Abstract
IMPORTANCE
There is an unmet need for interferon- and ribavirin-free treatment for chronic hepatitis C virus (HCV) infection in patients co-infected with human immunodeficiency virus (HIV).
OBJECTIVE
To evaluate the rates of sustained virologic response (SVR) and adverse events in previously untreated patients with HCV genotype 1 and HIV co-infection following a 12-week treatment of the fixed-dose combination of ledipasvir and sofosbuvir.
DESIGN, SETTING, AND PARTICIPANTS
Open-label, single-center, phase 2b pilot study of previously untreated, noncirrhotic patients with HCV genotype 1 and HIV co-infection conducted at the Clinical Research Center of the National Institutes of Health, Bethesda, Maryland, from June 2013 to September 2014. Patients included those receiving antiretroviral therapy with HIV RNA values of 50 copies/mL or fewer and a CD4 T-lymphocyte count of 100 cells/mL or greater or patients with untreated HIV infection with a CD4 T-lymphocyte count of 500 cells/mL or greater. Serial measurements of safety parameters, virologic and host immune correlates, and adherence were performed.
INTERVENTIONS
Fifty patients with HCV genotype 1 never before treated for HCV were prescribed a fixed-dose combination of ledipasvir (90 mg) and sofosbuvir (400 mg) once daily for 12 weeks.
MAIN OUTCOMES AND MEASURES
The primary study outcome was the proportion of patients with sustained viral response (plasma HCV RNA level <12 IU/mL) 12 weeks after end of treatment.
RESULTS
Forty-nine of 50 participants (98% [95% CI, 89% to 100%]) achieved SVR 12 weeks after end of treatment, whereas 1 patient experienced relapse at week 4 following treatment. In the patient with relapse, deep sequencing revealed a resistance associated mutation in the NS5A region conferring resistance to NS5A inhibitors, such as ledipasvir. The most common adverse events were nasal congestion (16% of patients) and myalgia (14%). There were no discontinuations or serious adverse events attributable to study drug.
CONCLUSIONS AND RELEVANCE
In this open-label, uncontrolled, pilot study enrolling patients co-infected with HCV genotype 1 and HIV, administration of an oral combination of ledipasvir and sofosbuvir for 12 weeks was associated with high rates of SVR after treatment completion. Larger studies that also include patients with cirrhosis and lower CD4 T-cell counts are required to understand if the results of this study generalize to all patients co-infected with HCV and HIV.
TRIAL REGISTRATION
clinicaltrials.gov Identifier: NCT01878799
Approximately 185 million people are infected with chronic hepatitis C (HCV) infection worldwide, and about 5 million are co-infected with human immunodeficiency (HIV).1 In the United States, co-infection occurs in approximately one-third of all patients with HIV-1 infection, with incidence rates as high as 75% to 90% among patients with reported history of intravenous drug use, and is associated with higher rates of end-stage liver disease, liver cancer, and mortality.2,3
Prior to 2013, treatment of HCV genotype-1 infection in individuals with HIV co-infection required 24 to 48 weeks of pegylated interferon, ribavirin, and a protease inhibitor (either boceprevir or telaprevir).4 Although this regimen was associated with improved rates of sustained virologic response (SVR) when compared with those observed with pegylated interferon and ribavirin alone,5–7 widespread use was limited because of high rates of adverse events, discontinuation rates, and complex drug-drug interactions.4,8,9
In 2013, 2 new directly acting antiviral agents—sofosbuvir, an HCV NS5B polymerase inhibitor, and simeprevir, an NS3/4A protease inhibitor—were licensed for treatment of HCV.10,11 Although sofosbuvir and ribavirin for 24 weeks was associated with high rates of SVR in HIV-negative and HIV-positive patients with HCV genotype 1,12,13 modest rates of hemoglobin decline were attributable to concomitant use of ribavirin. Recent studies evaluating the combination of ledipasvir, an HCV NS5A inhibitor, along with sofosbuvir, in a fixed-dose combination with or without ribavirin for 8 to 24 weeks, have demonstrated SVR rates of 91% to 100% in patients monoinfected with HCV genotype 1 who either have or have not previously received HCV treatment.5–7,14,15 In this study, we evaluated the rates of SVR following a 12-week treatment regimen of a fixed-dose combination of ledipasvir and sofosbuvir in patients co-infected with HCV genotype 1 and HIV who were not previously treated for HCV.
Methods
Participants
Participants were enrolled in a single-center, open-label, uncontrolled, nonrandomized phase 2b trial conducted at the Clinical Research Center of the National Institutes of Health (NIH), Bethesda, Maryland, from June 2013 to September 2014. Patients were recruited from existing HCV clinics in the District of Columbia as previously described.13 Patients were contacted for screening based on the order of initial communication with the study team and for start of study drugs based on completion of eligibility requirements. The study protocol is available in Supplement 1.
Eligible participants were at least 18 years old, had not received prior HCV treatment, and were co-infected with HCV genotype 1 and HIV. Participants could either not be receiving antiretroviral therapy with a CD4 T-lymphocyte count of 500 cells/mm3 or greater regardless of HIV viral load, a stable CD4 count with HIV viral load less than 500 copies/mL, or could be receiving a protocol-approved antiretroviral regimen with HIV RNA values of 50 copies/mL or fewer with a CD4 T-lymphocyte count of 100 cells/μL or greater. Participants receiving antiretroviral drugs were required to have been receiving a stable treatment-regimen for at least 8 weeks prior to starting study drug. Acceptable antiretroviral drugs were emtricitabine plus tenofovir and/or efavirenz, raltegravir, or rilpivirine.
Participants with cirrhosis, who may be less responsive to HCV treatment, were excluded from this investigational study. The presence or absence of cirrhosis was determined by liver biopsy within the previous 3 years of enrollment or by a combination of FibroSURE (Laboratory Corporation of America) score less than 0.48 and aspartate aminotransferase/platelet ratio index less than 1. A FibroSURE score less than 0.48 corresponds with a fibrosis stage from 0 to 2.
Race and ethnicity were self-reported. All eligible participants were required to have a primary medical provider and, if opioid-dependent, needed to be participating in supervised treatment. Full eligibility criteria are included in eAppendix 1 in Supplement 2.
Study Oversight
The study was approved by the National Institute of Allergy and Infectious Diseases (NIAID) institutional review board and was conducted in compliance with the Good Clinical Practice guidelines, the Declaration of Helsinki, and regulatory requirements. Written or oral informed consent approved by the NIAID institutional review board was obtained from all participants. The Regulatory Compliance and Human Participants Protection Branch of NIAID served as the study sponsor and medical monitor. Data collection, review and analysis, and writing were performed by NIH and Institute of Human Virology investigators.
Study Design
Participants from outpatient community clinics were prescribed a combination tablet containing ledipasvir (90 mg) and sofosbuvir (400 mg), administered orally for 12 weeks. Patients received instructions for proper administration of study drug. Criteria for stopping study medications were based on specific safety or virologic measures. Participants would be discontinued from therapy if they did not achieve a greater than 2 log10 decrease in HCV RNA values (unless a >2 log10 decrease would be below the lower limit of quantification [LLOQ] at their week 4 visit or if their serum RNA level was greater than or equal to the LLOQ at their week 8 visit [eAppendix 2 in Supplement 2]). Participants who did not achieve SVR 12 weeks after treatment completion (SVR12) were offered the option of additional treatment with the current standard of care. In addition to their screening visit, patients were reminded to return to the NIH for outpatient visits at days 0, 1, 3, 5, 7, and 10 and weeks 2, 3, 4, 8, 12, 14, 16, 20, and 24, which corresponds to an SVR12 visit.
Outcome Measures
Plasma HCV RNA levels were measured using the real-time HCV assay (Abbott), with an LLOQ of 12 IU/mL. The assay was used to measure HCV RNA levels in all participants at all time points. Serum HCV RNA levels were also measured using the COBAS TaqMan HCV RNA assay version 2.0 (Roche), with an LLOQ of 43 IU/mL at screening, week 4, week 8, end of treatment, 4 weeks after treatment, and 12 weeks after treatment. Plasma HIV RNA levels were measured at all points using reverse transcription polymerase chain reaction (real-time HIV assay), with an LLOQ of 40 copies/mL.
Hepatitis C viral relapse was defined as an HCV RNA level higher than the LLOQ at any posttreatment point after having an HCV RNA level lower than the LLOQ at the end of treatment. Hepatitis C viral breakthrough was defined as an HCV RNA level at the LLOQ or higher during treatment after having previously had an HCV RNA level lower than the LLOQ while taking study drugs, confirmed with 2 consecutive values. Renal function was assessed using both creatinine levels and estimated glomerular filtration rate (eGFR, calculated using the CKD-EPI equation).16
Safety and Adherence
Adverse events and clinical laboratory results were recorded throughout the study. Adverse events were graded from 1 (mild) to 4 (severe) according to the NIAID Division of AIDS toxicity table (version 1.0).17 Patient adherence was determined by pill counts at weeks 1, 4, 8, and end of treatment. Patients were determined to be fully adherent to the regimen if they missed no doses over the course of treatment.
Viral Kinetics
During the first 28 days of treatment, HCV RNA levels were measured at day 0, 1, 3, 5, 7, 10, 14, 21, and 28 in all patients. In 20 patients (10 not receiving antiretroviral treatment and 10 receiving antiretroviral treatment), HCV RNA levels were also obtained at 0, 1, 2, 4, 8, 12, 24, and 36 hours after administration of first dose of study.
IL28B and IFNL4 Genotyping
IL28B and IFNL4 single-nucleotide polymorphisms have previously been described as determinants of SVR.18,19 IL28B and IFNL4 genotype was determined on DNA specimens using 5' nuclease assay with IL28B and IFNL4 allele-specific TaqMan probes (ABI TaqMan allelic discrimination kit) and the ABI7500 Real-Time PCR system (Applied Biosystems). Genotyping of variants at the rs12979860 (referred to as IL28B genotype) and rs368234815 (IFNL4) loci was performed as previously described.18
Liver Biopsy
An optional additional liver biopsy was offered after treatment completion to participants who had an initial liver biopsy performed at the NIH Clinical Center. Assessments were performed by a single pathologist and staged according to the Knodell Histological Activity Index.20
Clinical End Points
The primary end point was sustained virologic response (HCV RNA level <12 IU/mL by real-time HIV assay) at 12 weeks after treatment completion (SVR12) among all patients enrolled in the study. Safety end points included frequency and severity of adverse events, discontinuations attributable to adverse events, and safety laboratory changes.
Secondary end points that have been completed and included here are the proportion of participants with unquantifiable HCV viral load at specified points during and after treatment, discontinuations attributable to adverse events, safety laboratory changes, and evaluation of HCV resistance mutations in the patient who experienced relapse. Data through SVR12 are included here.
Modeling Viral Kinetics
Viral kinetic modeling with a multiscale model was performed for all participants as previously described.21,22 A detailed version of the methodology is described in eAppendix 3 in Supplement 2.
Deep Sequencing
Deep sequencing of the HCV NS5A and NS5B regions was performed only for the patient who experienced relapse, from samples collected at baseline and at the time of virologic failure, using DDL (DDL Diagnostics Laboratory). The NS3, NS5A, and NS5B were amplified by RT-PCR using HCV genotype-specific primers. The PCR products were sequenced by Illumina MiSeq technology as described elsewhere.23 Deep sequencing that covers 5000 reads was performed for each sample.
Statistical Analysis
The primary virologic response and safety analyses were based on an intention-to-treat population (all patients who received at least 1 dose of study medication). In terms of safety, a sample size of 50 was calculated as sufficient to have a 96% chance of observing at least 1 adverse event having a probability of 10% or greater. With 50 participants, the study would be able to estimate the proportion of patients with suppression to below the limit of detection to within plus or minus 0.11, based on a 95% confidence interval.
Baseline demographics were compared using the Wilcoxon rank-sum test for continuous outcomes and χ2 tests for binary outcomes. Estimated decline in HCV viral load at different times was compared using a Wilcoxon rank-sum test. Analyses were performed using BiAS, PRISM 6.0 (GraphPad), SAS (SAS Institute Inc), STAT-CRUNCH, and S-Plus 8.0.
Results
Baseline Characteristics
Sixty-three potential participants were screened and 50 were enrolled (eFigure 1 in Supplement 2). Six participants screened and declined to participate, and 7 participants did not meet inclusion criteria (see eTable 1 in Supplement 2 for reasons for exclusion). Of the 50 enrolled, 37 were receiving antiretroviral treatment and 13 were not receiving antiretroviral treatment.
The demographic and baseline clinical characteristics are shown in Table 1. Median baseline CD4 count was 576 (95% CI, 453–657) cells/mm3 for patients receiving antiretroviral treatment and 687 (95% CI, 518–960) cells/mm3 for patients not receiving antiretroviral treatment. Participants were predominantly African American (84%), men (74%), IL28B non-CC genotype (84%), with genotype 1a infection (74%). Thirteen of 50 patients (26%) had stage 3 liver disease.
Table 1.
Baseline Demographics and Clinical Characteristics of Study Participants
| Antiretroviral Therapy |
||
|---|---|---|
| Characteristic | No (n = 13) | Yes (n = 37) |
| Age, median (IQR), y | 59 (54–62) | 58 (51–63) |
| Men, No. (%) | 7(54) | 30 (81) |
| Body mass index, median (IQR)a | 26 (23–29) | 26 (22–29) |
| BMI ≥30, No. (%) | 3 (23) | 8 (22) |
| Race/ethnicity, No. (%)b | ||
| White | 3(13) | 4 (11) |
| Black | 10 (77) | 32 (86) |
| Hispanic | 0 | 1 (3) |
| Fibrosis score, No. (%)c | ||
| 0 | 2(15) | 8 (22) |
| 1 | 6 (46) | 19(51) |
| 2 | 0 | 2 (5) |
| 3 | 5(38) | 8 (22) |
| HCV genotype 1 subtype, No. (%) | ||
| 1A | 9 (75) | 30 (81) |
| 1B | 3 (25) | 7 (19) |
| IL28B genotype, No. (%) | ||
| CC | 2 (15) | 6 (16) |
| CT | 6 (46) | 18 (39) |
| TT | 5 (38) | 13 (35) |
| IFNL4 genotype, No. (%) | ||
| TT/TT | 2 (15) | 6 (16) |
| ΔG/TT | 6 (46) | 15 (41) |
| ΔG/ΔG | 5 (38) | 16 (43) |
| Baseline HCV RNA,-log10IU/mL | ||
| Median (IQR) | 6.1 (5.3–6.5) | 6.0 (5.4–6.5) |
| Mean | 5.8 | 6.0 |
| HCV RNA >6 million IU/mL, No. (%) | 5 (50) | 14 (38) |
| Baseline creatinine, median (range), mg/dLd | 0.79 (0.69–0.98) | 0.91 (0.82–1.10) |
| Baseline eGFR, median (range), mL/min | 101 (94–115) | 93 (83–112) |
| CD4, cells/mm3, No. (%) | ||
| <200 | 0 | 1(3) |
| 200–350 | 1 (8) | 8 (22) |
| >350 | 12 (92) | 28 (76) |
| Antiretroviral use | ||
| Tenofovir/emtricitabine plus | 0 | 37 (100) |
| Efavirenz | 15 (41) | |
| Raltegravir | 10 (27) | |
| Rilpivirine | 8(21) | |
| Raltegravir/rilpivirine | 3 (8) | |
| Raltegravir/efavirenz | 1 (3) | |
Abbreviations: BMI, body mass index; eGFR, estimated glomerular filtration rate; HCV, hepatitis C virus; IQR, interquartile range.
SI conversion factor: To convert creatinine values to μmol/L, multiply by 88.4.
Calculated as weight in kilograms divided by height in meters squared.
Self-reported.
Fibrosis determined either by Knodell Histological Activity Index score (a low score ranges from 0–2; a high score from 3 to 4) or by FibroSURE test. Forty-four patients were classified based on liver biopsy. Six patients were classified based on FibroSURE score.
Baseline creatinine level (mg/dL) was the only significant difference in baseline characteristics between patients not receiving and receiving antiretroviral treatment (P = .01).
Virologic Response During and After Treatment
All 50 participants achieved virologic suppression lower than the LLOQ by week 4 of therapy (Table 2; eTable 2 in Supplement 2). All 13 participants not receiving antiretroviral treatment achieved SVR12 (100% [95% CI, 75%-100%]). Of the 37 participants receiving antiretroviral treatment, 36 (97% [95% CI, 89%-100%]) achieved SVR12.
Table 2.
Patients With HCV RNA Levels Below the Level of Quantification at Various Times of Treatment and Follow-upa
| Antiretroviral Therapy, No. (%) [95% CI]b |
||
|---|---|---|
| Week | No (n = 13) | Yes (n = 37) |
| Treatment Period | ||
| 4 | 13 (100) [75–100] | 37 (100) [91–100] |
| 8 | 13 (100) [75–100] | 37 (100) [91–100] |
| 12 | 13 (100) [75–100] | 37 (100) [91–100] |
| Postttreatment Period | ||
| 2 | 13 (100) [75–100] | 37 (100) [91–100] |
| 4 | 13 (100) [75–100] | 36 (97) [89–100] |
| 8 | 13 (100) [75–100] | 36 (97) [89–100] |
| 12 (SVR12) | 13 (100) [75–100] | 36 (97) [89–100] |
Abbreviation: SVR, sustained viral response.
Treatment response in all participants was included in the intention-to-treat analysis in all 50 participants. The RNA assay was used for these points, with a lower limit of quantification of 43 IU/mL.
The CI for proportion estimate for each treatment is obtained using the binomial distribution
Changes in Liver Function
Levels of alanine aminotransferase and aspartate aminotransferase normalized rapidly with treatment (eFigure 2 in Supplement 2). A comparison of pretreatment and posttreatment paired liver biopsies showed Knodell Histological Activity Index inflammation improved by a mean of 2.2 (from 8.07 to 5.86) with treatment (P = .01) (eFigure 3 in Supplement 2).
One patient did not achieve SVR12 and experienced relapse by week 4 after treatment completion. This was a 63-year-old African American woman, IL28B genotype TT, with HCV genotype 1b infection, a baseline HCV viral load of 5 338 580 IU/mL by real-time HIV assay, and stage 1 liver disease. She was receiving tenofovir plus emtricitabine and rilpivirine for HIV infection with viral suppression. She achieved viral suppression lower than the LLOQ by week 4, which was maintained through 12 weeks. At week 4 after treatment completion, HCV viral load was 64 749IU/mL by real-time HIV assay. This patient has been lost to follow-up.
Viral Resistance Testing
Deep sequencing was carried out for the 1 participant who experienced relapse. The only mutation was seen in the NS5A region. The participant had 58% of Y93H mutation at baseline, which was enriched to 89% at day 3 and to greater than 99% at relapse. This mutation confers resistance to NS5A inhibitors, such as ledipasvir in vitro.
HCV Viral Kinetic Response
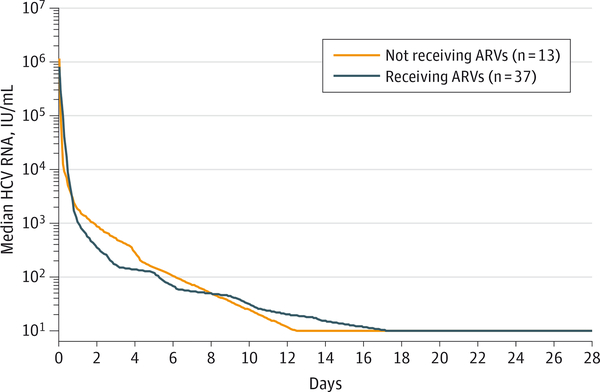
Viral kinetic modeling showed a rapid, sustained decline in HCV RNA in both groups (Figure). This multistage model was fitted in each participant shown in eFigure 4 in Supplement 2. Median overall treatment effect (ε) observed in participants receiving and not receiving antiretroviral treatment was similar (treated, 99.98%; not treated, 99.98%; P = .78). Clearance rate (c) of free virions (9.05 vs 10.3, P = .68) and loss of infected hepatocytes (δ) (0.13 vs 0.17, P = .25) were not significantly different between patients in the 2 groups. The virologic response rates were similar in patients receiving and not receiving antiretroviral treatment at the time of treatment.
Figure. Decline in Median HCV Viral Load After Initiation of Study Drugs, Days 0–28.
Baseline log hepatitis C virus (HCV) viral load was similar in both groups (6.0 [SD, 0.1] IU/mL in patients receiving antiretroviral (ARV) treatment and 6.1 in patients not receiving ARV treatment [SD, 0.3] in [P = .49]). The study drug combination of ledipasvir and sofosbuvir was equally effective in patients not receiving and receiving ARV treatment (P = .78 for total effectiveness). Lower limit of quantification is 12 IU/mL.
Adverse Events
There were no deaths or discontinuations observed in this study. Most adverse events were grade 1 in severity, with the most common being nasal congestion, myalgia, headache, and fatigue (Table 3). There were 4 grade 3 events, namely, decreased absolute neutrophil count, pneumonia, elevated aspartate aminotransferase level, and right upper quadrant pain. There was 1 grade 4 event (abnormal creatine phosphokinase level) in a participant who reported initiation of a vigorous exercise program, which resolved while the patient was still receiving study drugs. There was 1 significant adverse event reported in a smoker with a history of chronic bronchitis who was hospitalized with pneumonia in the winter. Symptoms resolved while the patient was receiving study drug, and the adverse event was determined to be unlikely related to the study drug.
Table 3.
Treatment Discontinuations, Adverse Events, and Hematologic Abnormalities
| Antiretroviral Therapy, No. (%) |
|||
|---|---|---|---|
| Variable | No (n = 13) | Yes (n = 37) | Overall (n = 50) |
| Treatment discontinuation | 0 | 0 | 0 |
| Any adverse event | 13 (100) | 37 (100) | 50 (100) |
|
Common adverse event | |||
| Nasal congestion | 2(15) | 6(16) | 8(16) |
| Myalgia | 5(38) | 2 (5) | 7(14) |
| Headache | 1(8) | 4(11) | 5 (10) |
| Fatigue | 3 (23) | 2 (5) | 5 (10) |
| Diarrhea | 2 (15) | 2 (5) | 4 (8) |
| Nausea | 1 (8) | 2 (5) | 3 (6) |
| Constipation | 2 (15) | 1 (3) | 3 (6) |
| Urinarytract infection | 1 (8) | 2 (5) | 3 (6) |
| Serious adverse event | |||
| Pneumonia | 1 (8) | 0 | 1 (2) |
| Hematologic abnormalitya | |||
| Increased aspartate aminotransferase (201–400 U/L) | 0 | 1 (3) | 1 (2) |
| Increased blood creatinine phosphokinase (≥6130 U/L) | 0 | 1 (3) | 1 (2) |
| Decreased neutrophilcount (0.500–0.749 K/uL) | 1 (8) | 0 | 1 (2) |
| Elevated lipase (181–300 U/L) | 0 | 1 (3) | 1 (2) |
Percentage represents proportion of patients who had at least 1 laboratory value above the upper limit of normal.
Changes in Renal Function
At baseline, median serum creatinine levels were significantly different between patients not receiving antiretroviral treatment (0.81 mg/dL [to convert to μmol/L, multiply by 88.4]) and those receiving treatment (0.91 mg/dL) (P = .01). There were no significant changes in estimated serum creatinine levels or eGFRs over time (Table 4, eFigure 5 in Supplement 2). No participants were identified as having a treatment-emergent eGFR less than 50 mL/min or a decrease in eGFR (mL/min) greater than 25%.
Table 4.
Changes in Renal Parameters Over Time
| Antiretroviral Therapy, Median (95% CI) |
|||
|---|---|---|---|
| Change at Week | No (n = 13) | Yes (n = 37) | P Value |
| eGFR, mL/mina | |||
| 4 | −1.0 (−9.0 to 6.0) | −2.0 (−5.0 to 0) | .46 |
| 8 | −3.0 (−8.0 to 2.0) | −1.0 (−7.0 to 1.0) | .86 |
| 12 | 0.0 (−11.0 to 1.0) | −4.0 (−6.0 to 1.0) | .92 |
| Creatinine, mg/dL | |||
| 4 | 0.01 (−0.08 to 0.11) | 0.02 (−0.01 to 0.08) | .51 |
| 8 | 0.04 (−0.04 to 0.11) | 0.02 (−0.01 to 0.11) | .98 |
| 12 | 0.06 (−0.02 to 0.16) | 0.03 (−0.05 to 0.08) | .26 |
| No Efavirenz (n = 21) | Efavirenz (n = 16) | ||
| eGFR, mL/mina | |||
| 4 | −1.0 (−10.0 to 2.0) | −5.0 (−7.0 to 1.0) | .66 |
| 8 | 0.0 (−3.0 to 4.0) | −6.5 (−13.0 to 2.0) | .22 |
| 12 | −4.0 (−12.0 to 3.0) | −4.0 (−8.0 to 3.0) | .76 |
| Creatinine, mg/dL | |||
| 4 | 0.01 (−0.06 to 0.10) | 0.05 (−0.01 to 0.10) | .51 |
| 8 | 0.0 (−0.04 to 0.03) | 0.09 (−0.03 to 0.13) | .14 |
| 12 | 0.01 (−0.06 to 0.08) | 0.04 (−0.09 to 0.17) | .53 |
Abbreviation: eGFR, estimated glomerular filtration rate.
SI conversion factor: To convert creatinine values to μmol/L, multiply by 88.4.
No patients were identified as having a treatment-emergent eGFR less than 50 mL/min or a decrease in eGFR greater than 25% mL/min.
One patient developed treatment-emergent hypophosphatemia (serum phosphorus level, 2.5 mg/dL [0.81 mmol/L]; grade 1) associated with a transient elevation in serum creatinine level, which resolved with treatment. This patient was receiving rilpivirine/tenofovir/emtricitabine and raltegravir. Two additional patients developed normoglycemic glycosuria that resolved with treatment.
Changes in HIV Parameters
Eight participants receiving antiretroviral treatment who experienced an increase in HIV viral load (HIV-1 RNA ≥40 copies/ mL, <1 log increase from nadir [eTable 3 in Supplement 2]) while in the study. All 8 participants achieved SVR12. One participant had a greater than 1 log increase in HIV viral load. This participant had missed 5 days of antiretrovirals (rilpivirine/ tenofovir/emtricitabine + raltegravir) and had an HIV viral load of 594 copies/mL at week 4. He continued the same regimen. By the next visit (week 8), his HIV viral load was less than 20 copies/mL and he also achieved SVR12. Participants not receiving antiretroviral treatment who had HIV RNA levels greater than 50 copies/mL at baseline did not experience any significant increases in their HIV viral load over the course of therapy (eTable 4 in Supplement 2). There were no significant changes in CD4 cell counts or CD4 cell percentages with treatment for participants both receiving and not receiving antiretroviral treatment (eTable 5 in Supplement 2).
Adherence
Adherence to ledipasvir and sofosbuvir, as measured by pill counts, was high over the course of treatment. Fifty percent of all participants had no missed doses (54% among participants not receiving antiretroviral treatment; 49% among participants receiving antiretroviral treatment). Forty percent of patients missed 1 to 4 doses of study drug, for an adherence rate greater than 95%. Five patients missed 5 or more doses of study drug over the course of treatment. As determined by pill count at the end of study, the participant who experienced HCV viral relapse by week 4 after treatment completion reported no missed doses.
Discussion
In this open-label, uncontrolled, nonrandomized trial, the combination of ledipasvir and sofosbuvir was associated with high rates of SVR in participants co-infected with HCV genotype 1 and HIV, similar to that observed inpatients monoinfected with HCV genotype 1.14 These results show for the first time, to our knowledge, that an interferon- and ribavirin-free therapy is associated with high SVR rates in patients co-infected with HCV and HIV.
Traditionally, interferon-based therapies have resulted in lower rates of SVR among patients co-infected with HCV and HIV as compared with patients monoinfected with HCV.5,6 However, recent studies have shown that the addition of directly acting antiviral agents (telaprevir, boceprevir, simeprevir, and sofosbuvir) has increased SVR in patients co-infected with HCV and HIV, reporting similar rates of SVR that were observed with patients monoinfected with HCV.4,8,24,25 Yet many of these studies used interferon, ribavirin, or both concomitantly with an exacerbation of adverse events in HIV-positive patients.4,8,24,25 Most recently, the once-daily, single-tablet regimen of ledipasvir and sofosbuvir, with or without ribavirin, has been shown to be an effective regimen to treat hepatitis C in HIV-negative individuals.14,15,26
The participants included in this study had many previously described poor prognostic factors, namely, African American race (84%), male sex (74%), IL28B non-CC genotype (84%), and genotype 1a infection (74%). We have hypothesized that many of these factors may no longer be relevant in determining responses to HCV treatment. All 13 patients not receiving antiretroviral treatment and 36 of 37 patients receiving antiretroviral treatment achieved SVR, regardless of HIV viral suppression. In the 1 participant who experienced relapse, HCV sequencing data showed that the Y93H mutation in the NS5A region was detected at baseline and was enriched during the study and at the time of relapse.
In this study, most adverse events associated with combined ledipasvir and sofosbuvir by participants co-infected with HCV and HIV were mild (grade 1–2) and clinically managed. There were no deaths, medication discontinuations, severe adverse events, or grade 4 adverse events attributable to the study drug. Given the potential drug interaction between ledipasvir and tenofovir resulting in increases in tenofovir levels, we performed additional safety evaluation in participants receiving antiretroviral treatment. Although there were no significant changes in eGFRs and serum creatinine levels throughout this study, larger studies are required to definitively determine the potential for renal toxicity.
Other studies have demonstrated that drug-drug interactions between certain directly acting antiviral agents such as boceprevir, telaprevir, and antiretrovirals could result in adverse events or antiretroviral failures, restricting the wider use of these directly acting antiviral agents in patients with HIV. We found no significant change in HIV viral load levels, CD4 T cells, or occurrence of opportunistic infections. The HIV viral breakthrough documented in 1 participant was associated with nonadherence to antiretroviral treatment.
Although only 10% of participants missed 5 or more doses of medications, adherence monitoring should remain a key aspect of treatment in the real world.
Study limitations include the lack of a control group and exclusion of participants with cirrhosis. In addition, use of antiretroviral therapy was limited to those for whom interactions with ledipasvir were predicted to be not clinically relevant. In particular, HIV protease inhibitors and nonnucleoside reverse transcriptase inhibitors are widely used in clinical practice; other than efavirenz or rilpivirine, these agents were not permitted in this study. Although participants with lower CD4 T-cell counts (>100 cells/mL for patients receiving antiretroviral therapy) were permitted, few of these participants enrolled (median baseline CD4 of participants receiving antiretroviral therapy was 576 cells/mL). Several large, ongoing studies of combined ledipasvir and sofosbuvir use among patients coinfected with HCV and HIV will help address some of these questions.
Conclusions
In this open-label, uncontrolled, pilot study enrolling patients co-infected with HCV genotype 1 and HIV, administration of an oral combination of ledipasvir and sofosbuvir for 12 weeks was associated with high rates of SVR after treatment completion. Larger studies that also include patients with cirrhosis and lower CD4 T-cell counts are required to understand if the results of this study generalize to all patients infected with HCV and HIV.
Supplementary Material
Acknowledgments
Funding/Support: The Regulatory Compliance and Human Participants Protection Branch of the National Institute of Allergy and Infectious Diseases (NIAID) served as the study sponsor. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (NIH), under contract HHSN261200800001E. This research was supported in part by the NIAID. The study was also supported in part by the German Research Foundation (DFG) by the clinical research unit KFO 129. The study was partially funded by a Collaborative Research and Development Agreement between NIH and Gilead Sciences.
Role of the Funder/Sponsors: Gilead Sciences provided the study drug. The Regulatory Compliance and Human Participants Protection Branch was involved in the review and approval of the study via the usual peer review process as well as the study management but had no role in the design of the study; data collection and analysis; interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. Drs Osinusi, Subramanian, Pang, and McHutchison, employees of Gilead Sciences Inc, participated in the writing of the manuscript.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Ms Gross reported receiving study medication from Gilead for another study and holding stock in Merck and Pfizer. Dr Hogan reported serving as a speaker or consultant for Gilead and VIV and serving as a speaker for Merck, Bristol-Myers Squibb, Abbott, and Boehringer Ingelheim. Dr Teferi reported receiving grants and personal fees from Gilead. Dr Talwani reported serving as a speaker for Gilead. No other disclosures are reported.
Additional Contributions: We would like to acknowledge the contributions of the following individuals: Katie Watkins, BS, Erin Rudzinski, BS, and Susan Vogel, RN, BSN (clinical monitoring support), Judith Starling, PharmD, William T. Symonds, PharmD, and Lori Gordon (pharmacy), Vanessa Eccard, BS, Jerome Pierson, PhD, John Tierney, BSN, MPM (regulatory support); William Ronnenberg, JD/MIP, MS, Richard Williams, PhD, and Mike Mowatt, PhD (technology transfer support), Marc Teitelbaum, MD, CPI (sponsor medical monitor), Mary Hall (protocol support), Cathy Rehm, Sarah Jones, David Wu, BS, Leighton Daigh, BS, and Jessica Johl, BS (laboratory support), Richard Kwan, PAC, Lisa L. Barrett, MD, PhD, Tim Jolley, RN, Gabbie Diaz, RN, Jose Chavez, MD, Stephen Abbott, MD, Colleen Kotb, NP, Angie Price, NP, Tess Petersen, BS, Josez Chavez, MD, and Senora Mitchell (clinic support).
Footnotes
Disclaimer: The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Contributor Information
Anu Osinusi, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland; Gilead Sciences Inc, Foster City, California; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore.
Kerry Townsend, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Anita Kohli, Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health, Bethesda, Maryland; Clinical Research Directorate/Clinical Monitoring Research Program, Leidos Biomedical Research, Inc (formerly SAIC-Frederick, Inc), Frederick National Laboratory for Cancer Research, Frederick, Maryland.
Amy Nelson, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore.
Cassie Seamon, Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health, Bethesda, Maryland.
Eric G. Meissner, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland; Department of Microbiology and Immunology, Medical University of South Carolina College of Medicine, Charleston.
Dimitra Bon, Institute of Biostatistics and Mathematical Modeling, Johann Wolfgang Goethe University, Frankfurt, Germany.
Rachel Silk, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Clinical Research Directorate/Clinical Monitoring Research Program, Leidos Biomedical Research, Inc (formerly SAIC-Frederick, Inc), Frederick National Laboratory for Cancer Research, Frederick, Maryland.
Chloe Gross, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Clinical Research Directorate/Clinical Monitoring Research Program, Leidos Biomedical Research, Inc (formerly SAIC-Frederick, Inc), Frederick National Laboratory for Cancer Research, Frederick, Maryland.
Angie Price, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Clinical Research Directorate/Clinical Monitoring Research Program, Leidos Biomedical Research, Inc (formerly SAIC-Frederick, Inc), Frederick National Laboratory for Cancer Research, Frederick, Maryland.
Mohammad Sajadi, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore.
Sreetha Sidharthan, Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health, Bethesda, Maryland.
Zayani Sims, Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health, Bethesda, Maryland.
Eva Herrmann, Institute of Biostatistics and Mathematical Modeling, Johann Wolfgang Goethe University, Frankfurt, Germany.
John Hogan, Unity Health Care Inc, Washington, DC.
Gebeyehu Teferi, Unity Health Care Inc, Washington, DC.
Rohit Talwani, Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore.
Michael Proschan, Biostatistics Research Branch, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Veronica Jenkins, Family and Medical Counseling Services, Washington, DC.
David E. Kleiner, Department of Pathology, National Cancer Institute, Rockville, Maryland.
Brad J. Wood, Center for Interventional Oncology, Radiology and Imaging Sciences, NIH Clinical Center and National Cancer Institute, Bethesda, Maryland.
G. Mani Subramanian, Gilead Sciences Inc, Foster City, California.
Phillip S. Pang, Gilead Sciences Inc, Foster City, California.
John G. McHutchison, Gilead Sciences Inc, Foster City, California.
Michael A. Polis, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Anthony S. Fauci, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland.
Henry Masur, Critical Care Medicine Department, NIH Clinical Center, National Institutes of Health, Bethesda, Maryland.
Shyam Kottilil, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland; Division of Infectious Diseases, Institute of Human Virology, University of Maryland, Baltimore.
REFERENCES
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013; 57(4):1333–1342. [DOI] [PubMed] [Google Scholar]
- 2.Limketkai BN, Mehta SH, Sutcliffe CG, et al. Relationship of liver disease stage and antiviral therapy with liver-related events and death in adults coinfected with HIV/HCV. JAMA. 2012;308 (4):370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber R, Sabin CA, Friis-Møller N, et al. Liver-related deaths in persons infected with the human immunodeficiency virus: the D:A:D study. Arch Intern Med. 2006;166(15):1632–1641. [DOI] [PubMed] [Google Scholar]
- 4.Sulkowski MS, Sherman KE, Dieterich DT, et al. Combination therapy with telaprevir for chronic hepatitis C virus genotype 1 infection in patients with HIV: a randomized trial. Ann Intern Med. 2013; 159(2):86–96. [DOI] [PubMed] [Google Scholar]
- 5.Chung RT, Andersen J, Volberding P, et al. ; AIDS Clinical Trials Group A5071 Study Team. Peginterferon alfa-2a plus ribavirin versus interferon alfa-2a plus ribavirin for chronic hepatitis C in HIV-coinfected persons. N Engl J Med. 2004; 351(5):451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torriani FJ, Rodriguez-Torres M, Rockstroh JK, et al. ; APRICOT Study Group. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med. 2004;351(5): 438–450. [DOI] [PubMed] [Google Scholar]
- 7.Carrat F, Bani-Sadr F, Pol S, et al. ; ANRS HCO2 RIBAVIC Study Team. Pegylated interferon alfa-2b vs standard interferon alfa-2b, plus ribavirin, for chronic hepatitis C in HIV-infected patients: a randomized controlled trial. JAMA. 2004;292 (23):2839–2848. [DOI] [PubMed] [Google Scholar]
- 8.Sulkowski M, Pol S, Mallolas J, et al. ; P05411 Study Investigators. Boceprevir versus placebo with pegylated interferon alfa-2b and ribavirin for treatment of hepatitis C virus genotype 1 in patients with HIV: a randomised, double-blind, controlled phase 2 trial. Lancet Infect Dis. 2013;13(7):597–605. [DOI] [PubMed] [Google Scholar]
- 9.Sulkowski MS. Current management of hepatitis C virus infection in patients with HIV co-infection. J Infect Dis. 2013;207(suppl 1):S26–S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olysio (R) (simeprevir) [package insert]. http://www.olysio.com/shared/product/olysio/prescribing-information.pdf. 2013. Accessed December 2, 2014.
- 11.Solvadi (R) (sofosbuvir) [package insert]. http://www.gilead.com/∼/media/Files/pdfs/medicines/liver-disease/sovaldi/sovaldi_pi.pdf. 2013. Accessed December 2, 2014.
- 12.Sulkowski MS, Naggie S, Lalezari J, et al. ; PHOTON-1 Investigators. Sofosbuvir and ribavirin for hepatitis C in patients with HIV coinfection. JAMA. 2014;312(4):353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osinusi A, Meissner EG, Lee YJ, et al. Sofosbuvir and ribavirin for hepatitis C genotype 1 in patients with unfavorable treatment characteristics: a randomized clinical trial [published correction appears in JAMA. 2013;310(18):1987]. JAMA. 2013; 310(8):804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afdhal N, Reddy KR, Nelson DR, et al. ; ION-2 Investigators. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med. 2014;370(16):1483–1493. [DOI] [PubMed] [Google Scholar]
- 15.Afdhal N, Zeuzem S, Kwo P, et al. ; ION-1 Investigators. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med. 2014;370(20):1889–1898. [DOI] [PubMed] [Google Scholar]
- 16.Levey AS, Stevens LA, Schmid CH, et al. ; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009; 150(9):604–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Division of AIDS Table for Grading the Severity of Adult and Pediatric Adverse Events. Version 1.0. National Institute of Allergy and Infectious Diseases website. http://www.niaid.nih.gov/LabsAndResources/resources/DAIDSClinRsrch/Documents/daidsaegradingtable.pdf. 2009. Accessed February 9, 2015.
- 18.Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009; 461(7262):399–401. [DOI] [PubMed] [Google Scholar]
- 20.Knodell RG, Ishak KG, Black WC, et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;1(5):431–435. [DOI] [PubMed] [Google Scholar]
- 21.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267(3):330–340. [DOI] [PubMed] [Google Scholar]
- 22.Guedj J, Rong L, Dahari H, Perelson AS. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17(12):825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninomiya M, Ueno Y, Funayama R, et al. Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J Clin Microbiol. 2012;50(3):857–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dieterich D, Rockstroh JK, Orkin C, et al. Simeprevir (TMC435) with pegylated interferon/ribavirin in patients coinfected with HCV genotype 1 and HIV-1: a phase 3 study. Clin Infect Dis. 2014;59(11):1579–1587. [DOI] [PubMed] [Google Scholar]
- 25.Lawitz E, Lalezari JP, Hassanein T, et al. Sofosbuvir in combination with peginterferon alfa-2a and ribavirin for non-cirrhotic, treatment-naive patients with genotypes 1, 2, and 3 hepatitis C infection: a randomised, double-blind, phase 2 trial. Lancet Infect Dis. 2013;13(5):401–408. [DOI] [PubMed] [Google Scholar]
- 26.Kowdley KV, Gordon SC, Reddy KR, et al. ; ION-3 Investigators. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med. 2014;370(20):1879–1888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.