic cell activation, and asthma.23 Tumor necrosis factor-inducible gene 6 (TSG6) is the secreted enzyme catalyzing the HC transfer from IaI to C6 hydroxyl of GlcNAc residues of HA.24,25
Alterations of HA content, size, and binding partners are common factors in many biological processes including development, differentiation, immune response, senescence, apoptosis, and angiogenesis. Similarly, HA microenvironment alterations lead to cardiovascular diseases, kidney, liver, intestinal and lung pathologies, musculoskeletal disorders, neurodegenerative diseases, neoplasia, and metastasis.26-33 Although the role of HA in all these aspects is not the topic of this review, the cellular production of HA is critical for all the processes mentioned above and the following chapters will focus on the mechanism of HA synthesis and its regulation.
HA Metabolizing Enzymes
Differently from other GAGs that are synthetized in the Golgi, HA is polymerized on the plasma membrane by a family of glycosyl-transferases named HA synthases (HASes).34 These enzymes are unique under different points of view. They recognize two distinct uridine diphosphate (UDP)-sugar precursors, and they catalyze the two different glycosidic bonds on the growing HA chain that can be extruded through the plasma membrane.35 Although the 3D structures of HASes proteins are still not resolved due to structural complexity, HASes have genetic similarity in their sequences with cellulose and chitin synthases.36
Human genome contains three HAS genes coding for three different isoenzymes named HAS1, 2, and 3.37 The precise role of each HAS isoenzyme in different biological processes is still under investigation, but it is widely accepted that HAS2 is the most important HA synthetic protein for several reasons: (1) HAS2 null mice are not viable for severe cardiac malformation, (2) HAS2 is highly expressed in several tissues and cell lines, and (3) the catalytic properties of HAS2 allow a very efficient HA synthesis.10,38,39 In plasma membrane, HAS2 is active as homo- and heterodimers with HAS3 and it is in a cholesterol-rich lipid microenvironment that increases its activity.40,41 HASes do not directly use ATP, but utilize the two cytosolic sugar nucleotides UDP-glucuronic acid (UDP-GlcUA) and UDP-N-acetylglucosamine (UDP-GlcNAc) as precursors which contain high energy bonds42 (Fig. 1). As HA deposition is mainly due to HAS2,43 the study of its regulation is fundamental to understand HA biology. HAS2 displays a multistep regulation that involves the action of different growth factors and cytokines. These molecules include TGF-β, insulin-like growth factor (IGF), fibroblast growth factor (FGF), prostaglandins, and also oxidized sterol-related species delivered through oxLDL that, upon the interaction with specific receptors, increase HAS2 and therefore HA deposition.44,45 Lastly, HAS2 undergoes post-translational and epigenetic modifications, that will be described in detail in the following chapters.
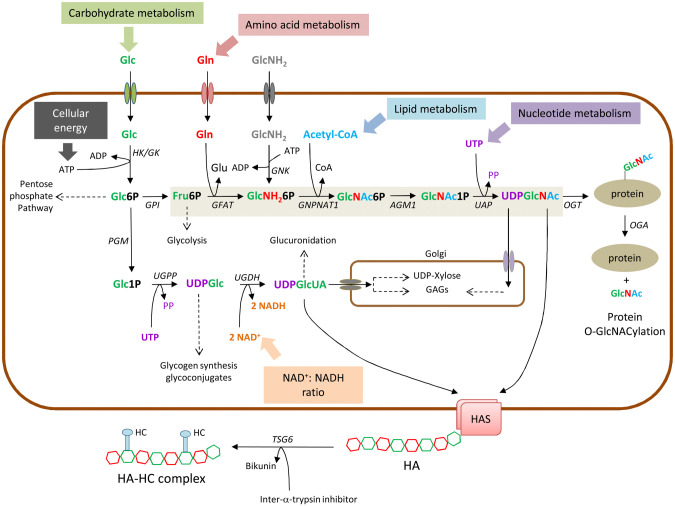
Figure 1.
Schematic representation of the biosynthesis of UDP-GlcUA and UDP-GlcNAc (through the hexosamine biosynthetic pathway, shaded in light brown). Connections with the main biochemical pathways are indicated with dotted lines. Protein O-GlcNAcylation in the cytosol and GAGs synthesis in the Golgi are also shown. The mechanism of TSG6 adding HCs from IaI to form the HC-HA matrix is shown in the extracellular space. Abbreviation: Glc, glucose; Gln, glucosamine; GlcNH2, glucosamine; Glc6P, glucose-6-phosphate; Glc1P, glucose-1-phosphate; UDPGlc, UDP-glucose, UDPGlcUA, UDP-glucuronic acid; Fru6P, Fructose-6-phosphate, GlcNH26P, Glucosamine-6 phosphate; GlcNAc6P, N-acetyl-glucosamine-6-phosphate; GlcNAc1P, N-acetyl-glucosamine-1-phosphate; UDPGlcNAc, UDP-N-acetyl-glucosamine; Glu, glutamate; Acetyl-CoA, acetyl-coenzyme A; PP, pyrophosphate. HK; hexokinase; GK, glucokinase; GPI, glucose-6-phosphate isomerase; PGM, phospho glucomutase; UGPP, UDP-Glucose pyrophosphorylase; UGDH, UDP-Glucose dehydrogenase; GFAT, glutamine: fructose-6-phosphate amidotransferase; GNK, glucosamine kinase; GNPNAT1, GlcNH 2-6-phosphate N-acetyltransferase; AGM1, phospho-GlcNAc mutase; UAP, UDP-GlcNAc pyrophosphorylase; OGT, O-GlcNAc transferase; OGA; O-GlcNAcase.
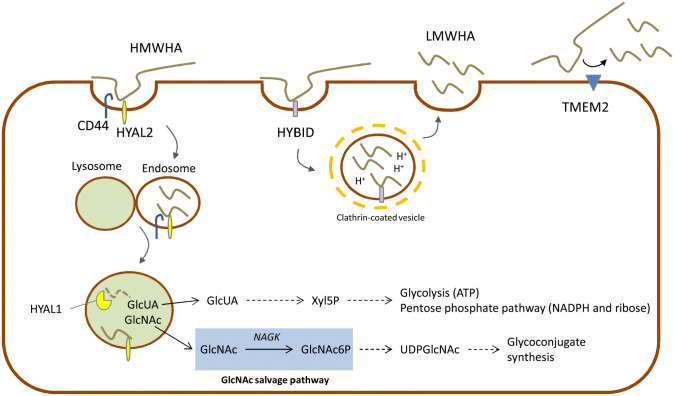
The degradation of HA is due to several hyaluronidases (HYALs) that chop HMW-HA into fragments at the plasma membrane level (HYAL2) that, in turn, are internalized in the cell via endocytosis and further degraded in the lysosome (HYAL1)46 (Fig. 2). HYALs require an acidic pH to catalyze HA cleavage.47 Recently, other HA degrading enzymes have been described to have hyaluronidase activity like, such as the Hyaluronan Binding Protein Involved in Hyaluronan Depolymerization (HYBID, also known with alias KIAA1199/CEMIP) or the transmembrane protein 2 (TMEM2)48,49 which is able to generate LMW-HA with different functions in biological processes.50-52
Figure 2.
Schematic representation of three main degradation pathways of HA through HYALs, HYBID and TMEM2. Connections with the main biochemical pathways are indicated with dotted lines. GlcNAc salvage shaded in light blue.
The complete intracellular degradation of HA, due to the coordinated action of HYALs, β-glucuronidase and hexosaminidase, leads to free GlcUA and GlcNAc (Fig. 2). Differently from plants and probably from some microorganisms that recycle GlcUA to GlcUA-1-phosphate and then to UDP-GlcUA, in vertebrates GlcUA is converted by several redox and decarboxylation reactions into xylulose-5-phosphate. Cells with a very high rate of HA turnover could alternatively use xylulose-5-phosphate to sustain pentose phosphate pathway (i.e., for the synthesis of NADPH and ribose) or, by shunting xylulose-5-phosphate to the non-oxidative part of pentose phosphate pathway, by increasing glycolysis and energy metabolism.53 In cancer cells, that typically have high expression of HYALs,54 these points could contribute to the metabolic reprogramming of cancer metabolism increasing the flux through glycolysis to obtain ATP, and through pentose phosphate pathway to have biosynthetic reducing power and ribose for anabolism.55
The cytoplasmic pool of UDP-sugars influences also HA chain length in some HA producing bacteria. Bacteria usually do not synthesize HA but some pathological species acquired the capacity to produce an HA coat to evade the host immune defenses.56 In these organisms, the length of the final polysaccharide depends on the availability of UDP-sugars: when one precursor becomes limiting the chain stops growing.57
GlcNAc is also converted in GlcNAc-6-phosphate by the GlcNAc kinase in the GlcNAc salvage pathway.58,59 Concluding, GlcNAc recycling is important to sustain the synthesis of complex glycoconjugates without affecting other biochemical pathways as carbohydrate, amino acid, lipid, and nucleotide metabolisms.
Influence of UDP-sugars Availability on HA Synthesis
HA biosynthesis is a dynamic process strictly influenced by cell metabolism and the availability of energy substrates, as it requires glucose- and glutamine-utilizing pathways for the biosynthesis of its precursors GlcNAc and GlcUA, respectively. GlcNAc is produced through the hexosamine biosynthetic pathway (HBP), a branch of glycolysis that metabolizes from 2% to 5% of the glucose that enters the cell.60 HBP provides UDP-GlcNAc units for the synthesis of several glycoconjugates and uses substrates coming from amino acids (glutamine), nucleotides (uridine), carbohydrates (glucose), and fatty acids (Acetyl-CoA) metabolism. Therefore, it is not surprising that UDP-GlcNAc is considered a “sensing molecule” being one of the most represented UDP-sugars with an intracellular concentration comparable to ATP (from 100 µM to low millimolar).61,62 In this regard, it has been shown that an increased availability of glucosamine or glucose in cell culture media stimulated the production of GlcNAc as happens in different in vivo conditions, such as hyperinsulinemia and cancer.63-66 Besides glycoconjugates, UDP-GlcNAc is also the substrate for O-glycosylation of nuclear, cytoplasmic and mitochondrial proteins. This reaction, named O-GlcNAcylation, is a reversible post-translational modification on serine/threonine residues controlled by the enzymes O-GlcNAc transferase (OGT) and O-GlcNAc hydrolase (OGA).67 This post-translational modification competes with protein phosphorylation, modulating many cellular functions including gene expression, signaling, degradation, and trafficking.68
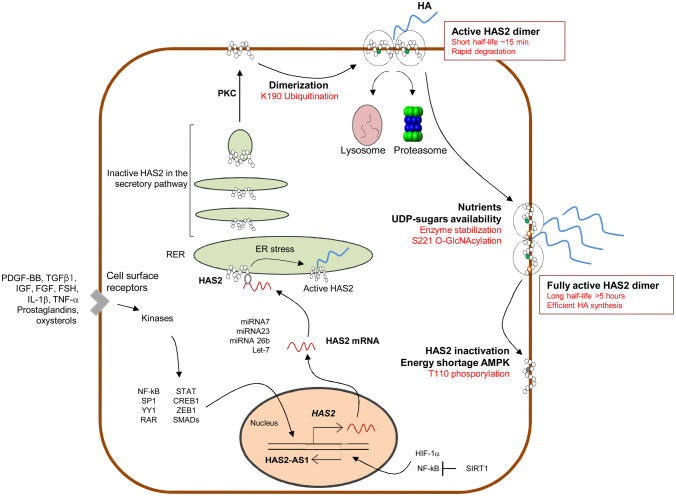
Interestingly, in vitro experiments conducted on Aortic Smooth Muscle Cells (AoSMCs) demonstrated that the treatment with glucosamine increased the synthesis of both HA and chondroitin sulfate (CS),69 suggesting that elevated UDP-GlcNAc levels can increase general GAG synthesis. However, the same study showed that the chemical inhibition of OGA significantly induced only HA production, without altering CS levels. This specific effect on HA is due to HAS2 O-GlcNAcylation on serine 221, which stabilizes the enzyme in the membrane avoiding its proteasomal degradation and increasing its half-life from 17 min to 5 hr69 (Fig. 3). Such a modification can have important consequences from a pathological point of view considering that a high glucose availability can induce a dramatic UDP-GlcNAc and HA increase, especially in cells where the uptake of glucose is insulin independent. Indeed, clinical and experimental evidences showed an accumulation of HA both in plasma and vascular walls of hyperglycemic patients and streptozotocin-induced diabetes animal models. The stimulation of HA production, as a consequence of increased UDP-GlcNAc, can be considered as an effort to reduce the cytosolic pool of UDP-GlcNAc to acceptable levels. This is consistent with the fact that when HA synthesis is significantly increased by overexpression of HASes, UDP-GlcNAc pools decrease considerably70 and negatively controls O-GlcNAcylation.71 HASes respond in a different way to cellular UDP-GlcNAc levels, as HAS1 requires higher UDP-GlcNAc amounts than HAS2 and HAS3 to produce HA.70 The enhancement of HBP flux and O-GlcNAcylation has been observed in many tumors,72 where the nutrient request is sustained by an increased glucose uptake and aerobic glycolysis (Warburg effect). Multidimensional analysis of microarray datasets demonstrated that HBP enzymes are up-regulated in breast cancer and that the co-expression of HAS2 and the HBP rate limiting enzyme glutamine-fructose-6-phosphate transaminase (GFAT) is associated to a poor prognosis in advanced cancer patients.73 Moreover, the enhancement of HBP flux is related to the activation of pro-tumorigenic pathways and elevated overall levels of O-GlcNAcylation.73 The over-production of HA correlates to plasticity of breast cancer cells, where the acceleration of the metabolism of hexosamine, along with a metabolic shift toward glycolysis, reverts cancer cells phenotype to stem cell states.74 The analysis of human breast cancer biopsies showed a dramatic increment of UDP-GlcNAc and UDP-GlcUA in the tumors, elevated GFAT levels and a strong accumulation of HA as compared to normal glandular tissue obtained from breast reductions.75 All these evidences clearly demonstrate that glucose flux into UDP-sugars offers a new mechanistic insight into tumor HA accumulation, as well as a new potential therapeutic approach.
Figure 3.
Schematic representation of HAS2 regulation. Several soluble molecules induce activation of transcription factors able to activate HAS2 transcription; HAS2 mRNA interacts with several miRNA altering stability and translation; HAS2 protein follows the secretory pathway to reach the plasma membrane; HAS2 ubiquitination leads to dimerization which represent the active HAS2 on the membrane. In this state, HAS2 is a very rapidly degraded enzyme; HAS2 O-GlcNAcylation strongly increases half-life of the enzyme in the membrane; HAS2 phosphorylation by AMPK blocks HAS2 activity. NF-kB and HIF-1α modulate HAS2-AS1 transcription; SIRT1 inhibits HAS2-AS1 that, in turn, reduces HAS2 transcription.
The other critical sugar nucleotide for the synthesis of HA (and other GAGs) is UDP-GlcUA. Besides being a substrate for GAGs synthesis, UDP-GlcUA is also necessary for the formation of PGs as its decarboxylation to UDP-xylose is necessary for the linking of GAGs to core proteins. In addition, it is the donor substrate for detoxification reactions such as glucuronidation, which takes place in the liver to detoxify the organism and in other cell types. The overexpression of UDP-glucose dehydrogenase (UGDH) in AoSMCs helped to understand the role of UDP-GlcUA in the synthesis of GAGs. UGDH catalyzes the oxidation of UDP-glucose into UDP-GlcUA using two molecules of NAD+ as a cofactor and strongly influences HA synthesis. In fact, UGDH up-regulation is associated to increased HA in the cell culture media and in the pericellular coat, without any changes in the other GAGs levels. On the contrary, the knockdown of UGDH does not alter HASes expression and reduces UDP-GlcUA levels with a corresponding decrease of HA amounts in cell culture media.76 These experiments suggest that HA is the most sensitive GAG to UDP-GlcUA concentration inside the cytoplasm and that the availability of this UDP-sugar can regulate specifically HA synthesis (as also demonstrated for UDP-GlcNAc). Moreover, the activity of UGDH is sensitive to oxygen fluctuations and the ratio of NAD+/NADH, demonstrating that HBP flux and the biosynthesis of HA are strongly influenced by the energy status of the cell.
Interestingly, the cytoplasmic pool of UDP sugar substrates can also regulate the intracellular trafficking and the extracellular shedding of HAS3 via O-GlcNAcylation. In melanoma, the depletion of UDP-GlcUA and UDP-GlcNAc from the cytosolic pool is able to stimulate HAS3 endocytosis and to inhibit HA production, while a surplus of UDP-sugars favors HAS3 retention in the plasma membrane, increasing HA synthesis and sustaining cancer progression.77
Another intermediate of HBP that influences HA synthesis is UDP-glucose (UDP-Glc),78 obtained from the reaction of UDP-glucose pyrophosphorylase (UPP) that uses glucose-1-phosphate (Glc1P) and UTP as substrates. UDP-Glc amount has to be finely tuned, as it is a critical substrate for glycogen synthesis and a key component of cellular energy homeostasis.
Metabolic Inhibition of HA Synthesis by 4-MU
Another example demonstrating that UDP-sugars availability controls HA synthesis is the use of 4-methylumbelliferone (4-MU) as an inhibitor of HA production. 4-MU is a fluorescent derivative of coumarin widely used in experimental conditions to specifically inhibit HA production both in vitro and in vivo models.31,79-82 Although the mechanism of action is still under debate, some evidences demonstrated that 4-MU is metabolized to 4-MU-glucuronide (4-MUG) functioning as a competitive substrate for UDP-glucuronosyltransferase (UGT) with the consequent decline of UDP-GlcUA concentration and HA levels.83 Other mechanisms proposed are based on HAS2 and HAS3 inhibition,83 as well on UPP and dehydrogenase (UGDH) mRNA reduction.84 As 4-MU acts on different enzymes and intermediates of the HBP pathway, its inhibitory activity has been investigated also for other GAGs such as chondroitin and heparan sulfates. Differently from HA, these GAGs are less sensitive to UDP-GlcUA deficiency, probably because they are synthesized in the Golgi apparatus, which has transporters with a very high affinity that pump in UDP-sugars from the cytosol. This mechanism might render the inhibition by a competitive substrate such as 4-MU less efficient. In contrast, HA is synthesized at the cytoplasmic membrane. Lastly, it has been proposed that 4-MU, altering the equilibrium among cytosolic sugars, can affect protein O-GlcNAcylation.69
Role of AMPK in HA Metabolism and Cell Homeostasis
As showed in Fig. 1 the synthesis of HA is an energy-consuming process and requires the equivalent of two molecules of ATP (five from double oxidation of NADPH, minus three from the oxidation of UDP-glucose for GlcNAc production),71 UTP and other metabolic molecules like glucose, glucosamine and acetyl-CoA. Therefore, it is not surprising that HA production should be regulated by the energy status of the cell and nutrients availability. Our group demonstrated that the AMP activated protein kinase (AMPK) and the availability of nutrients control HA synthesis through post-translational modification on HAS2.85 AMPK is the main cellular energy sensor and contributes to the restoration of the metabolic balance activating various genes involved in cellular homeostasis.86 The enzyme is a trimeric complex composed by a catalytic (α) and two regulatory (β and γ) subunits, that are particularly sensitive to intracellular changes of the ATP/AMP ratio. In fact, AMP and ADP allosterically activate AMPK through the binding to the γ-subunit, which causes, in turn, the phosphorylation of Thr172 by upstream kinases. The main upstream kinase responsible of AMPK phosphorylation is the Liver-Kinase-B1 (LKB1),87,88 which can increase AMPK activity up to 100-fold in in vitro models. Moreover, the binding of AMP or ADP to the regulatory subunit promotes a conformational change that protects Thr172 from the activity of phosphatases. As a result, AMPK increases ATP levels promoting glucose uptake and controlling some enzymes involved in glycolysis, fatty acid metabolism, gluconeogenesis, and protein synthesis.89 On the contrary, ATP inhibits the mechanism described above. Besides the fluctuations of adenine nucleotide, other mechanisms have been proposed that can control the AMPK activity, like the phosphorylation of Thr172 by calcium/calmodulin-dependent kinase kinase 2 (CAMKK) in response of intracellular Ca2+ increase.90 Recent studies have demonstrated that AMPK activity is related to its localization at the plasma membrane upon myristylation of the β-subunit.91 Interestingly, HAS2 (which is a plasma membrane protein) undergoes a great reduction of its enzymatic activity after AMPK activation,42 with a consequent depletion of HA levels and without any changes in the other GAGs amount. This is due to the phosphorylation of Thr110 (Fig. 3), a very well conserved residue, from zebrafish to mouse, that it is not present neither in HAS1 nor in HAS3 sequences. These evidences, along with the fact that Thr110 is situated in the cytoplasmic loop of HAS2, indicate that AMPK can directly phosphorylate HAS2 decreasing HA levels without any alteration in the amount of the other GAGs, whose synthesis occurs in the Golgi. Moreover, after the activation of AMPK significant changes in HAS2 mRNA were also detected. This line of regulation of HAS2 is a further step that helps understanding how the synthesis of HA strictly depends on the energetic homeostasis of the cell and glucose availability. For example, in hypoglycemic conditions, cells have a low energy charge and tend to switch off non vital-anabolic processes (e.g., HA synthesis) and to switch on lipid catabolism. In this way, AMPK can increase the ATP/AMP ratio and overcome stress periods. On the contrary, in hyperglycemic conditions, cells inactivate AMPK and initiate several intracellular stress responses included HA biosynthesis. This would help to maintain the balance of intracellular concentration of UDP sugars and the HBP flux.
During nutrient deprivation, AMPK is able to induce autophagy,92-94 a lysosomal-dependent self-digestive process whereby cytoplasmic organelles, proteins, and even invading pathogens are hydrolyzed to recycle and generate nutrients to maintain cell homeostasis. A study of Chen et al. demonstrated that the C-terminus part of perlecan (endorepellin)95 induces HAS2 degradation via autophagy and suppresses extracellular HA deposition,96 confirming that nutrient starvation negatively regulates HA synthesis. Interestingly, a recent paper reports that the activation of AMPK by salicylate affects the expression levels of HASes as well as the deposition of secreted and cell-associated HA, probably affecting HAS2 phosphorylation or salicylate glucuronidation and therefore, altering the availability of UDP-GlcUA.97
Regulation of HAS2 by SIRT1 and HAS2-AS1
Metabolic homeostasis is a delicate balance among energy intake, storage, and utilization. These processes are finely regulated by different signaling pathways, such as those revolving around insulin, target of rapamycin (mTOR) and food restriction. Besides AMPK, sirtuins are another class of proteins considered important metabolic sensors. Only recently, it has become evident that sirtuins and AMPK have similar effects on cellular metabolism, inflammation and mitochondrial function98,99 Moreover, it has been shown that they both regulate each other and share many common target molecules.98,100 Sirtuins are a class of NAD+ dependent deacetylases, which received particular attention after the discovery that the yeast sirtuin silent information regulator 2 (Sir2) extends lifespan.101 In mammals the sirtuin family comprises 7 members that are classified according to their catalytic activity (mono-ADP-ribosyltransferase or deacetylase), subcellular localization (cytoplasmic, nuclear or mitochondrial) and tissue specificity. The activation of sirtuins is beneficial not only for metabolic diseases, such as type 2 diabetes and obesity,102,103 but also in cancer and neurodegenerative disorders.104,105 In general, sirtuins are sensitive to cytoplasmic fluctuations of NAD+, which are directly linked to nutritional state of the cells via NAD+/NADH ratio, the absolute levels of NAD+ and NADH, or the levels of nicotinamide (NAM), which at high concentrations can non-competitively bind and inhibit sirtuins.106
In this review article, we gave particular attention to sirtuin1 (SIRT1), which is the most important metabolic regulator of the family, as it can target several genes involved in mitochondrial biogenesis, glucose and lipid metabolism. SIRT1 has a deacetylase activity and is mainly localized in the nucleus. It also possesses export signals that allow its shuttling to the cytosol under specific conditions. Although the physiological importance of this shuttling is still unclear, it has been shown that SIRT1 can deacetylase both nuclear (including histones) and cytoplasmic targets, like peroxisome proliferator-activated receptor-gamma coactivator 1-α (PGC1-α), tumor protein 53 (p53), nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), hypoxia-inducible factor 1-α (HIF1α), liver X receptor (LXR) and sterol regulatory element-binding protein 1-c (SREBP1c).107
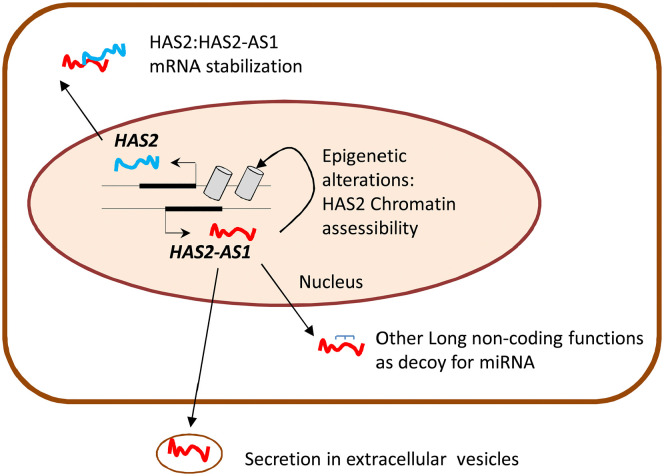
The NAD+ dependent activity of SIRT1 makes this enzyme very interesting for the study of HA synthesis regulation. Moreover, since HA production is likely a high energy demanding process and sirtuins are activated during calorie restriction and energy stress, there could be a competition between HA metabolism and SIRT1 activity. Interestingly, UGDH catalyzes the oxidation of UDP-Glc into UDP-GlcUA by using two molecules of the cofactor NAD+, influencing the NAD+/NADH ratio and, therefore, the activity of sirtuins. With experiments conducted in vitro our group demonstrated for the first time that SIRT1 can prevent inflammation through inhibition of HA metabolism. The activation of SIRT1 by the treatment with resveratrol or SRT1720 decreased HAS2 expression and reduced the amount of pericellular HA in AoSMCs. The combined treatment with tumor necrosis factor α (TNFα) and SIRT1 activators inhibited AoSMCs migration and immune cells adhesion to the HA-rich ECM. From a mechanistic point of view, we demonstrated that the activation of SIRT1 prevented NF-kB/p65 nuclear translocation and decreased the levels of HAS2-AS1, which in turn regulates HAS243 (Fig. 3). HAS2-AS1 is a long non coding RNA (lncRNA) belonging to the class of natural antisense transcripts, which controls HAS2 expression via epigenetic modifications (Fig. 4). lncRNAs are non-protein-coding RNA molecules longer than 200 base pairs that can regulate gene expression at multiple levels.108 HAS2-AS1 is synthesized from the opposite strand of HAS2 and shares about 200 base pairs with HAS2 exon 1.109 This peculiarity allows the formation of an RNA: RNA duplex between HAS2 and HAS2-AS1, which, under particular stimuli like NF-kB/p65 O-GlcNAcylation, stabilizes HAS2 expression and increases HA production.110 In addition to the nuclear function of HAS2-AS1 as an epigenetic regulator, some groups demonstrated its ability to interact with other RNA species (e.g., miRNAs) in the cytoplasm.111,112
Figure 4.
Schematic representation of cytoplasmic and nuclear functions of HAS2-AS1.
These data are in line with the findings that SIRT1 controls inflammation altering histones and transcription factors like NF-kB113 and that HA plays a pivotal role in tissue inflammation.26 Several studies suggest that the sequential course of inflammation is linked with metabolism.114 The concentration of NAD+ and the levels of sirtuins are strongly reduced in certain tissues during chronic inflammation, which is sustained by a high glycolytic flux. This condition, along with a decreased mitochondrial glucose oxidation, is dependent upon HIF-1α115,116 that can be regulated by NF-kB117 providing a link between glucose metabolism and inflammation. Interestingly, HA production and HAS2 expression can be stimulated under hypoxic conditions, as HAS2-AS1 promoter contains an hypoxia-responsive element (HRE) that responds to HIF-1α.118
In conclusion, cell metabolism greatly affects the synthesis of HA acting at different levels; many cytosolic processes involving energetic homeostasis, sugar precursor availability and protein modifications can directly regulate HAS enzymes transcription, activity, and stability. Epigenetic factors such as miRNAs or the long non-coding HAS2-AS1 can add further levels of regulation. Moreover, the possibility of such molecules to be spread in extracellular vesicles (EVs) could allow a coordinated expression of critical enzymes also in distant body district, influencing ECM composition and signaling pathways. Interestingly, the spreading of EVs coated with HA or containing other GAGs is an important event for tissue regeneration, allowing ECM turnover and remodeling.119,120 These mechanisms make HA synthesis a finely tuned system that integrates almost all the signals deriving from inside the cells but also from the cellular microenvironment. Remarkably, the Golgi synthesis of other GAGs is not affected by such cytosolic processes as kinases and O-GlcNAcylation. The synthesis of such polysaccharides is therefore regulated in different manners involving the regulation of levels of polysaccharide synthetic enzymes, of the PG core proteins and of the organelles sugars transporters. This is a very important point as an ad hoc pharmaceutical approach could be specific for HA without altering the other GAGs functions.
Acknowledgments
Ar.P. is a PhD student of the “Life Science and Biotechnology” course at Università degli studi dell’Insubria.
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: I.C. and D.V. planned the review structure; I.C., Ar.P, M.V., and E.K. wrote the manuscript; D.V. prepared the figures; I.C., D.V., and A.P. finalized the manuscript. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by PRIN2017 to E.K. (prot. 2017T8CMCY), FAR-University of Insubria, and EU grant RISE-HORIZON 2020 (ID645756) to A.P.
Contributor Information
Ilaria Caon, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Arianna Parnigoni, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Manuela Viola, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Evgenia Karousou, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Alberto Passi, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Davide Vigetti, Department of Medicine and Surgery, University of Insubria, Varese, Italy.
Literature Cited
- 1. Lu P, Takai K, Weaver VM, Werb Z. Extracellular Matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol. 2011;3:a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karamanos NK, Theocharis AD, Neill T, Iozzo RV. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 2019;75–76:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sainio A, Järveläinen H. Extracellular matrix-cell interactions: focus on therapeutic applications. Cell Signal. 2020;66:109487. [DOI] [PubMed] [Google Scholar]
- 4. Maquart FX, Pasco S, Ramont L, Monboisse J-C. An introduction to matrikines: extracellular matrix—derived peptides which regulate cell activity—implication in tumor invasion. Crit Rev Oncol/Hematol. 2004;49:199–202. [DOI] [PubMed] [Google Scholar]
- 5. Wells JM, Gaggar A, Blalock JE. MMP generated matrikines. Matrix Biol. 2015;44–46:122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iijima J, Konno K, Itano N. Inflammatory alterations of the extracellular matrix in the tumor microenvironment. Cancers. 2011;3:3189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tavianatou AG, Caon I, Franchi M, Piperigkou Z, Galesso D, Karamanoset NK. Hyaluronan: molecular size-dependent signaling and biological functions in inflammation and cancer. FEBS J. 2019;286:2883–908. [DOI] [PubMed] [Google Scholar]
- 8. Monslow J, Govindaraju P, Puré E. Hyaluronan—a functional and structural sweet spot in the tissue microenvironment. Front Immunol. 2015;6:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bartolini B, Caravà E, Caon I, Parnigoni A, Moretto P, Passi A, Vigetti D, Viola M, Karousou E. Heparan sulfate in the tumor microenvironment. Adv Exp Med Biol. 2020;1245:147–61. [DOI] [PubMed] [Google Scholar]
- 10. Karamanos NK, Piperigkou Z, Theocharis AD, Watanabe H, Franchi M, Baud M, Brézillon S, Götte M, Passi A, Vigetti D, Ricard-Blum S, Sanderson RD, Neill T, Iozzo RV. Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chem Rev. 2018;118:9152–232. [DOI] [PubMed] [Google Scholar]
- 11. Cowman MK, Lee H-G, Schwertfeger KL, McCarthy JB, Turley EA. The content and size of hyaluronan in biological fluids and tissues. Front Immunol. 2015;6:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cyphert JM, Trempus CS, Garantziotis S. Size matters: molecular weight specificity of hyaluronan effects in cell biology. Int J Cell Biol. 2015;2015:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Passi A, Vigetti D. Hyaluronan as tunable drug delivery system. Adv Drug Deli Rev. 2019;146:83–96. [DOI] [PubMed] [Google Scholar]
- 14. Viola M, Vigetti D, Karousou E, D’Angelo ML, Caon I, Moretto P, De Luca G, Passi A. Biology and biotechnology of hyaluronan. Glycoconj J. 2015;32:93–103. [DOI] [PubMed] [Google Scholar]
- 15. Stern R, Asari A, Sugahara K. Hyaluronan fragments: an information-rich system. Eur J Cell Biol. 2006;85:699–715. [DOI] [PubMed] [Google Scholar]
- 16. Collins SL, Black KE, Chan-Li Y, Ahn Y-H, Cole PA, Powell JD, Horton MR. Hyaluronan fragments promote inflammation by down-regulating the anti-inflammatory A2a receptor. Am J Respir Cell Mol Biol. 2011;45:675–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kessler SP, Obery DR, Nickerson KP, Petrey AC, McDonald C, de la Motte CA. Multifunctional role of 35 kilodalton hyaluronan in promoting defense of the intestinal epithelium. J Histochem Cytochem. 2018;66:273–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Day AJ, Prestwich GD. Hyaluronan-binding proteins: tying up the giant. J Biol Chem. 2002;277:4585–8. [DOI] [PubMed] [Google Scholar]
- 19. Toole BP. Hyaluronan and its binding proteins, the hyaladherins. Curr Opin Cell Biol. 1990;2:839–44. [DOI] [PubMed] [Google Scholar]
- 20. Turley EA, Noble PW, Bourguignon LYW. Signaling properties of hyaluronan receptors. J Biol Chem. 2002;277:4589–92. [DOI] [PubMed] [Google Scholar]
- 21. Bano F, Tammi MI, Kang DW, Harris EN, Richter RP. Single-molecule unbinding forces between the polysaccharide hyaluronan and its binding proteins. Biophys J. 2018;114:2910–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagy N, De La Zerda A, Kaber G, Johnson PY, Hu KH, Kratochvil MJ, Yadava K, Zhao W, Cui Y, Navarro G, Annes GP, Wight TN, Heilshorn SC, Bollyky PL, Butte ML. Hyaluronan content governs tissue stiffness in pancreatic islet inflammation. J Biol Chem. 2018;293:567–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Day AJ, Milner CM. TSG-6: a multifunctional protein with anti-inflammatory and tissue-protective properties. Matrix Biol. 2019;78–79:60–83. [DOI] [PubMed] [Google Scholar]
- 24. Zhao M, Yoneda M, Ohashi Y, Iwata H, Ohnuki Y, Kimata K. Evidence for the covalent binding of SHAP, heavy chains of inter-α- trypsin inhibitor, to hyaluronan. J Biol Chem. 1995;270:26657–63. [DOI] [PubMed] [Google Scholar]
- 25. Spinelli FM, Vitale DL, Icardi A, Caon I, Brandone A, Giannoni P, Saturno V, Passi A, García M, Sevic I, Alaniz L. Hyaluronan preconditioning of monocytes/macrophages affects their angiogenic behavior and regulation of TSG -6 expression in a tumor type-specific manner. FEBS J 2019;286:3433–49. [DOI] [PubMed] [Google Scholar]
- 26. Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev. 2011;91:221–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang J, Jiang D, Noble PW. Hyaluronan as a therapeutic target in human diseases. Adv Drug Deli Rev. 2016;97:186–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filpa V, Bistoletti M, Caon I, Moro E, Grimaldi A, Moretto P, Baj A, Giron MC, Karousou E, Viola M, Crema F, Frigo G, Passi A, Giaroni C, Vigetti D. Changes in hyaluronan deposition in the rat myenteric plexus after experimentally-induced colitis. Sci Rep. 2017;7:17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knudson W, Ishizuka S, Terabe K, Askew EB, Knudson CB. The pericellular hyaluronan of articular chondrocytes. Matrix Biol. 2019;78–79:32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manou D, Caon I, Bouris P, Triantaphyllidou IE, Giaroni C, Passi A, Karamanos NK, Vigetti D, Theocharis AD. The complex interplay between extracellular matrix and cells in tissues. Methods Mol Biol. 2019;1952:1–20. [DOI] [PubMed] [Google Scholar]
- 31. Caon I, Bartolini B, Parnigoni A, Caravà E, Moretto P, Viola M, Karousou E, Vigetti D, Passi A. Revisiting the hallmarks of cancer: the role of hyaluronan. Sem Cancer Biol. 2019;62:9–19. [DOI] [PubMed] [Google Scholar]
- 32. Perkins KL, Arranz AM, Yamaguchi Y, Hrabetova S. Brain extracellular space, hyaluronan, and the prevention of epileptic seizures. Rev Neurosci. 2017;28:869–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Viola M, Karousou E, Luisa D’, Angelo M, Caon I, Luca G, Passi A, Vigetti D. Extracellular matrix in atherosclerosis: hyaluronan and proteoglycans insights. Curr Med Chem. 2016;23:2958–71. [DOI] [PubMed] [Google Scholar]
- 34. Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem. 1997;272:13997–4000. [DOI] [PubMed] [Google Scholar]
- 35. Weigel PH, DeAngelis PL. Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J Biol Chem. 2007;282:36777–81. [DOI] [PubMed] [Google Scholar]
- 36. Bi Y, Hubbard C, Purushotham P, Zimmer J. Insights into the structure and function of membrane-integrated processive glycosyltransferases. Curr Opin Struct Biol. 2015;34:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Itano N, Kimata K. Mammalian hyaluronan synthases. IUBMB Life. 2002;54:195–9. [DOI] [PubMed] [Google Scholar]
- 38. Itano N, Sawai T, Yoshida M, Lenas P, Yamada Y, Imagawa M, Shinomura T, Hamaguchi M, Yoshida Y, Ohnuki Y, Miyauchi S, Spicer AP, McDonald JA, Kimata K. Three isoforms of mammalian hyaluronan synthases have distinct enzymatic properties. J Biol Chem. 1999;274:25085–92. [DOI] [PubMed] [Google Scholar]
- 39. Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A, Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106:349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karousou E, Kamiryo M, Skandalis SS, Ruusala A, Asteriou T, Passi T, Yamashita H, Hellman U, Heldin C-H, Heldin P. The activity of hyaluronan synthase 2 is regulated by dimerization and ubiquitination. J Biol Chem. 2010;285:23647–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ontong P, Hatada Y, Taniguchi S, Kakizaki I, Itano N. Effect of a cholesterol-rich lipid environment on the enzymatic activity of reconstituted hyaluronan synthase. Biochem Biophys Res Commun. 2014;443:666–71. [DOI] [PubMed] [Google Scholar]
- 42. Vigetti D, Viola M, Karousou E, De Luca G, Passi A. Metabolic control of hyaluronan synthases. Matrix Biol. 2014;35:8–13. [DOI] [PubMed] [Google Scholar]
- 43. Caon I, Bartolini B, Moretto P, Parnigoni A, Caravà E, Vitale DL, Alaniz L, Viola M, Karousou E, De Luca G, Hascall VC, Passi A, Vigetti D. Sirtuin 1 reduces hyaluronan synthase 2 expression by inhibiting nuclear translocation of NF-κB and expression of the long-noncoding RNA HAS2-AS1. J Biol Chem. 2020;295:3485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tammi RH, Passi AG, Rilla K, Karousou E, Vigetti D, Makkonen K, Tammi MI. Transcriptional and post-translational regulation of hyaluronan synthesis. FEBS J. 2011;278:1419–28. [DOI] [PubMed] [Google Scholar]
- 45. Viola M, Bartolini B, Vigetti D, Karousou E, Moretto P, Deleonibus S, Sawamura T, Wight TN, Hascall VC, De Luca G, Passi A. Oxidized low density lipoprotein (LDL) affects hyaluronan synthesis in human aortic smooth muscle cells. J Biol Chem. 2013;288:29595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Csoka AB, Frost GI, Stern R. The six hyaluronidase-like genes in the human and mouse genomes. Matrix Biol. 2001;20:499–508. [DOI] [PubMed] [Google Scholar]
- 47. Stern R. Hyaluronan catabolism: a new metabolic pathway. Eur J Cell Biol. 2004;83:317–25. [DOI] [PubMed] [Google Scholar]
- 48. Yoshida H, Nagaoka A, Kusaka-Kikushima A, Tobiishi M, Kawabata K, Sayo T, Sakai S, Sugiyama Y, Enomoto H, Okada Y, Inoue S. KIAA1199, a deafness gene of unknown function, is a new hyaluronan binding protein involved in hyaluronan depolymerization. Proc Natl Acad Sci U S A. 2013;110:5612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamamoto H, Tobisawa Y, Inubushi T, Irie F, Ohyama C, Yamaguchi Y. A mammalian homolog of the zebrafish transmembrane protein 2 (TMEM2) is the long-sought-after cell-surface hyaluronidase. J Biol Chem. 2017;292:7304–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Angelis JE, Lagendijk AK, Chen H, Tromp A, Bower NI, Tunny KA, Brooks AJ, Bakkers J, Francois M, Yap AS, Simons C, Wicking C, Hogan BM, Smith KA. Tmem2 regulates embryonic vegf signaling by controlling hyaluronic acid turnover. Dev Cell. 2017;40:123–36. [DOI] [PubMed] [Google Scholar]
- 51. Schinzel RT, Higuchi-Sanabria R, Shalem O, Moehle EA, Webster BM, Joe L, Bar-Ziv R, Frankino PA, Durieux J, Pender C, Kelet N, Kumar SS, Savalia N, Chi H, Simic M, Nguyen N-T, Dillin A. The hyaluronidase, TMEM2, promotes ER homeostasis and longevity independent of the UPRER. Cell. 2019;179:1306–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yoshida H, Okada Y. Role of HYBID (Hyaluronan binding protein involved in hyaluronan depolymerization), alias KIAA1199/CEMIP, in hyaluronan degradation in normal and photoaged skin. Int J Mol Sci. 2019;20:5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McAtee CO, Barycki JJ, Simpson MA. Emerging roles for hyaluronidase in cancer metastasis and therapy. Adv Cancer Res. 2014;123:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boroughs LK, Deberardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Csoka AB, Stern R. Hypotheses on the evolution of hyaluronan: a highly ironic acid. Glycobiology. 2013;23:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jing W, DeAngelis PL. Synchronized chemoenzymatic synthesis of monodisperse hyaluronan polymers. J Biol Chem. 2004;279:42345–9. [DOI] [PubMed] [Google Scholar]
- 58. Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code.” Sci STKE. 2005;2005:re13. [DOI] [PubMed] [Google Scholar]
- 59. MacNicholl AD, Wusteman FS, Winterburn PJ. Deg-radation of [3H]chondroitin 4-sulphate and re-utilization of the [3H]hexosamine component by the isolated perfused rat liver. Biochem J. 1980;186:279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McClain DA, Crook ED. Hexosamines and insulin resistance. Diabetes. 1996;45:1003–9. [DOI] [PubMed] [Google Scholar]
- 61. Marshall S, Nadeau O, Yamasaki K. Dynamic actions of glucose and glucosamine on hexosamine biosynthesis in isolated adipocytes: differential effects on glucosamine 6-phosphate, UDP-N-acetylglucosamine, and ATP levels. J Biol Chem. 2004;279:35313–9. [DOI] [PubMed] [Google Scholar]
- 62. Tomiya N, Ailor E, Lawrence SM, Betenbaugh MJ, Lee YC. Determination of nucleotides and sugar nucleotides involved in protein glycosylation by high-performance anion-exchange chromatography: sugar nucleotide contents in cultured insect cells and mammalian cells. Anal Biochem. 2001;293:129–137. [DOI] [PubMed] [Google Scholar]
- 63. Marshall S, Nadeau O, Yamasaki K. Dynamic actions of glucose and glucosamine on hexosamine biosynthesis in isolated adipocytes. J Biol Chem. 2004;279:35313–9. [DOI] [PubMed] [Google Scholar]
- 64. Schleicher ED, Weigert C. Role of the hexosamine biosynthetic pathway in diabetic nephropathy. Kidney Int Suppl. 2000;77:S13–8. [DOI] [PubMed] [Google Scholar]
- 65. Vasconcelos-Dos-Santos A, Loponte HFBR, Mantuano NR, Oliveira IA, de Paula IF, Teixeira LK, de-Freitas-Junior JCM, Gondim KC, Heise N, Mohana-Borges R, Morgado-Díaz JA, Dias WB, Todeschini AR. Hyperglycemia exacerbates colon cancer malignancy through hexosamine biosynthetic pathway. Oncogenesis. 2017;6:e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Akella NM, Ciraku L, Reginato MJ. Fueling the fire: emerging role of the hexosamine biosynthetic pathway in cancer. BMC Biol. 2019;17:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dias WB, Hart GW. O-GlcNAc modification in diabetes and Alzheimer’s disease. Mol Biosyst. 2007;3:766–72. [DOI] [PubMed] [Google Scholar]
- 69. Vigetti D, Deleonibus S, Moretto P, Karousou E, Viola M, Bartolini B, Hascall VC, Tammi M, De Luca G, Passi A. Role of UDP-N-acetylglucosamine (GlcNAc) and O-GlcNacylation of hyaluronan synthase 2 in the control of chondroitin sulfate and hyaluronan synthesis. J Biol Chem. 2012;287:35544–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rilla K, Oikari S, Jokela TA, Hyttinen JMT, Kärnä R, Tammi RH, Tammi MI. Hyaluronan Synthase 1 (HAS1) requires higher cellular UDP-GlcNAc concentration than HAS2 and HAS3. J Bol Chem. 2013;288:5973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hascall VC, Wang A, Tammi M, Oikari S, Tammi R, Passi A, Vigetti D, Hanson RW, Hart GW. The dynamic metabolism of hyaluronan regulates the cytosolic concentration of UDP-GlcNAc. Matrix Biol. 2014;35:14–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chiaradonna F, Ricciardiello F, Palorini R. The nutrient-sensing hexosamine biosynthetic pathway as the hub of cancer metabolic Rewiring. Cells. 2018;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chokchaitaweesuk C, Kobayashi T, Izumikawa T, Itano N. Enhanced hexosamine metabolism drives metabolic and signaling networks involving hyaluronan production and O-GlcNAcylation to exacerbate breast cancer. Cell Death Dis. 2019;10:803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chanmee T, Ontong P, Izumikawa T, Higashide M, Mochizuki N, Chokchaitaweesuk C, Khansai M, Nakajima K, Kakizaki I, Kongtawelert P, Taniguchi N, Itano N. Hyaluronan production regulates metabolic and cancer stem-like properties of breast cancer cells via hexosamine biosynthetic pathway-coupled HIF-1 signaling. J Biol Chem. 2016;291:24105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Oikari S, Kettunen T, Tiainen S, Häyrinen J, Masarwah A, Sudah M, Sutela A, Vanninen R, Tammi M, Auvinen P. UDP-sugar accumulation drives hyaluronan synthesis in breast cancer. Matrix Biol. 2018;67:63–74. [DOI] [PubMed] [Google Scholar]
- 76. Vigetti D, Ori M, Viola M, Genasetti A, Karousou E, Rizzi M, Pallotti F, Nardi I, Hascall VC, De Luca G, Passi A. Molecular cloning and characterization of UDP-glucose dehydrogenase from the amphibian Xenopus laevis and its involvement in hyaluronan synthesis. J Biol Chem. 2006;281:8254–63. [DOI] [PubMed] [Google Scholar]
- 77. Deen AJ, Arasu UT, Pasonen-Seppänen S, Hassinen A, Takabe P, Wojciechowski S, Kärnä R, Rilla K, Kellokumpu S, Tammi R, Tammi M, Oikari S. UDP-sugar substrates of HAS3 regulate its O-GlcNAcylation, intracellular traffic, extracellular shedding and correlate with melanoma progression. Cell Mol Life Sci. 2016;73:3183–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Magee C, Nurminskaya M, Linsenmayer TF. UDP-glucose pyrophosphorylase: up-regulation in hypertrophic cartilage and role in hyaluronan synthesis. Biochem J. 2001;360:667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nagy N, Kuipers HF, Frymoyer AR, Ruppert SM, Marshall PL, Xie BJ, Sun W, Malkovskiy AV, Rajadas J, Grandoch M, Fischer JW, Frymoyer AR, Kaber G, Bollyky PL. 4-methylumbelliferone treatment and hyaluronan inhibition as a therapeutic strategy in inflammation, autoimmunity, and cancer. Front Immunol. 2015;6:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nagy N, Gurevich I, Kuipers HF, Ruppert SM, Marshall PL, Xie BJ, Sun W, Malkovskiy AV, Rajadas J, Grandoch M, Fischer JW, Frymoyer AR, Kaber G, Bollyky PL. 4-Methylumbelliferyl glucuronide contributes to hyaluronan synthesis inhibition. J Biol Chem. 2019;294:7864–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kuipers HF, Nagy N, Ruppert SM, Ruppert SM, Marshall PL, Xie BJ, Sun W, Malkovskiy AV, Rajadas J, Grandoch M, Fischer JW, Frymoyer AR, Kaber G, Bollyky PL. The pharmacokinetics and dosing of oral 4-methylumbelliferone for inhibition of hyaluronan synthesis in mice. Clin Exp Immunol. 2016;185:372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li F, Hao P, Liu G, Wang W, Han R, Jiang Z, Li X. Effects of 4-methylumbelliferone and high molecular weight hyaluronic acid on the inflammation of corneal stromal cells induced by LPS. Graefe’s Arch Clin Exp Ophthalmol. 2017;255:559–66. [DOI] [PubMed] [Google Scholar]
- 83. Kultti A, Pasonen-Seppänen S, Jauhiainen M, Rilla KJ, Kärnä R, Pyöriä E, Tammi RH, Tammi MI. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp Cell Res. 2009;315:1914–23. [DOI] [PubMed] [Google Scholar]
- 84. Vigetti D, Rizzi M, Viola M, Genasetti A, Clerici M, Bartolini B, Hascall VC, De Luca G, Passi A. The effects of 4-methylumbelliferone on hyaluronan synthesis, MMP2 activity, proliferation, and motility of human aortic smooth muscle cells. Glycobiology. 2009;19:537–46. [DOI] [PubMed] [Google Scholar]
- 85. Vigetti D, Clerici M, Deleonibus S, Karousou E, Viola M, Moretto P, Heldin P, Hascall VC, De Luca G, Passi A. Hyaluronan synthesis is inhibited by adenosine monophosphate-activated protein kinase through the regulation of HAS2 activity in human aortic smooth muscle cells. J Biol Chem. 2011;286:7917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Garcia D, Shaw RJ. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell. 2017;66:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Mäkelä TP, Alessi DR, Hardie DG. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Herzig S, Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat Rev Mol Cell Biol. 2018;19:121–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. [DOI] [PubMed] [Google Scholar]
- 91. Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, Macaulay SL, Kemp BE. β-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK). Proc Natl Acad Sci U S A. 2010;107:19237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jang M, Park R, Kim H, Namkoong S, Jo D, Huh YH, Jang IS, Lee JI, Park Y. AMPK contributes to autophagosome maturation and lysosomal fusion. Sci Rep. 2018;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hardie DG. AMPK and autophagy get connected. EMBO Journal. 2011;30:634–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Mongiat M, Sweeney SM, San Antonio JD, Fu J, Iozzo RV. Endorepellin, a novel inhibitor of angiogenesis derived from the C terminus of perlecan. J Biol Chem. 2003;278:4238–49. [DOI] [PubMed] [Google Scholar]
- 96. Chen CG, Gubbiotti MA, Kapoor A, Han X, Yu Y, Linhardt RJ, Iozzo RV. Autophagic degradation of HAS2 in endothelial cells: a novel mechanism to regulate angiogenesis. Matrix Biol. Epub 19 February2020. doi: 10.1016/j.matbio.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 97. Karalis TT, Chatzopoulos A, Kondyli A, Aletrasa Aj, Karamanos NK, Heldin P, Skandalisa SS. Salicylate suppresses the oncogenic hyaluronan network in metastatic breast cancer cells. Matrix Biol Plus. 2020;5:100031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Price NL, Gomes AP, Ling AJY, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, Hubbard BP, Varela AT, Davis JG, Varamini B, Hafner A, Moaddel R, Rolo AP, Coppari R, Palmeira CM, de Cabo R, Baur JA, Sinclair DA. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fulco M, Sartorelli V. Comparing and contrasting the roles of AMPK and SIRT1 in metabolic tissues. Cell Cycle. 2008;7:3669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y. AMPK and SIRT1: a long-standing partnership? Am J Physiol Endocrinol Metab. 2010;298:E751–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kitada M, Ogura Y, Monno I, Koya D. Sirtuins and type 2 diabetes: role in inflammation, oxidative stress, and mitochondrial function. Front Endocrinol. 2019;10:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kurylowicz A. In search of new therapeutic targets in obesity treatment: sirtuins. Int J Mol Sci. 2016;17:572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bosch-Presegué L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2:648–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Min SW, Sohn PD, Cho SH, Swanson RA, Gan L. Sirtuins in neurodegenerative diseases: an update on potential mechanisms. Front Aging Neurosci. 2013;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nakagawa T, Guarente L. Sirtuins at a glance. J Cell Sci. 2011;124:833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Molecul Cell Biol. 2012;13:225–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Michael DR, Phillips AO, Krupa A, Martin J, Redman JE, Altaher A, Neville RD, Webber J, Kim M-Y, Bowen T. The human hyaluronan synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. J Biol Chem. 2011;286:19523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vigetti D, Deleonibus S, Moretto P, Bowen T, Fischer JW, Grandoch M, Oberhuber A, Love DC, Hanover JA, Cinquetti R, Karousou E, Viola M, D’Angelo ML, Hascall VC, De Luca G, Passi A. Natural antisense transcript for hyaluronan synthase 2 (HAS2-AS1) induces transcription of HAS2 via protein O-GlcNAcylation. J Biol Chem. 2014;289:28816–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tong L, Wang Y, Ao Y, Sun X. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis. Biomed Pharmacother. 2019;115:108891. [DOI] [PubMed] [Google Scholar]
- 112. Zhang L, Wang H, Xu M, Chen F, Li W, Hu H, Yuan Q, Su Y, Liu X, Wuri J, Yan T. Long noncoding RNA HAS2-AS1 promotes tumor progression in glioblastoma via functioning as a competing endogenous RNA. J Cell Biochem. 2020;121:661–71. [DOI] [PubMed] [Google Scholar]
- 113. Xie J, Zhang X, Zhang L. Negative regulation of inflammation by SIRT1. Pharmacol Res. 2013;67:60–7. [DOI] [PubMed] [Google Scholar]
- 114. Vachharajani VT, Liu T, Wang X, Hoth JH, Yoza BK, McCall CE. Sirtuins link inflammation and metabolism. J Immunol Res. 2016;2016:8167273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. [DOI] [PubMed] [Google Scholar]
- 116. Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. [DOI] [PubMed] [Google Scholar]
- 117. Ignazio LD, ’Bandarra D, Rocha S. NF-kB and HIF crosstalk in immune responses. FEBS J. 2016;283;413–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhu G, Wang S, Chen J, Wang Z, Liang X, Wang X, Jiang J, Lang J, Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56:2210–22. [DOI] [PubMed] [Google Scholar]
- 119. Arasu UT, Kärnä R, Härkönen K, Oikari S, Koistinen A, Kröger H, Qu C, Lammi MJ, Rilla K. Human mesenchymal stem cells secrete hyaluronan-coated extracellular vesicles. Matrix Biol. 2017;64:54–68. [DOI] [PubMed] [Google Scholar]
- 120. Coulson-Thomas VJ, Caterson B, Kao WW-Y. Trans-plantation of human umbilical mesenchymal stem cells cures the corneal defects of mucopolysaccharidosis VII mice. Stem Cells. 2013;31:2116–26. [DOI] [PMC free article] [PubMed] [Google Scholar]